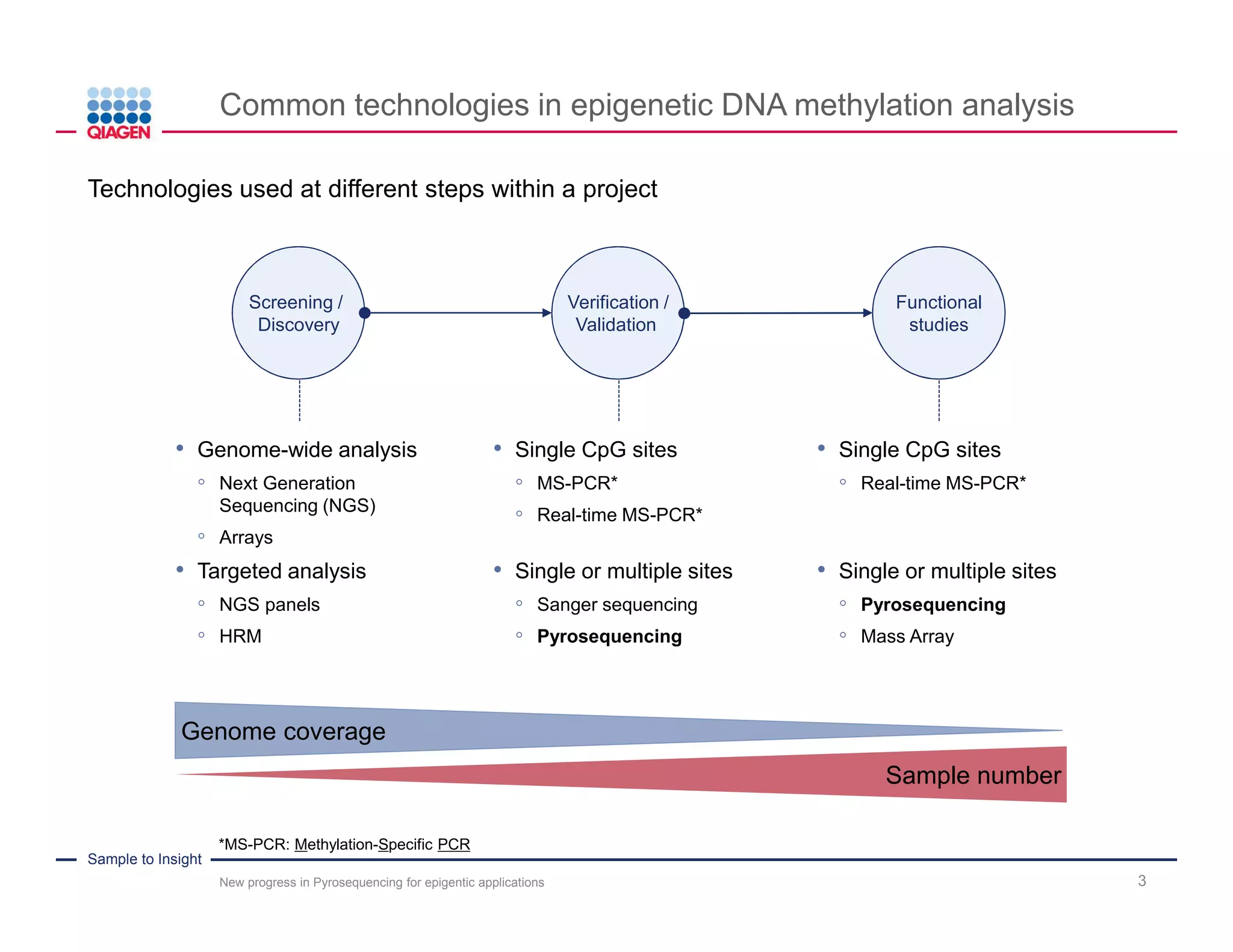
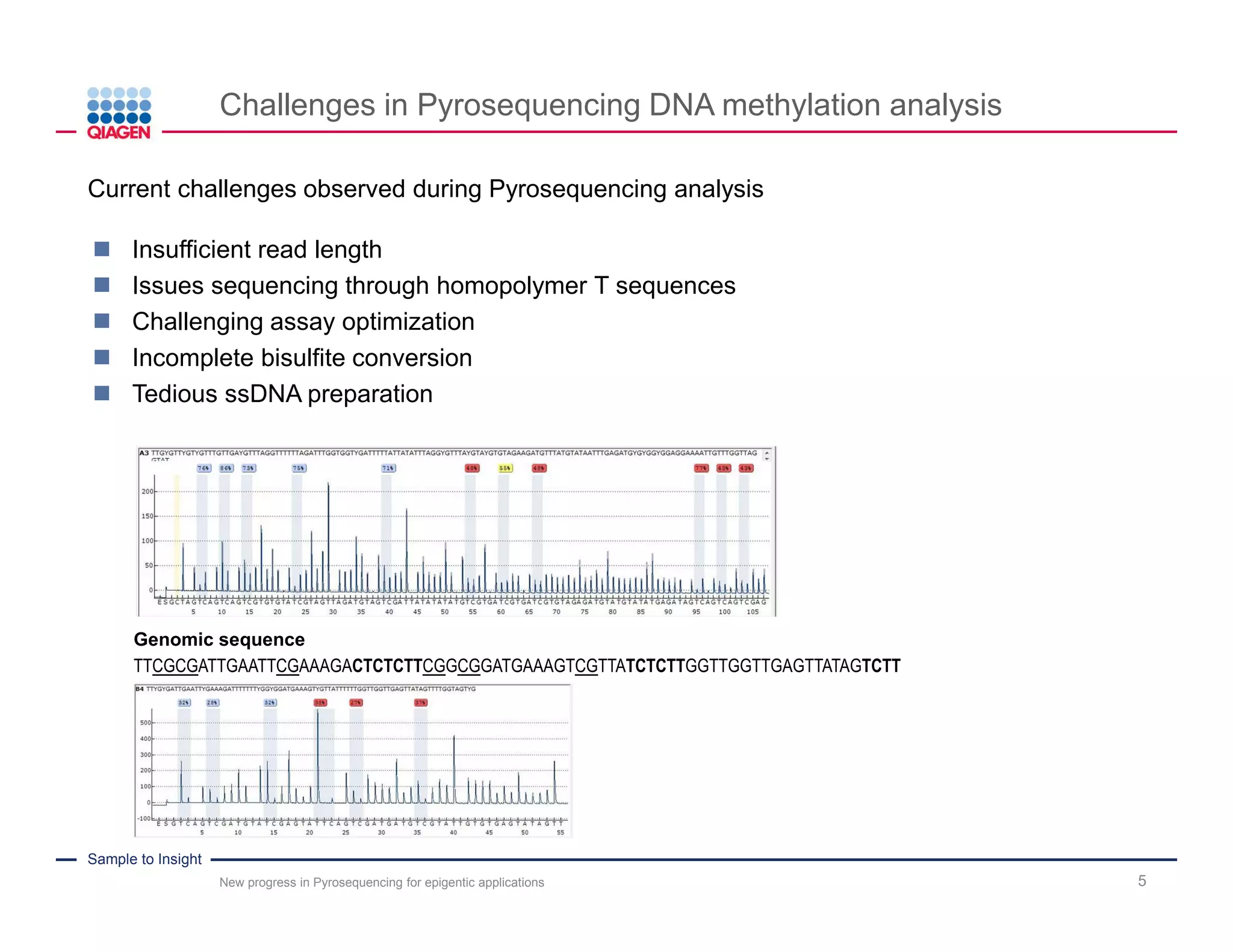
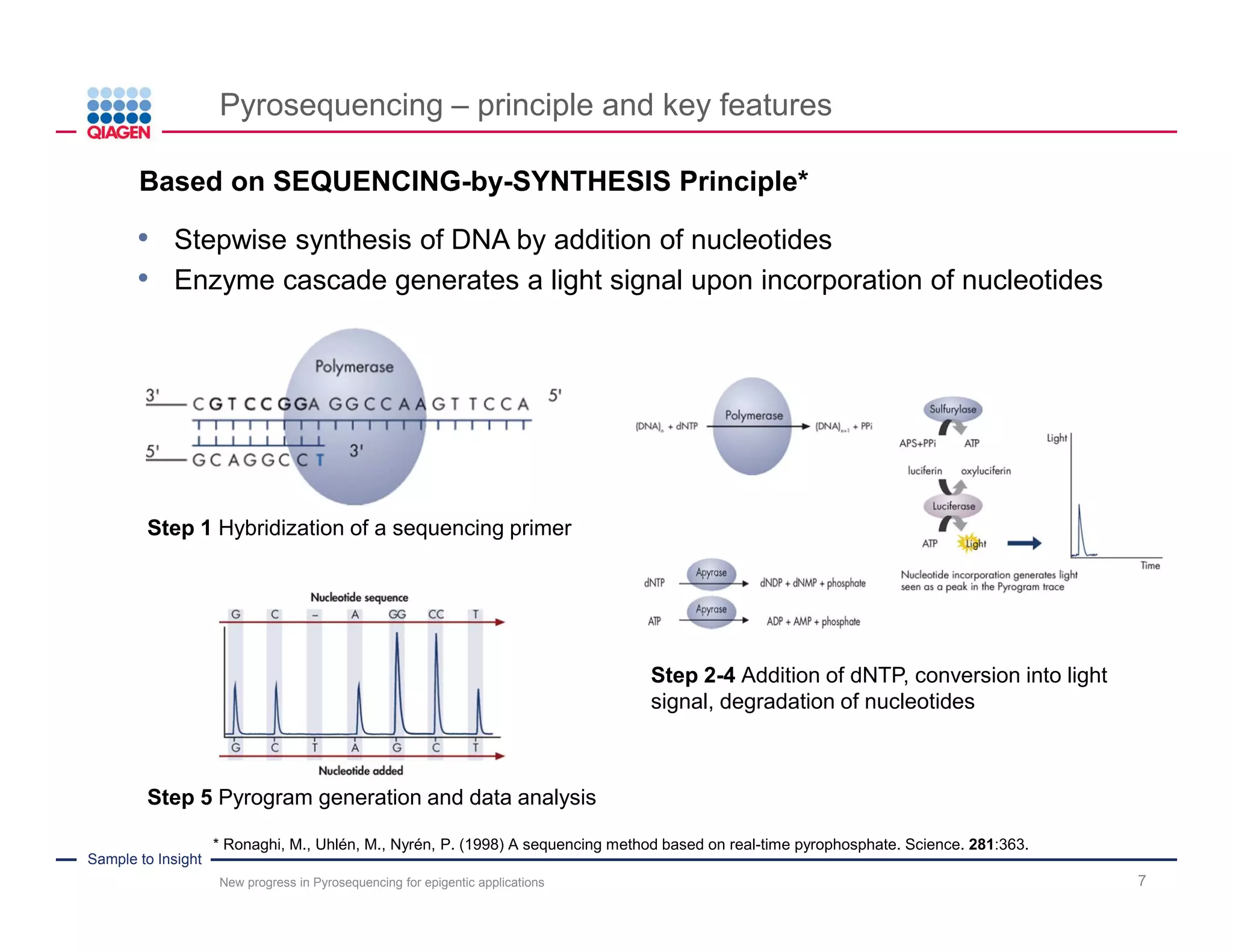
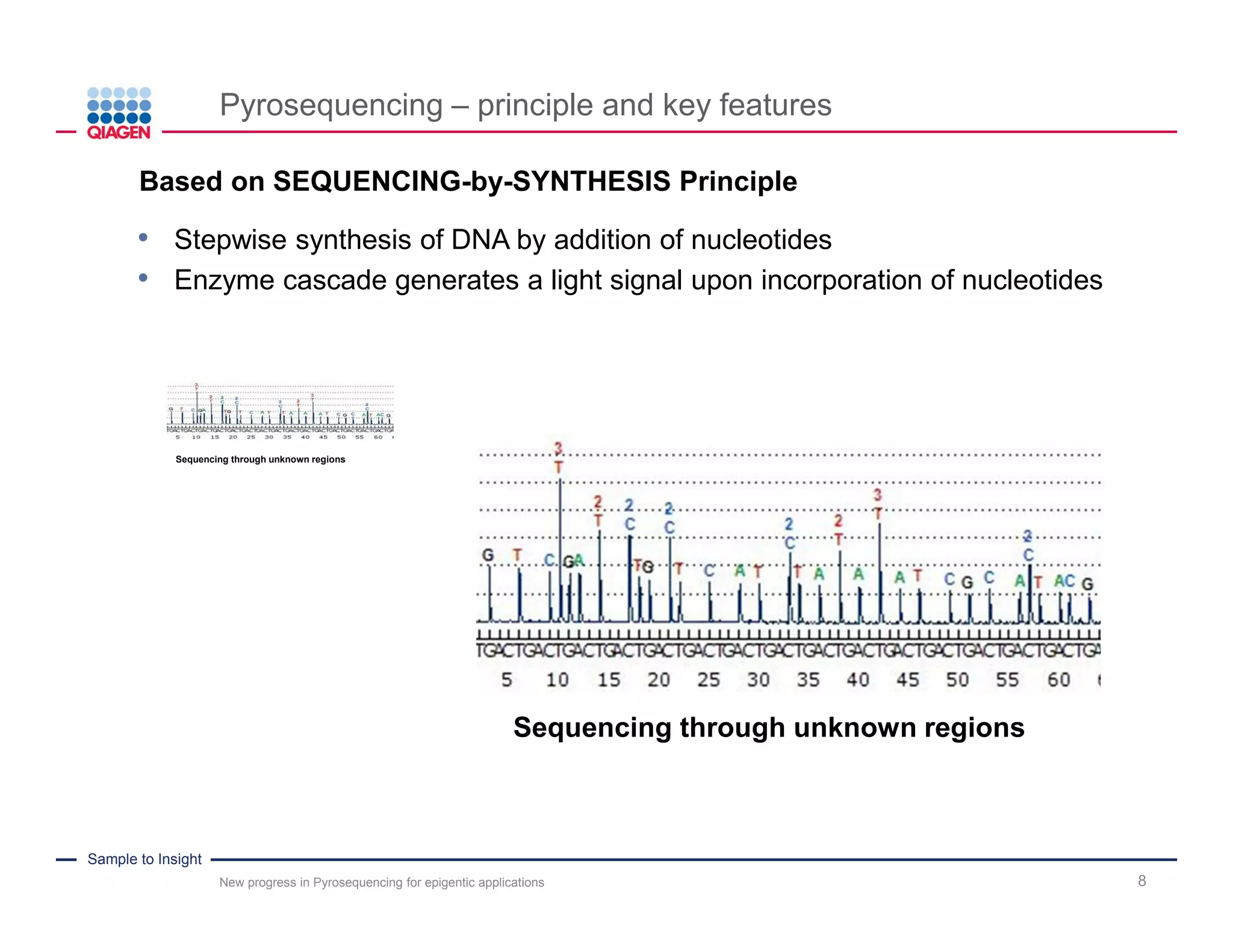
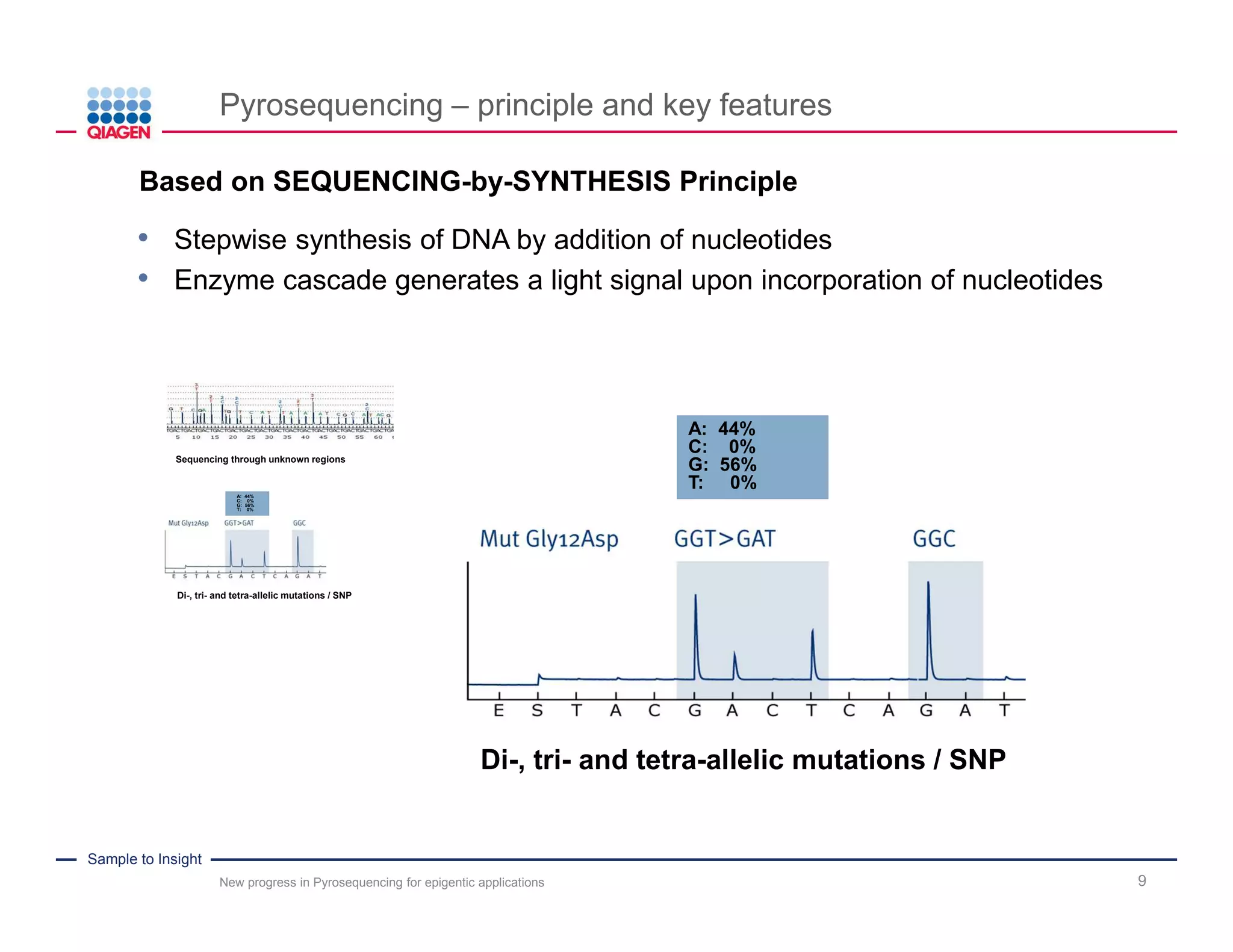
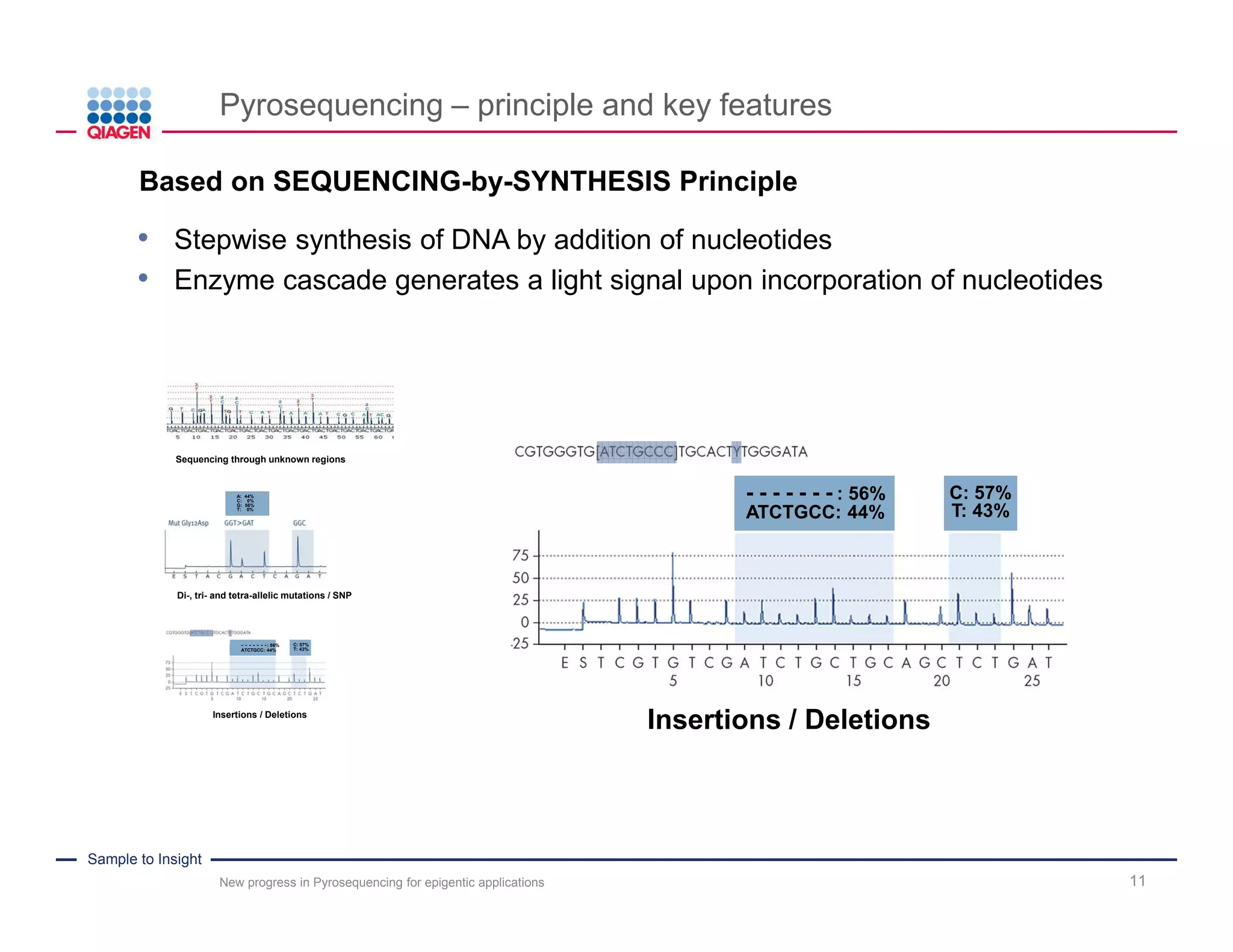
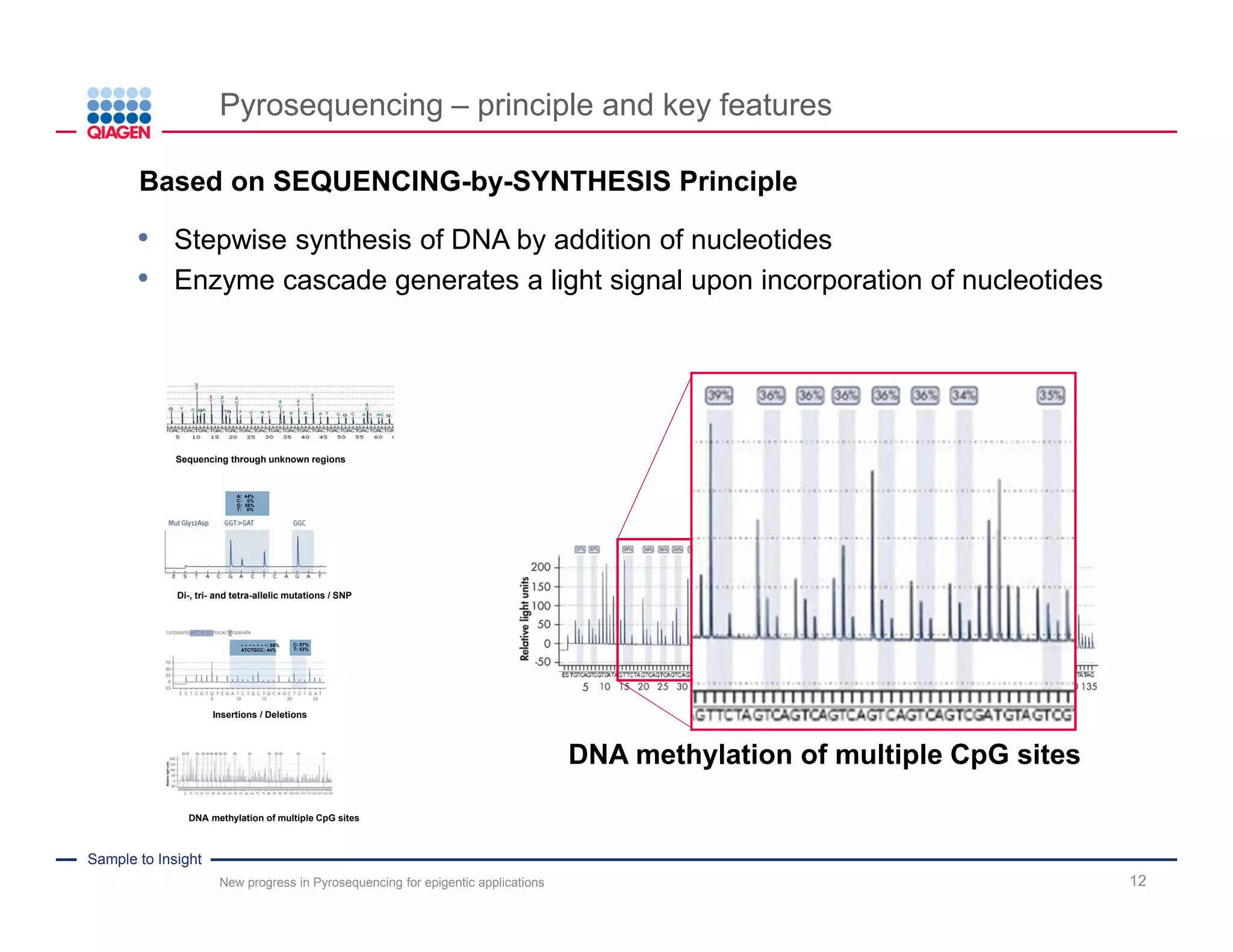
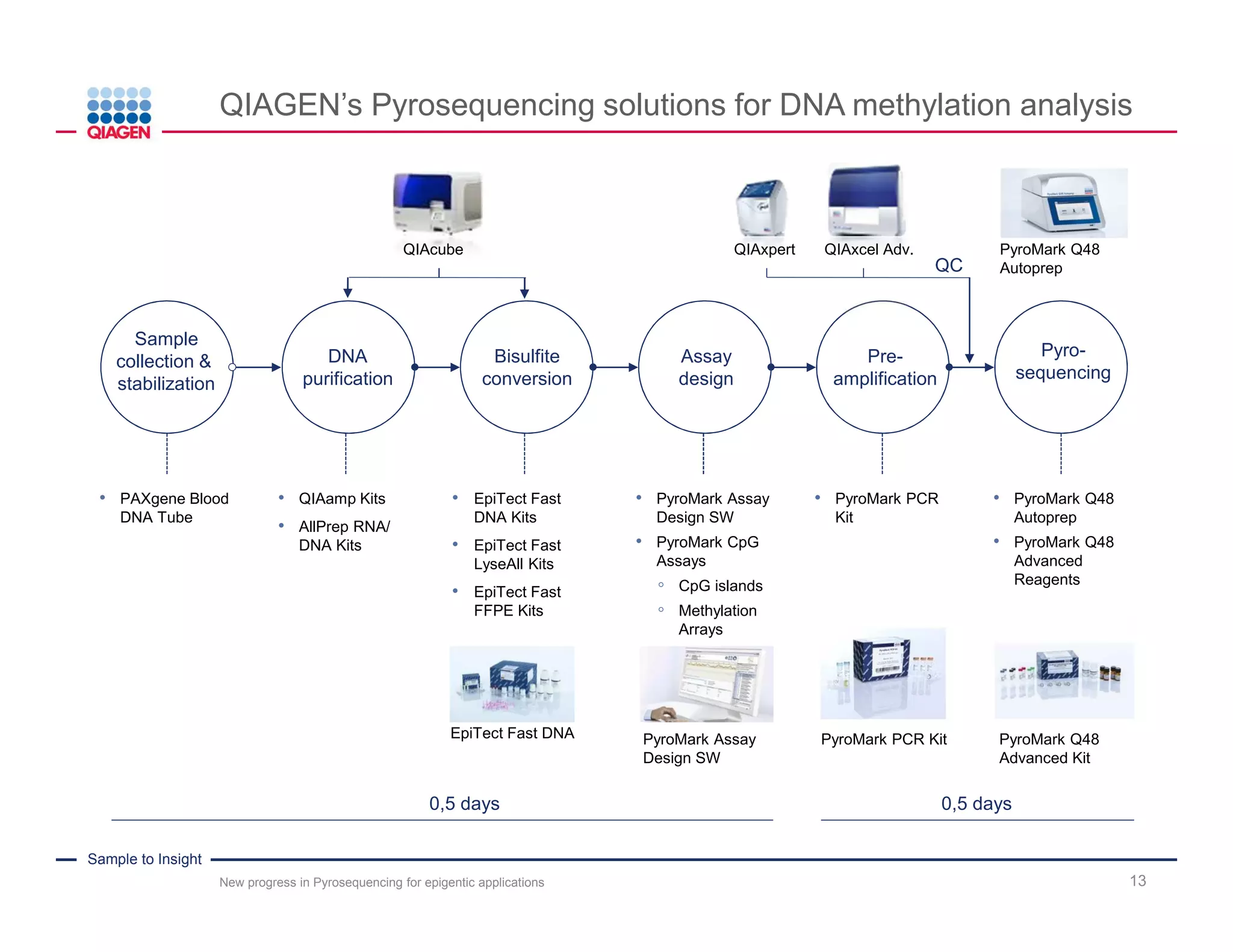
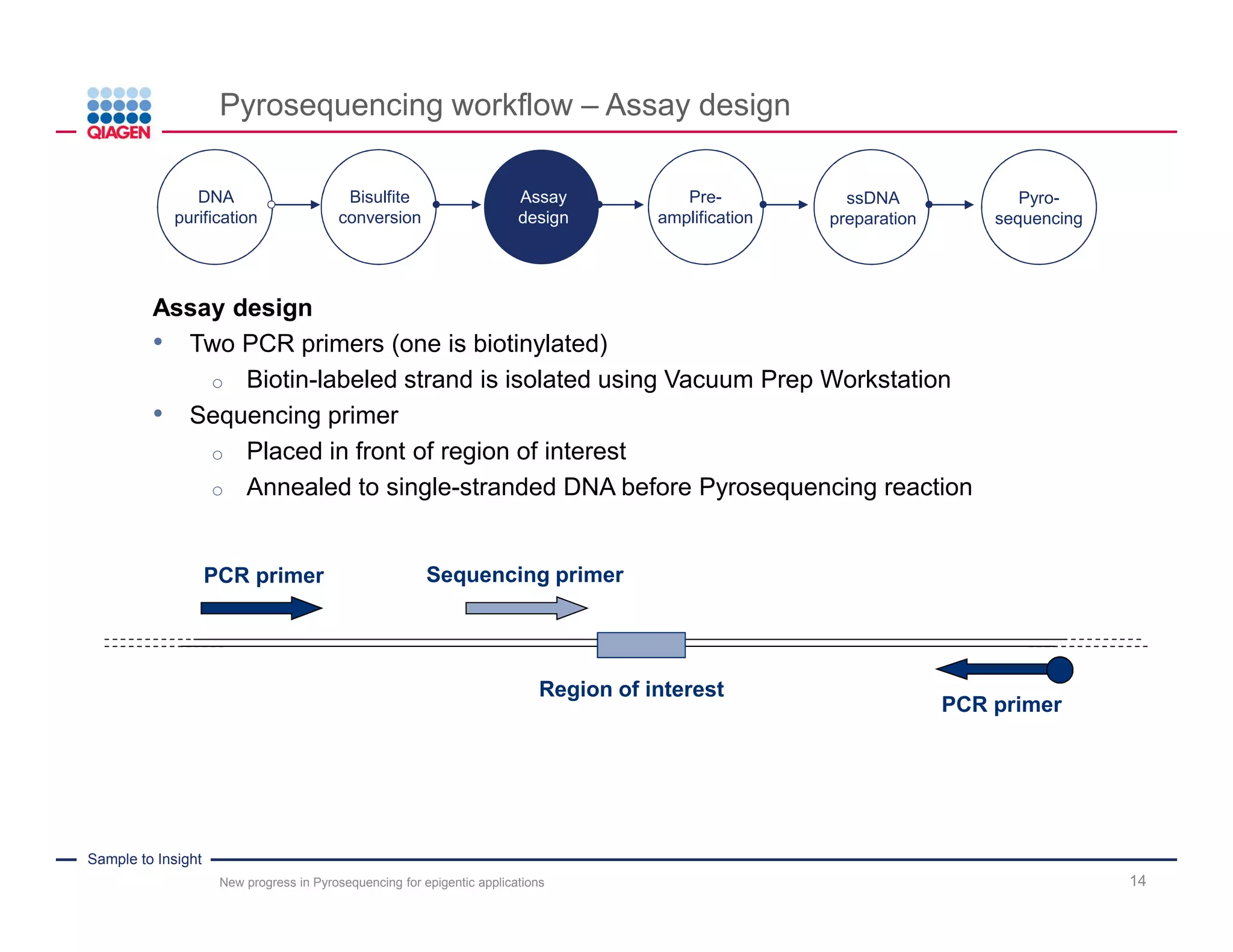
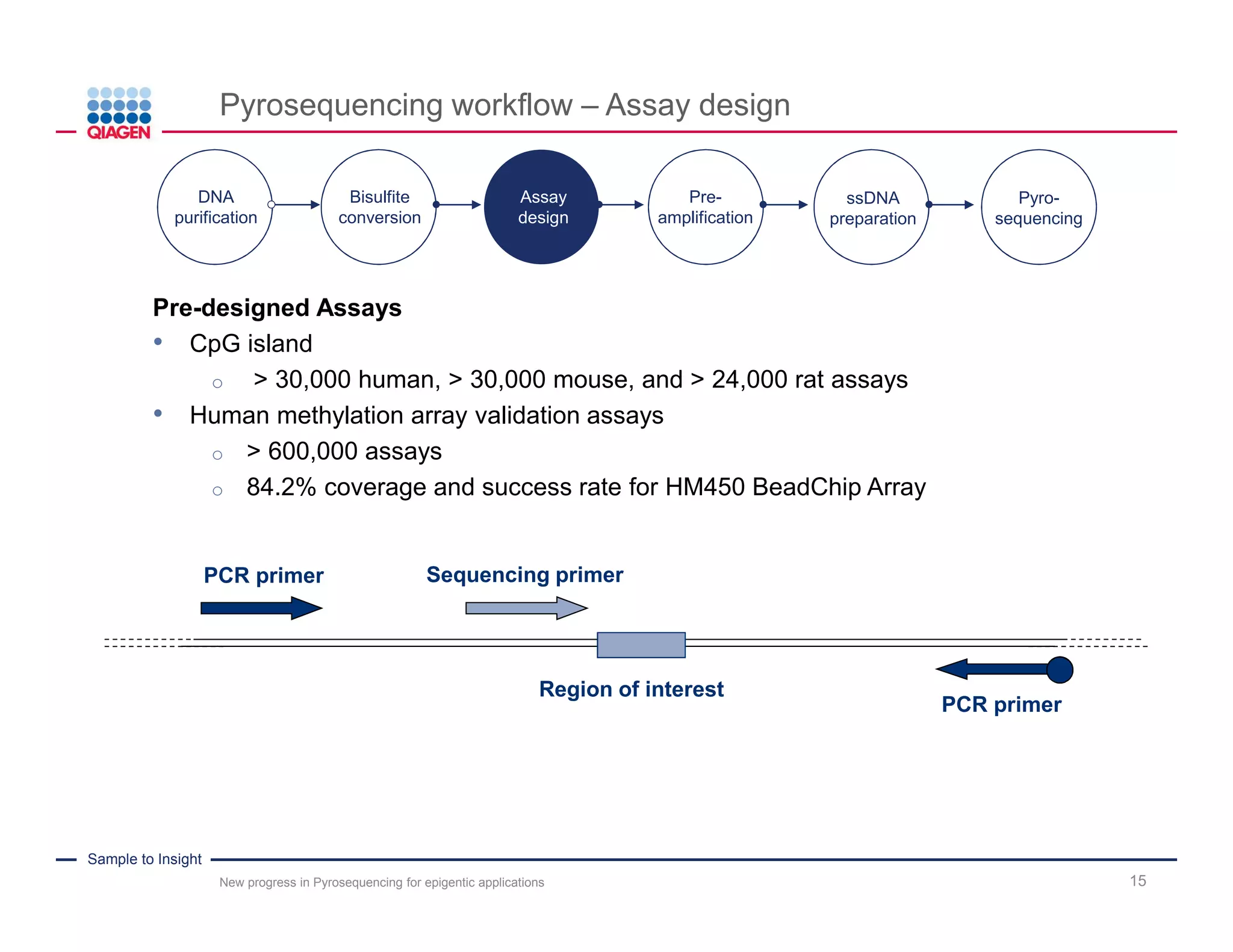
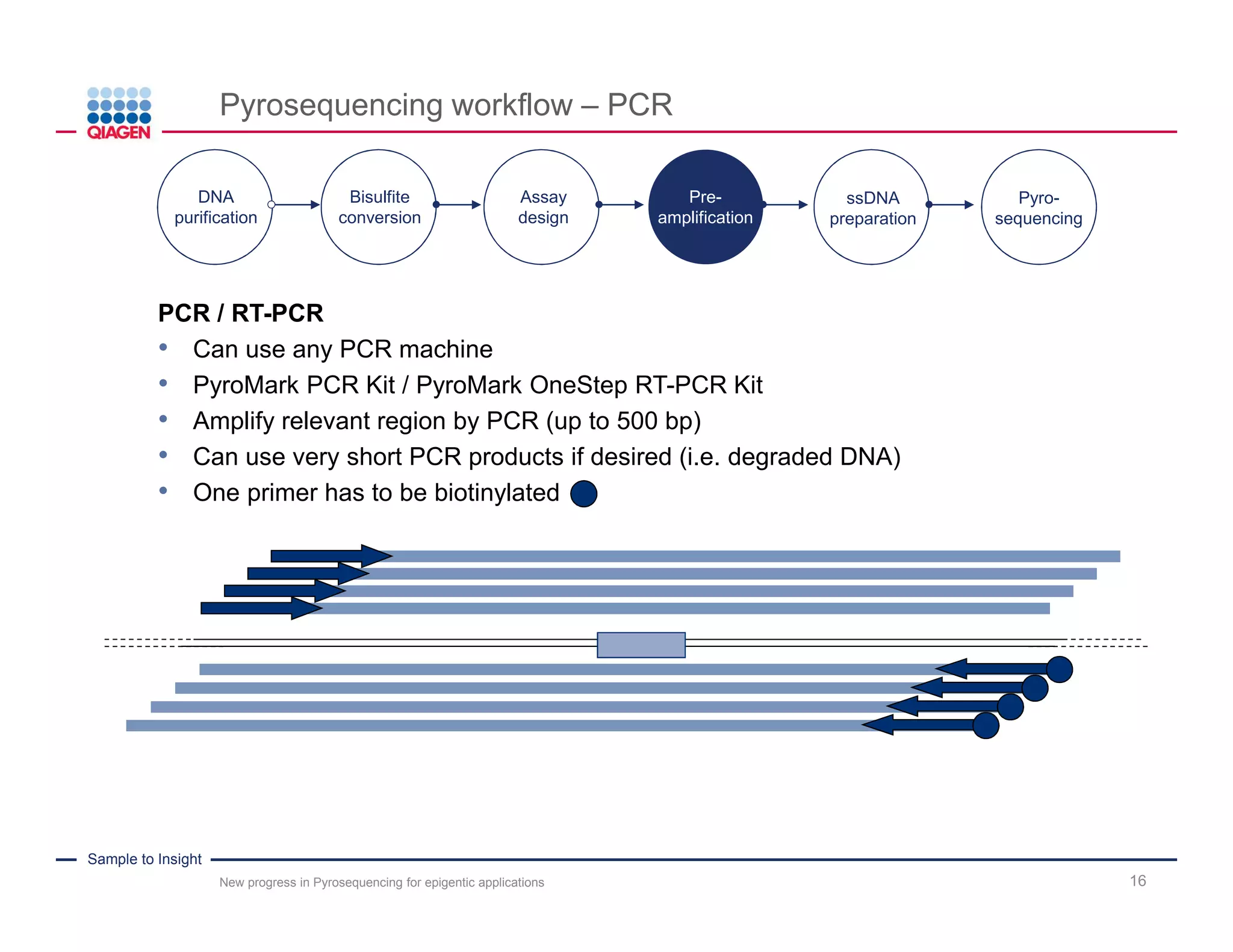
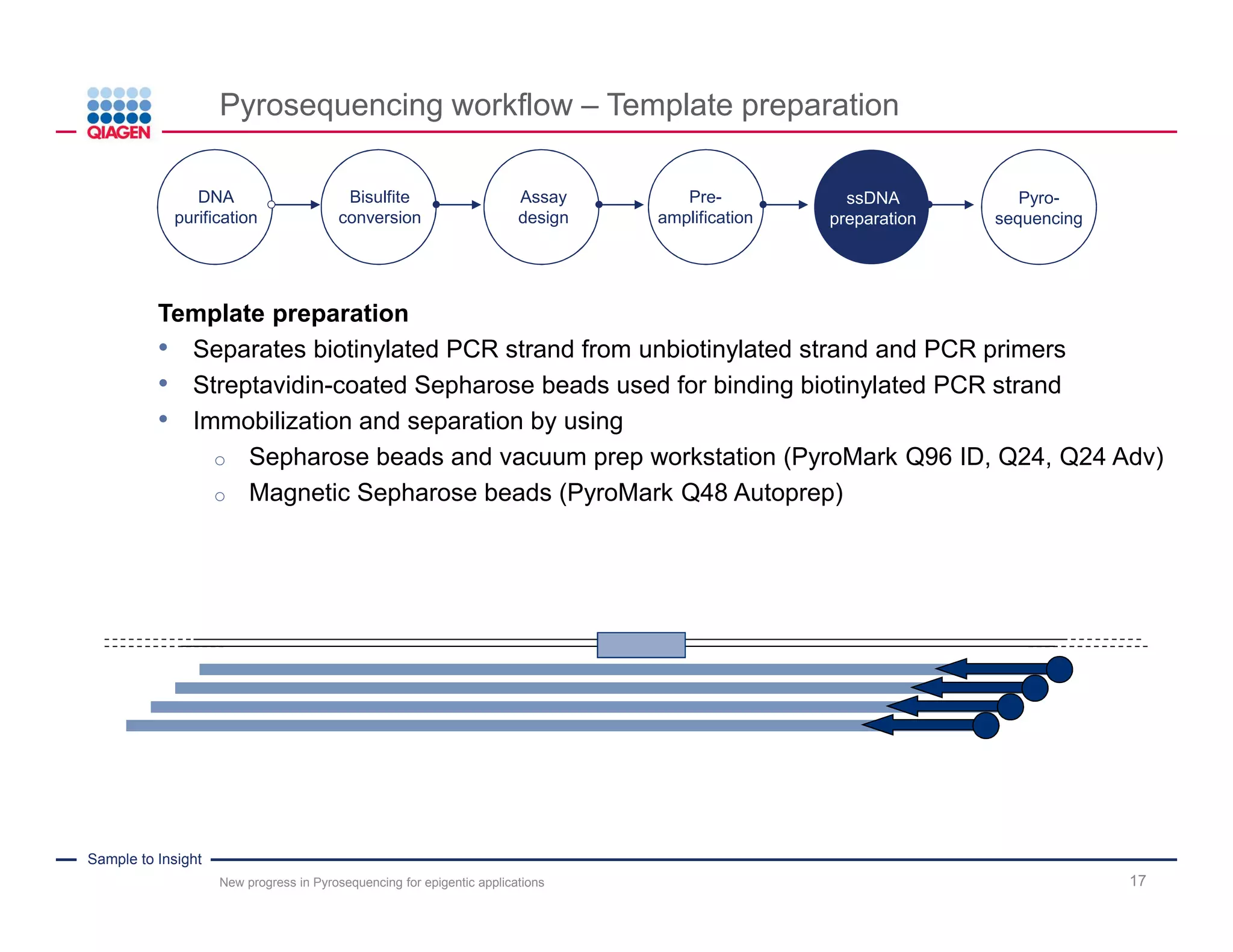
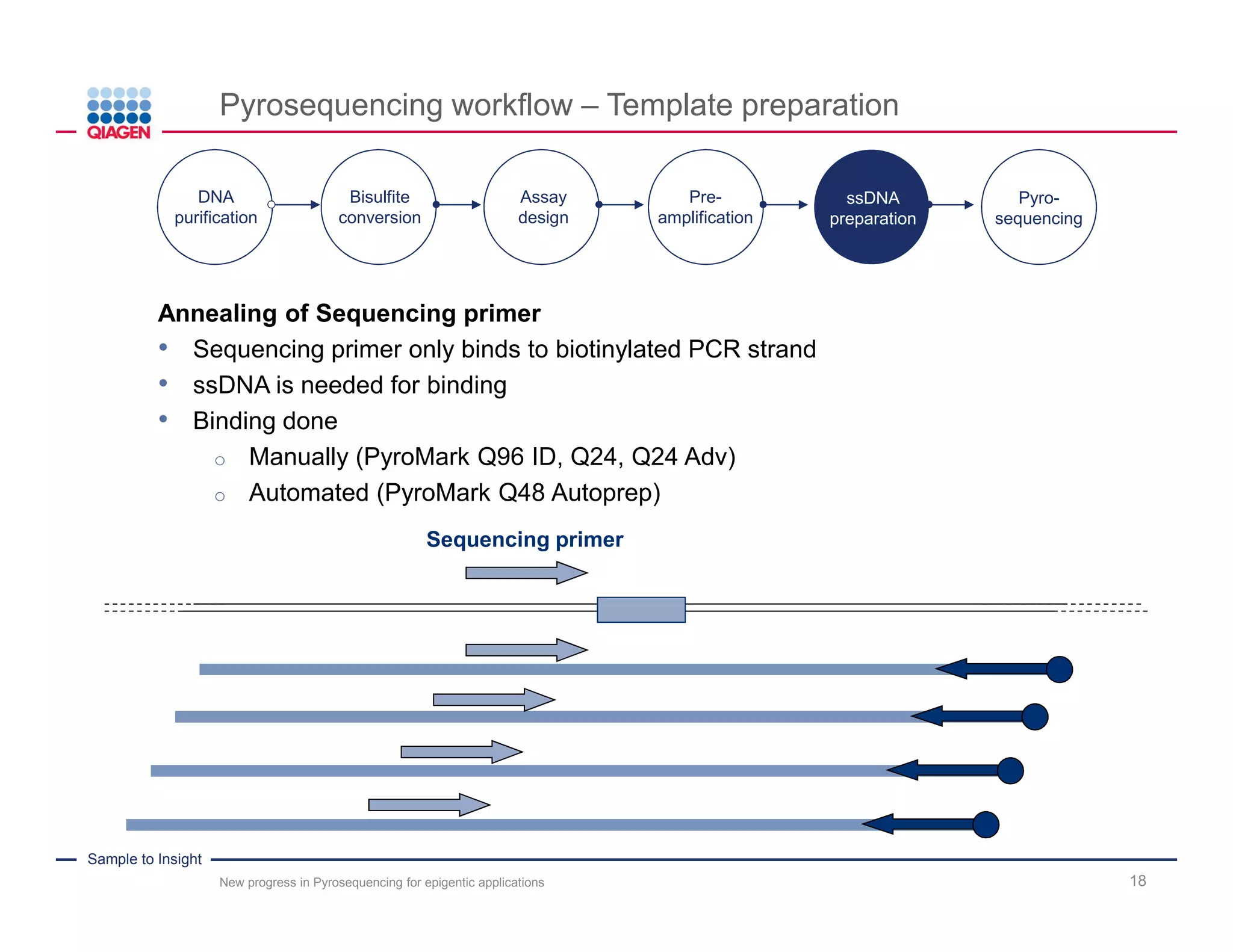
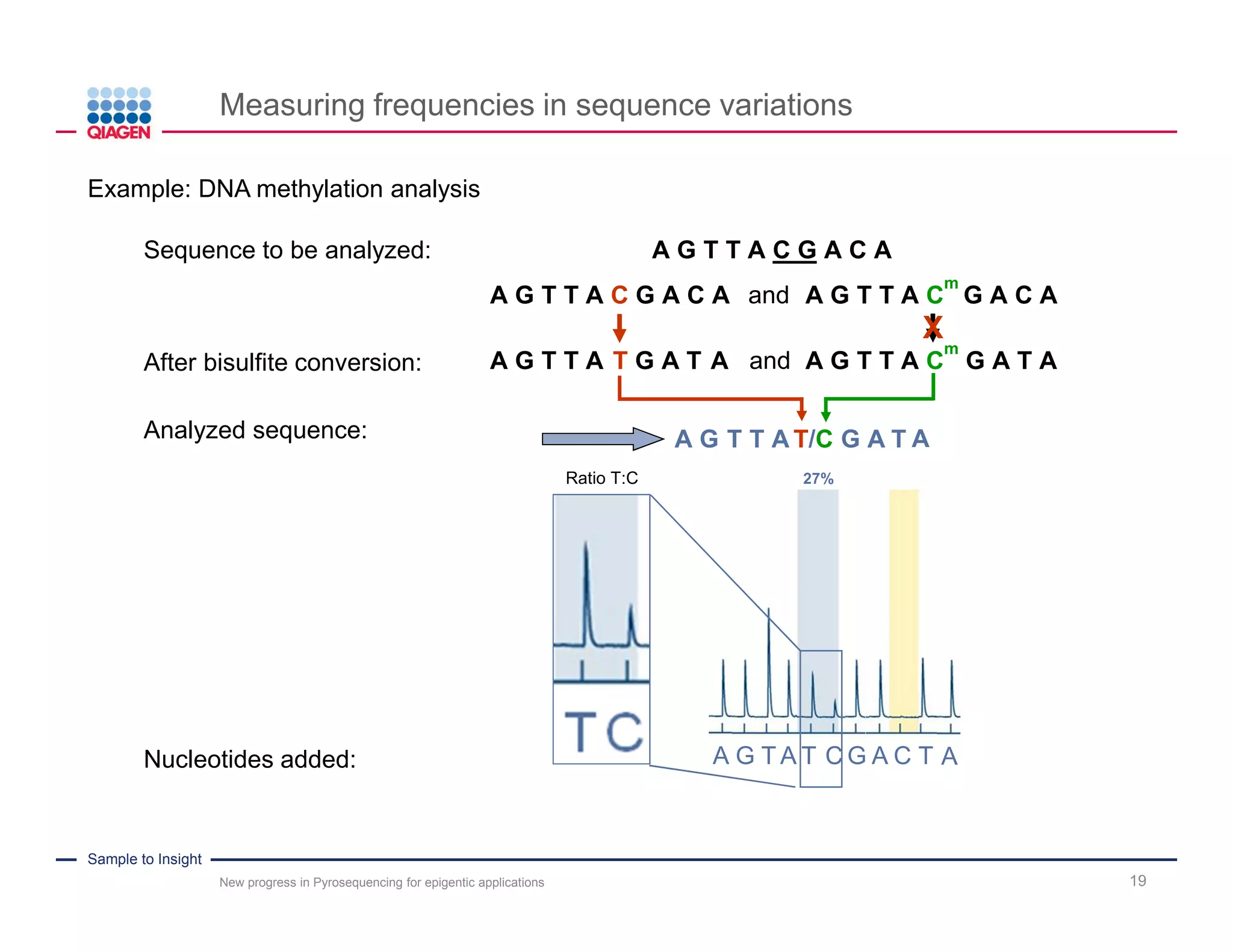
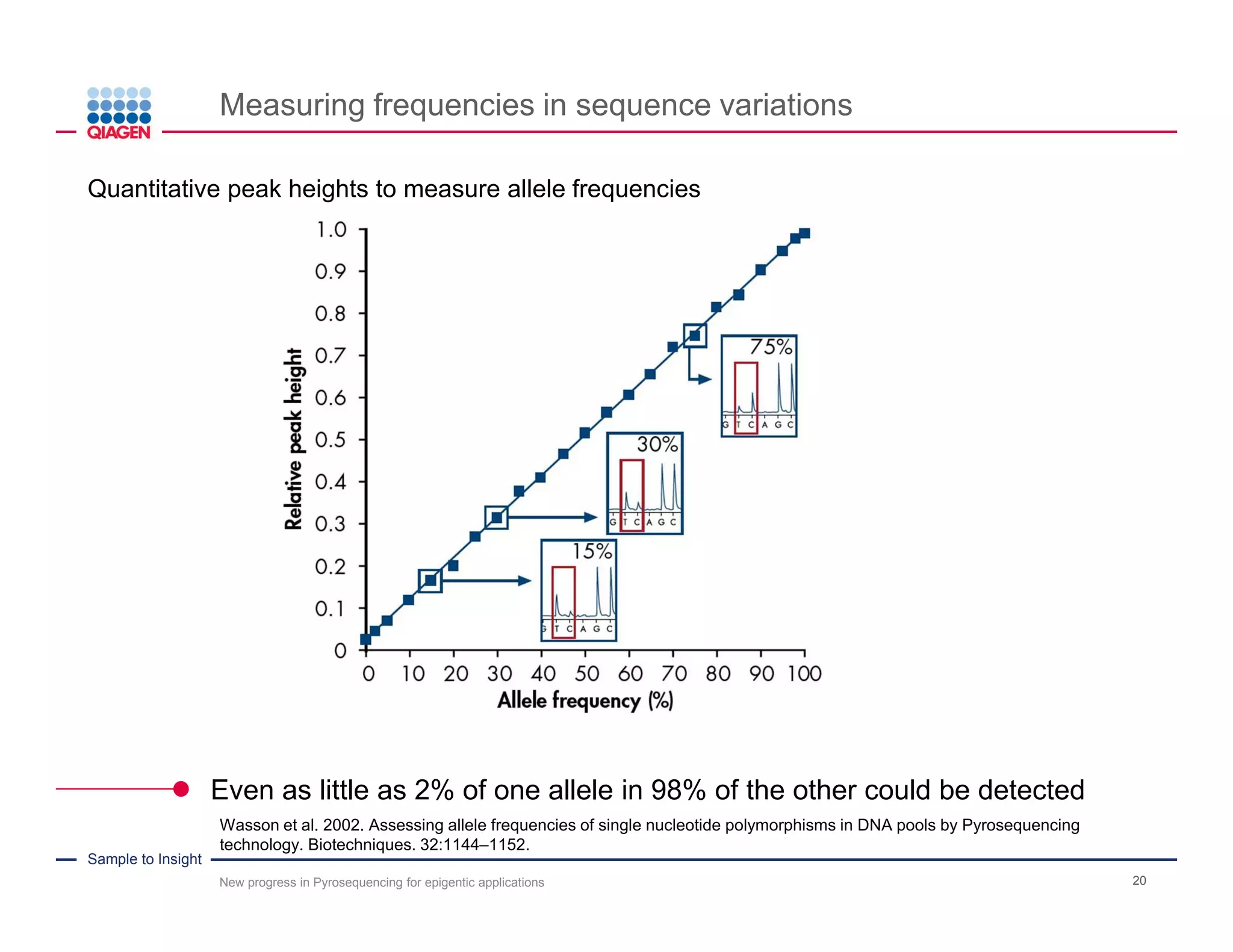
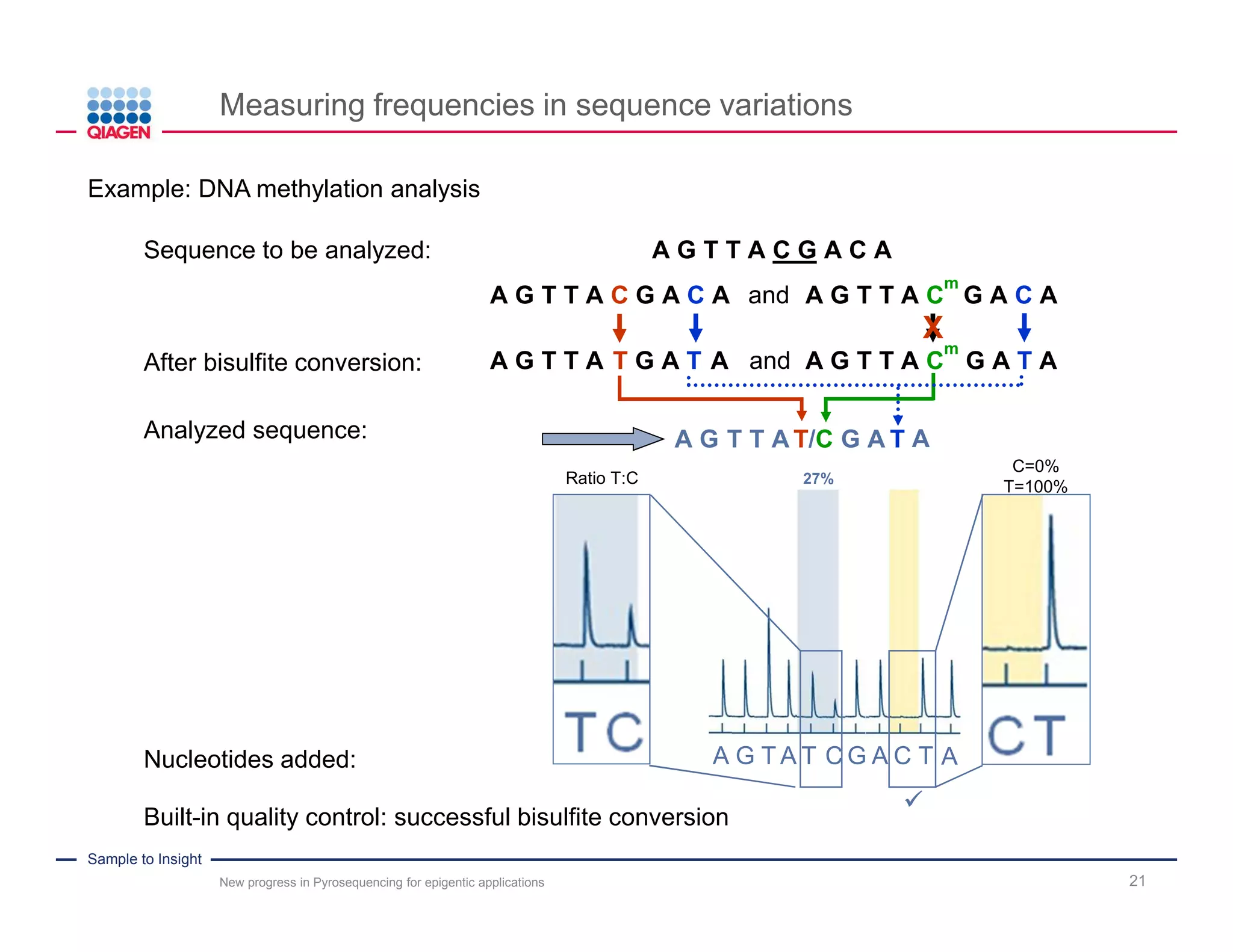
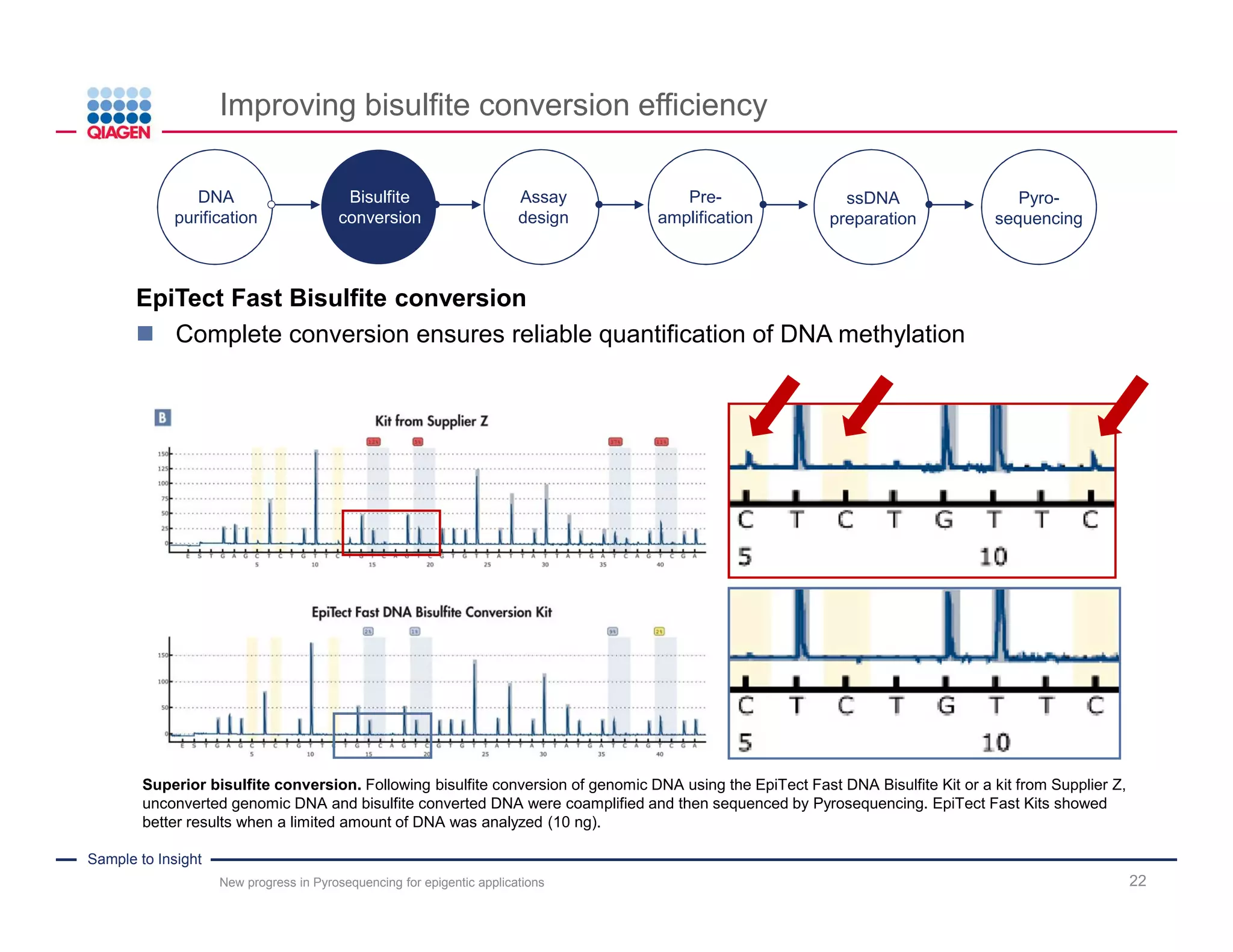
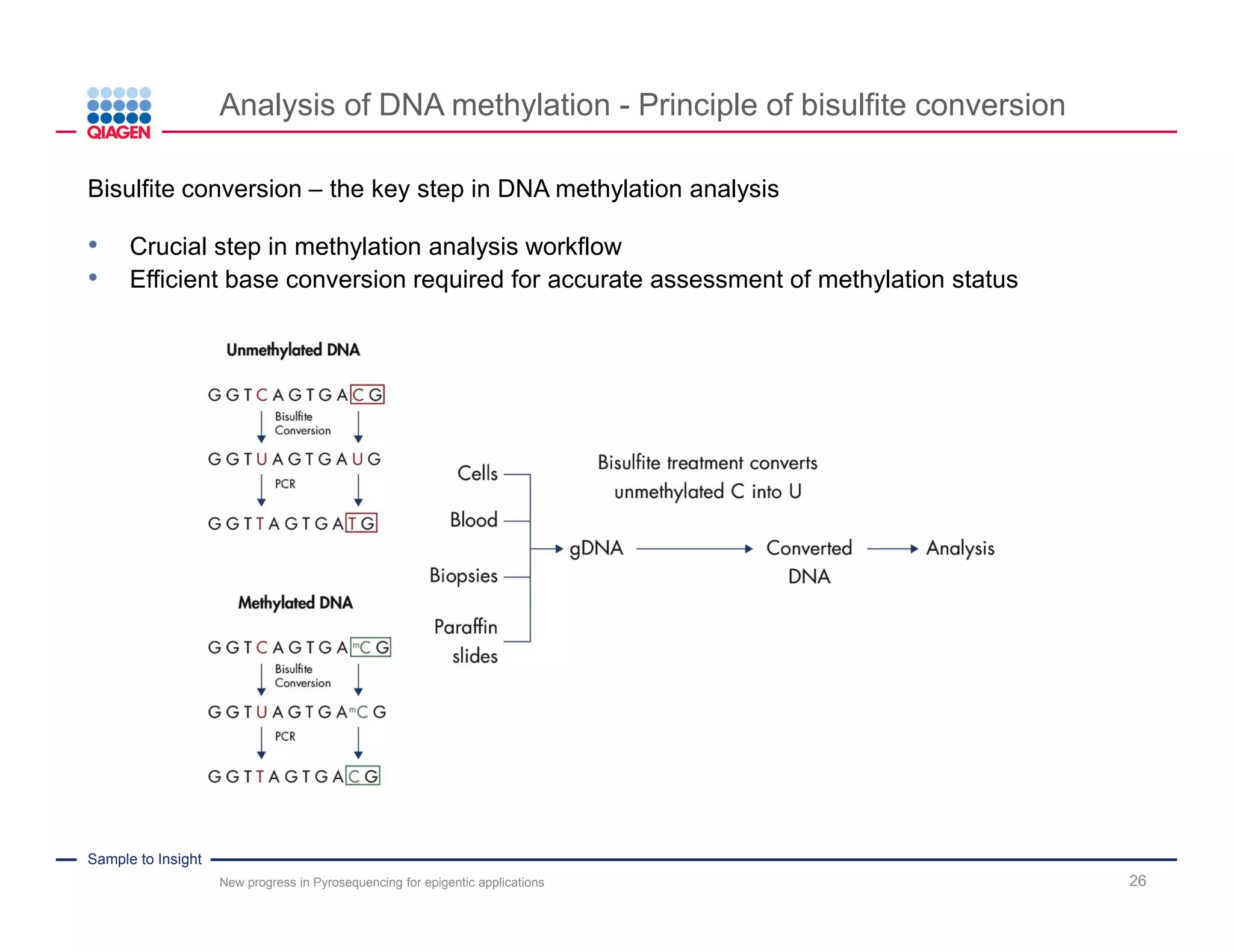
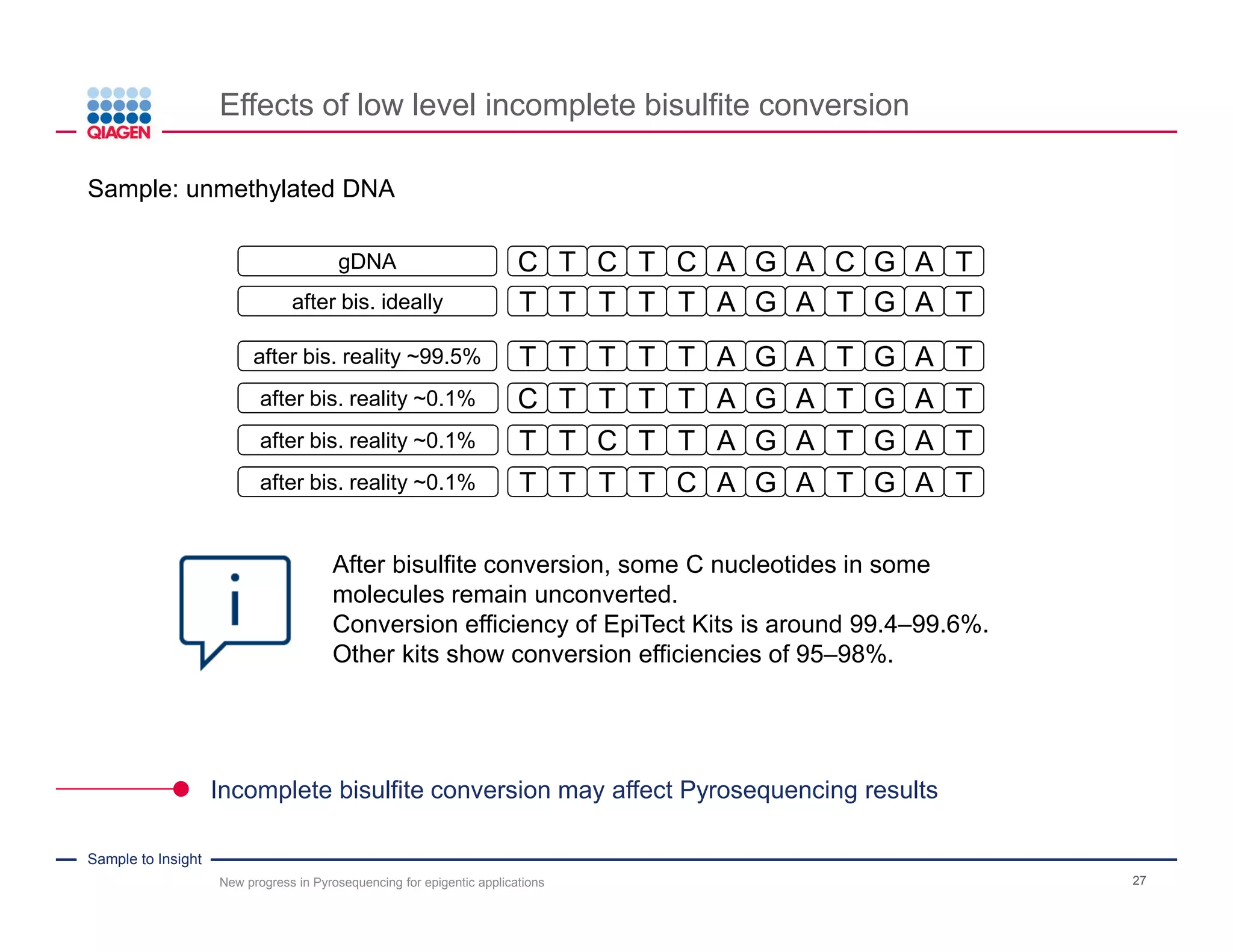
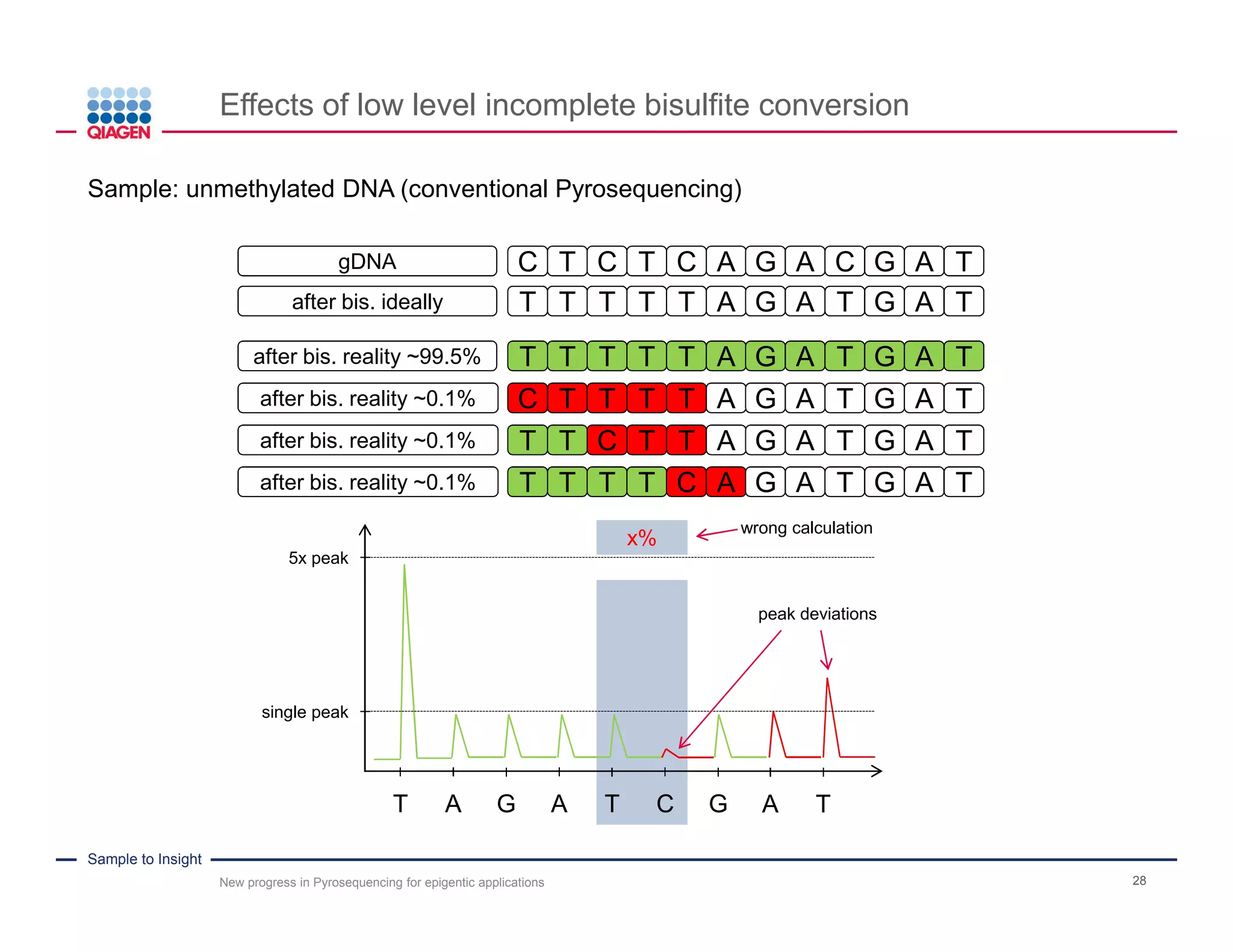
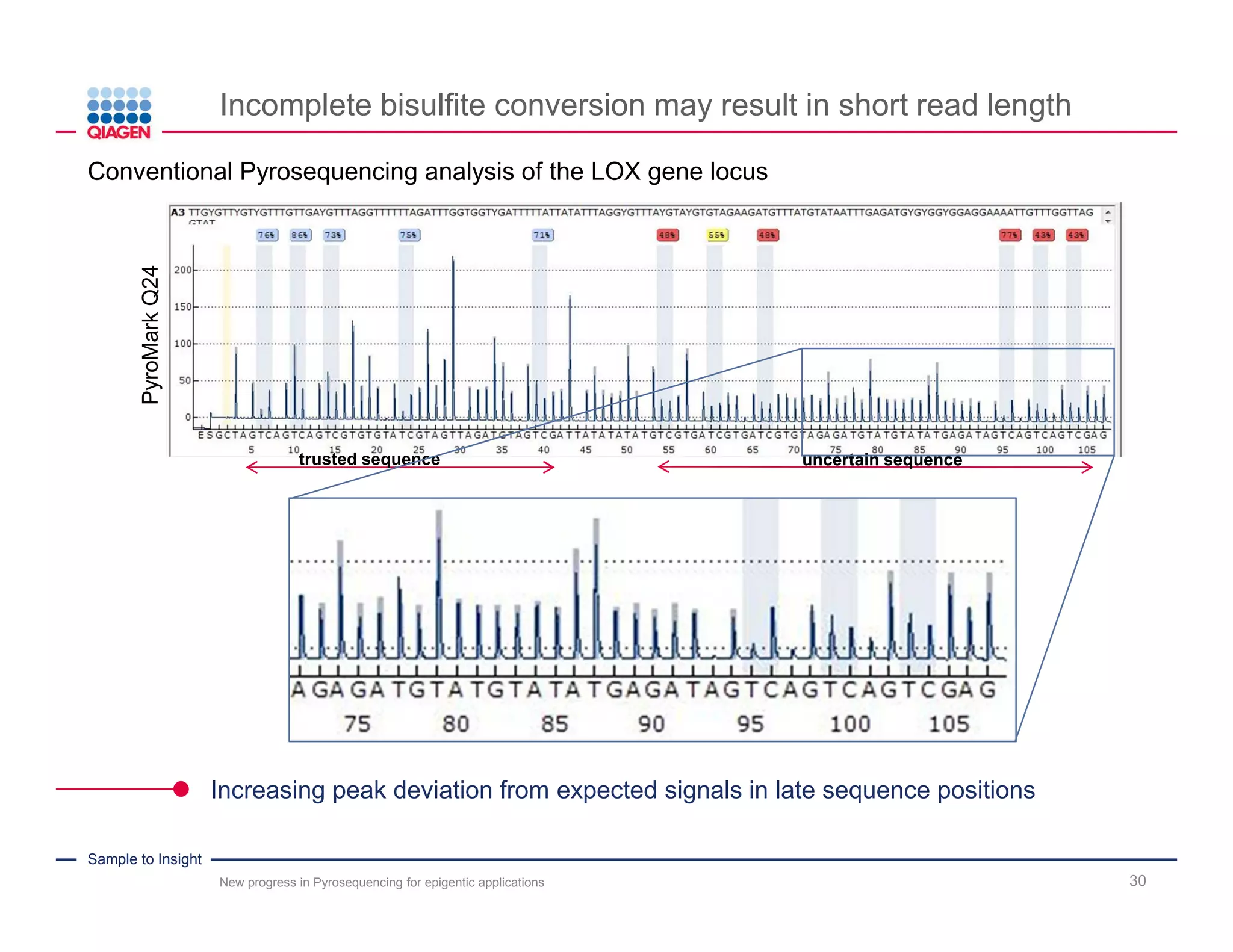
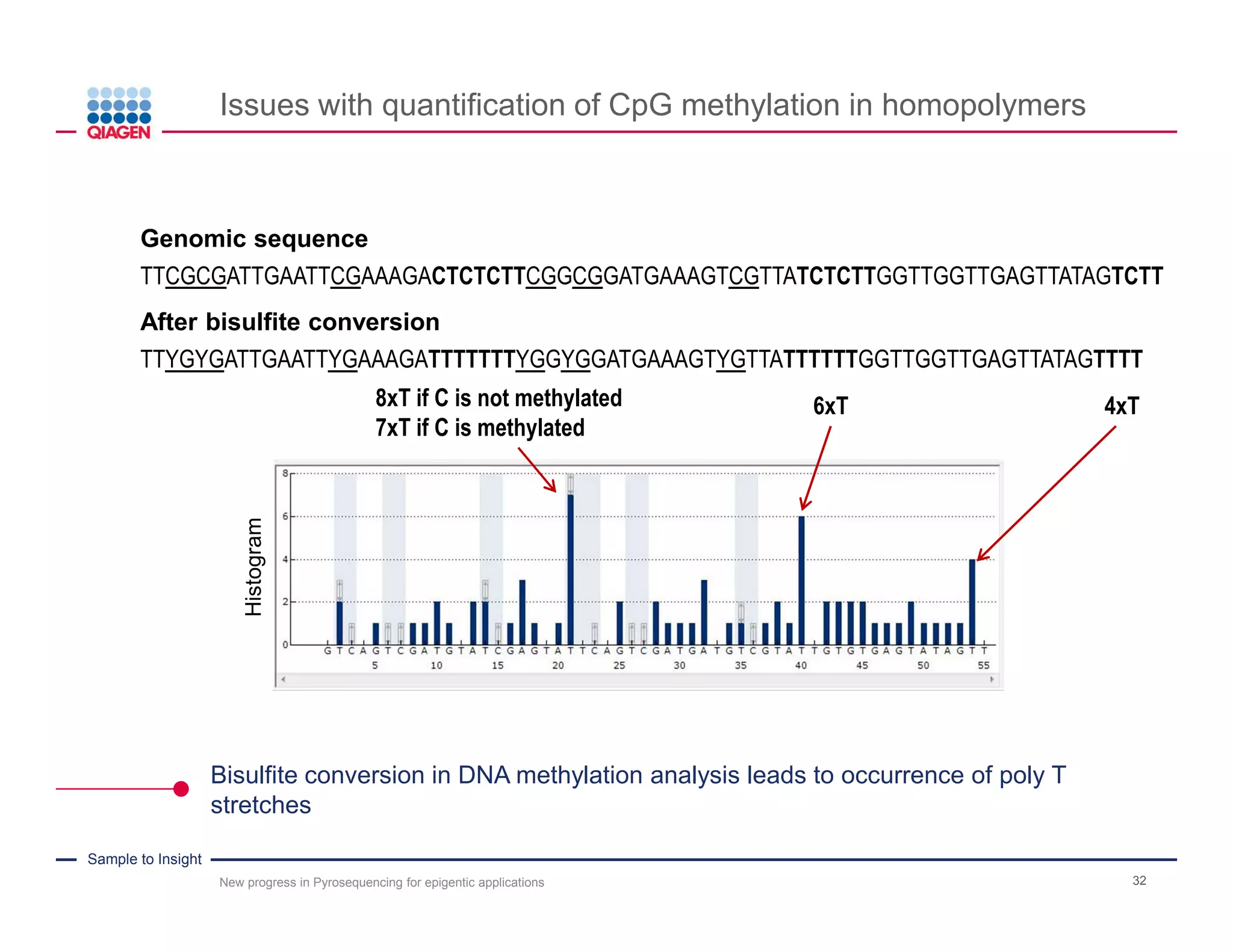
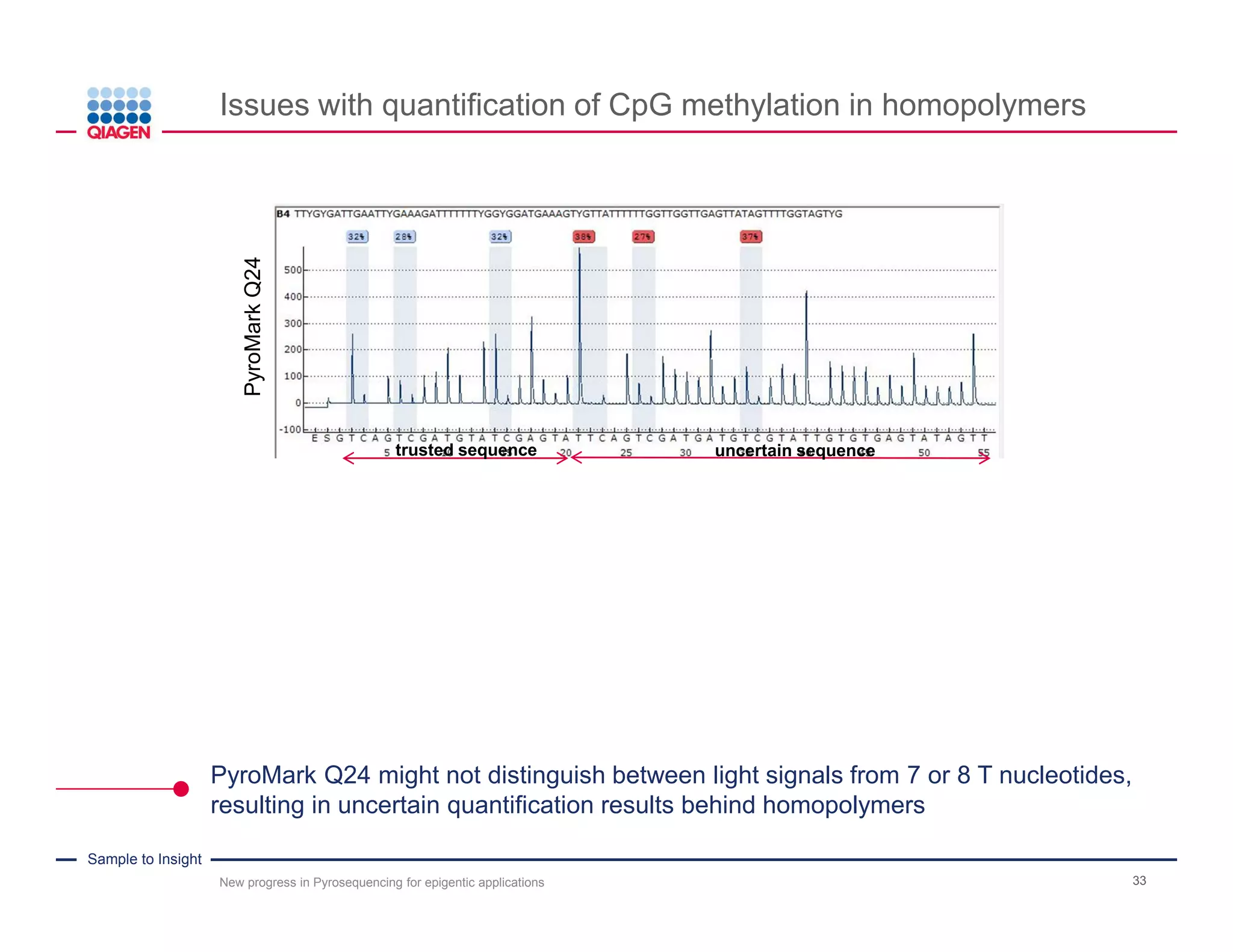
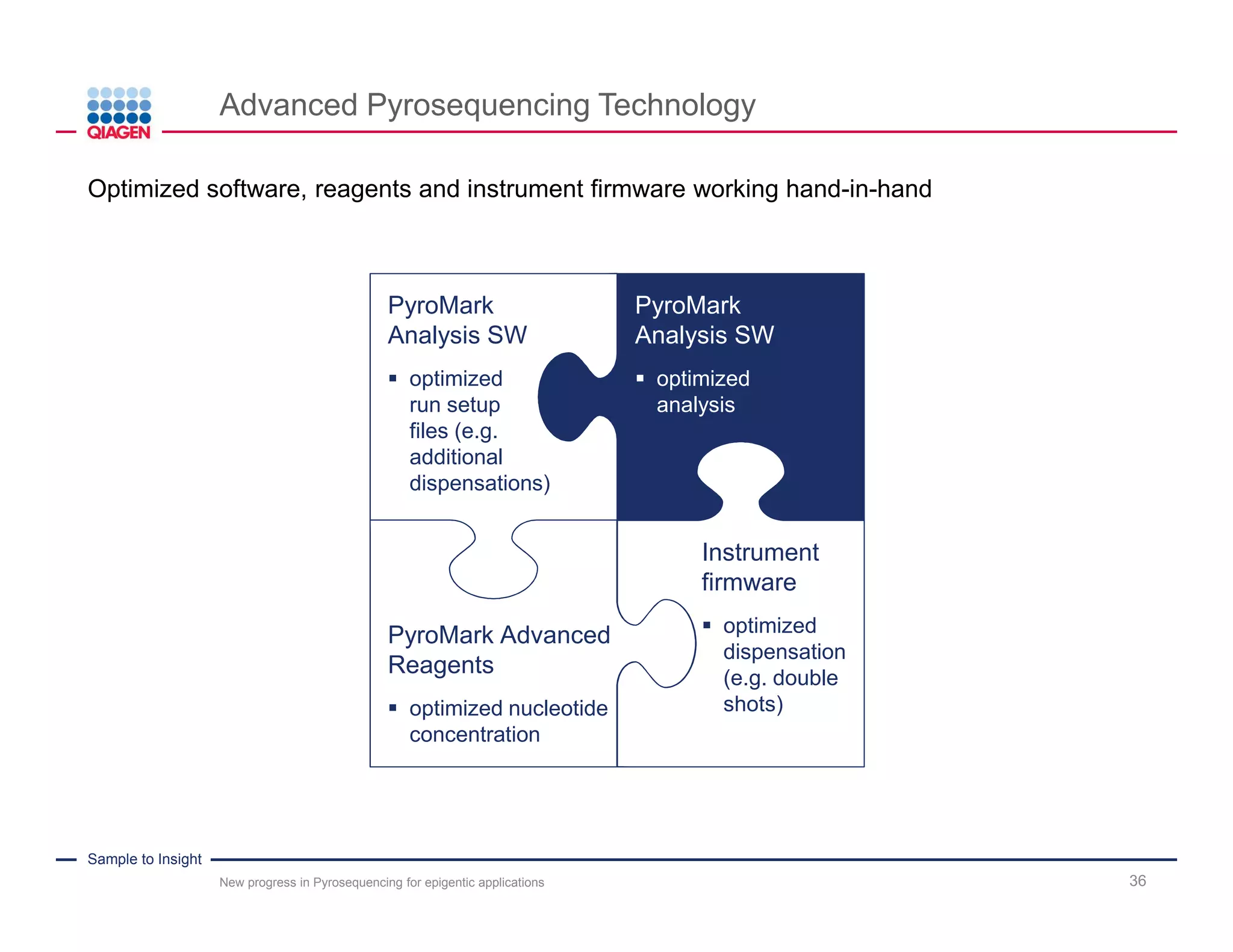
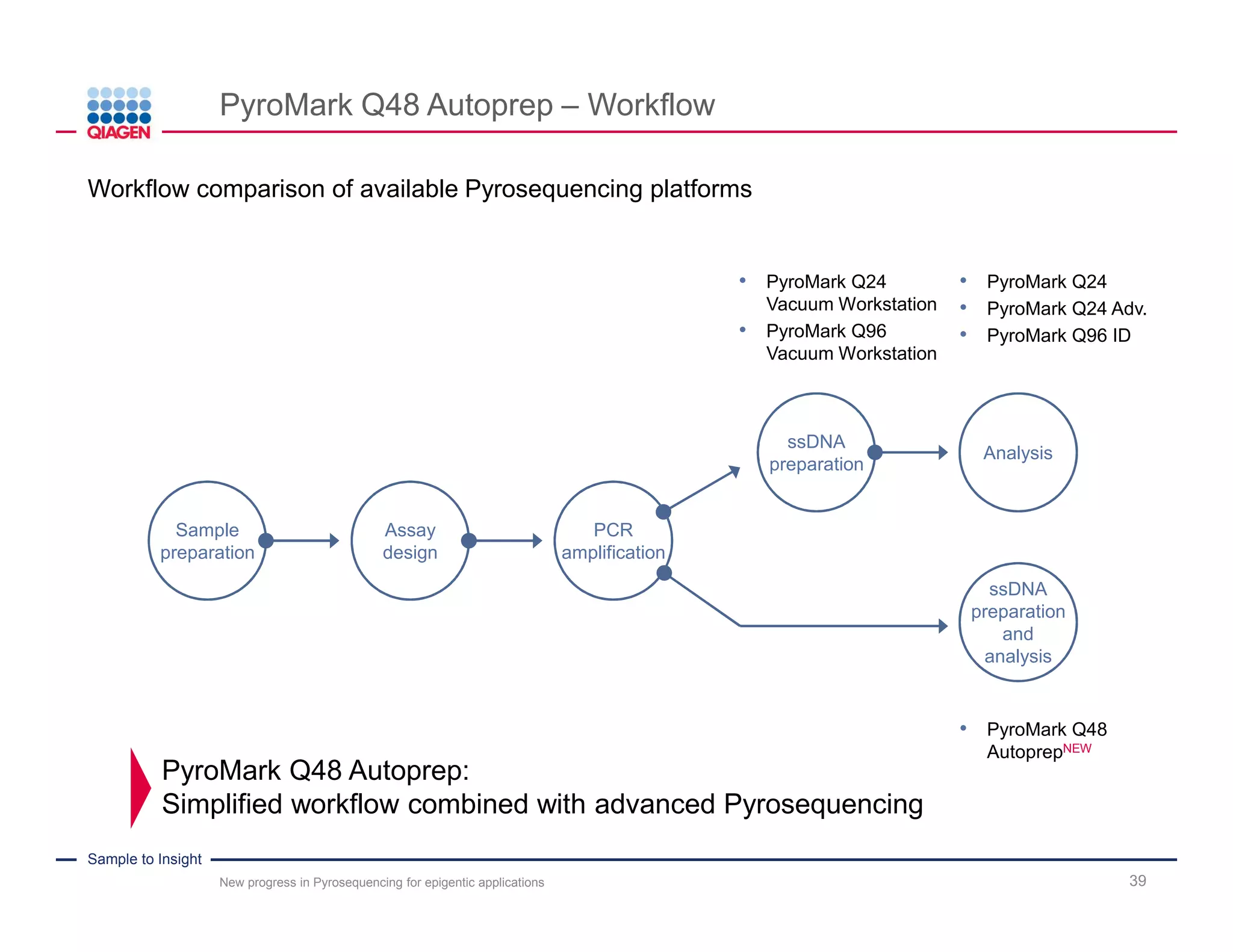
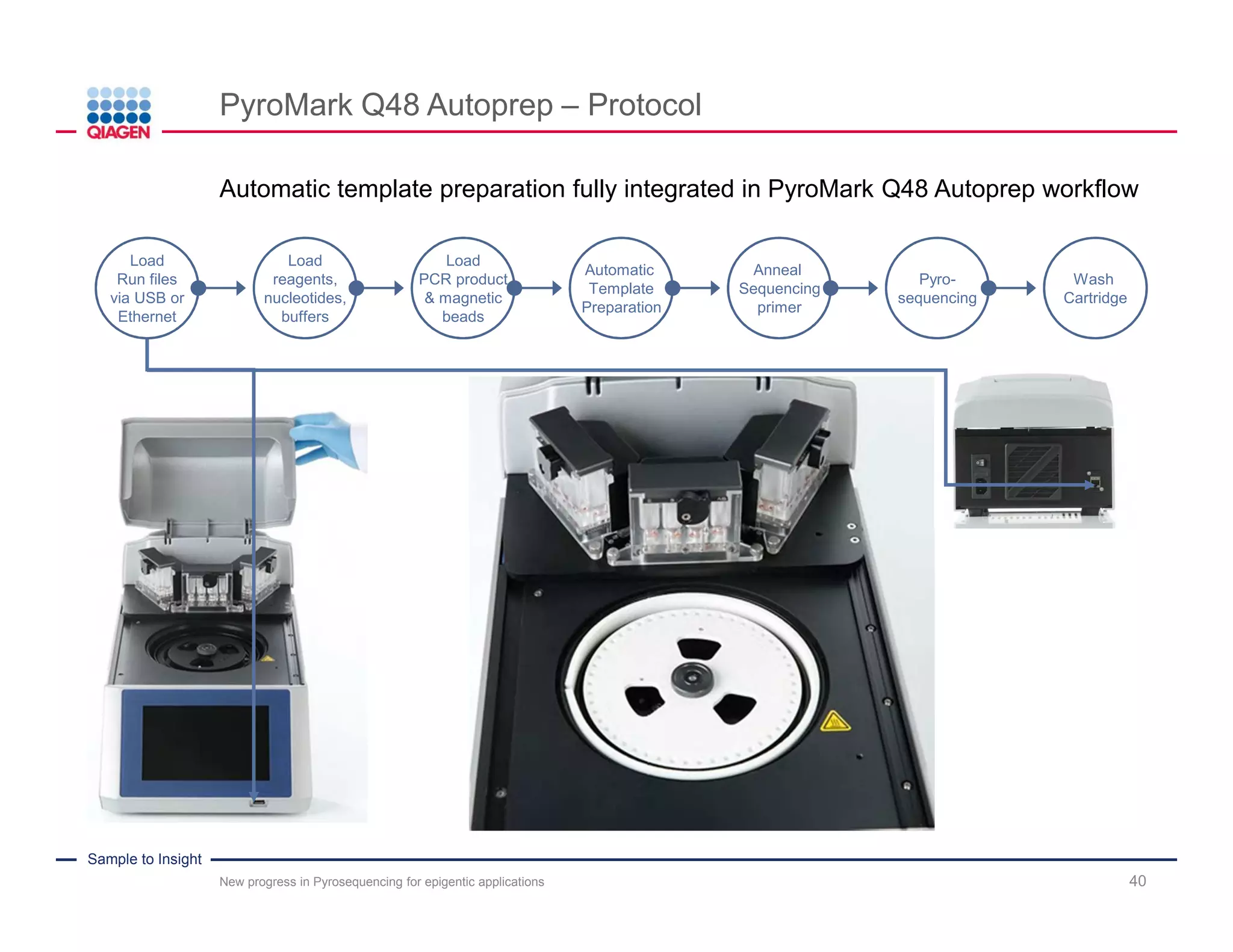
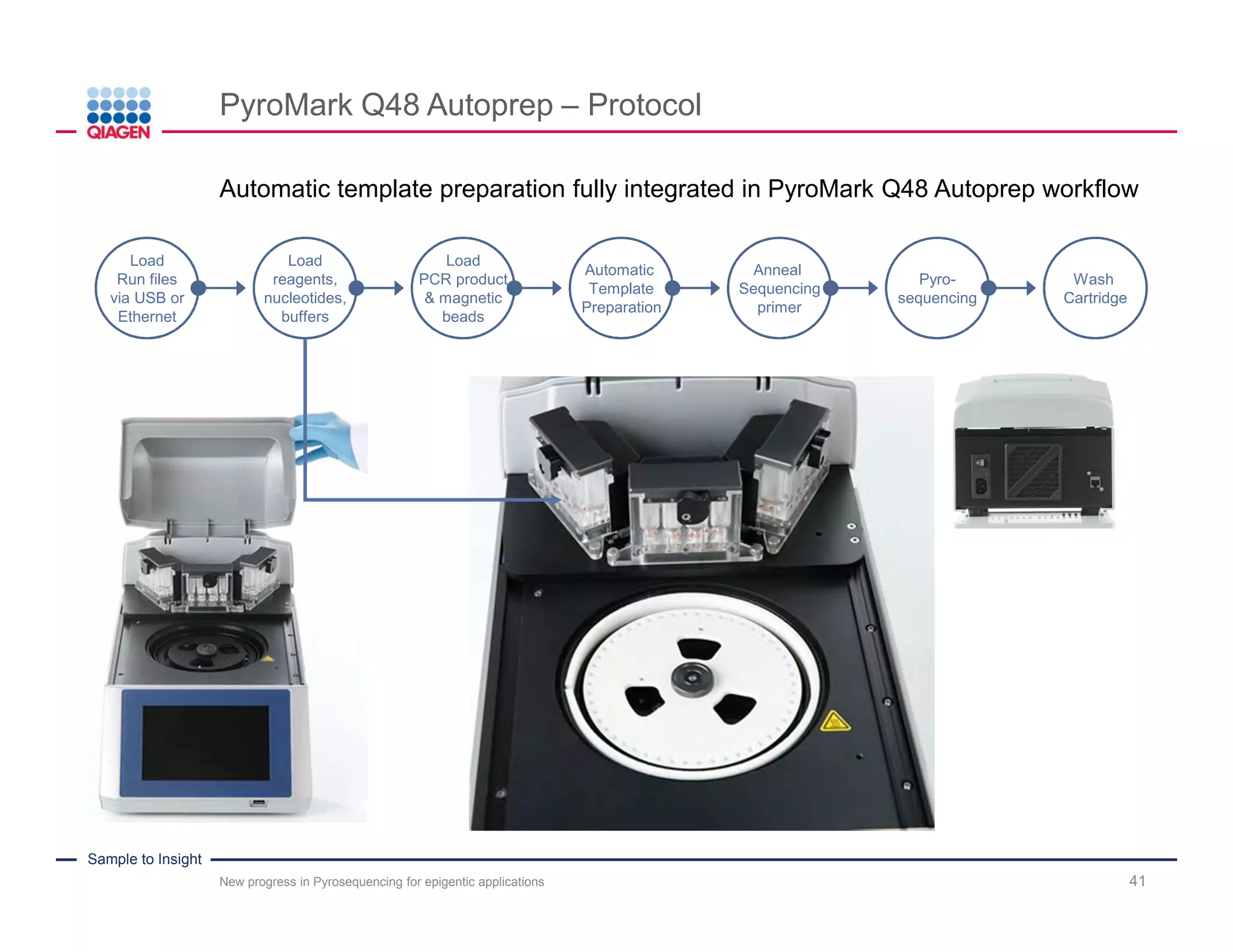
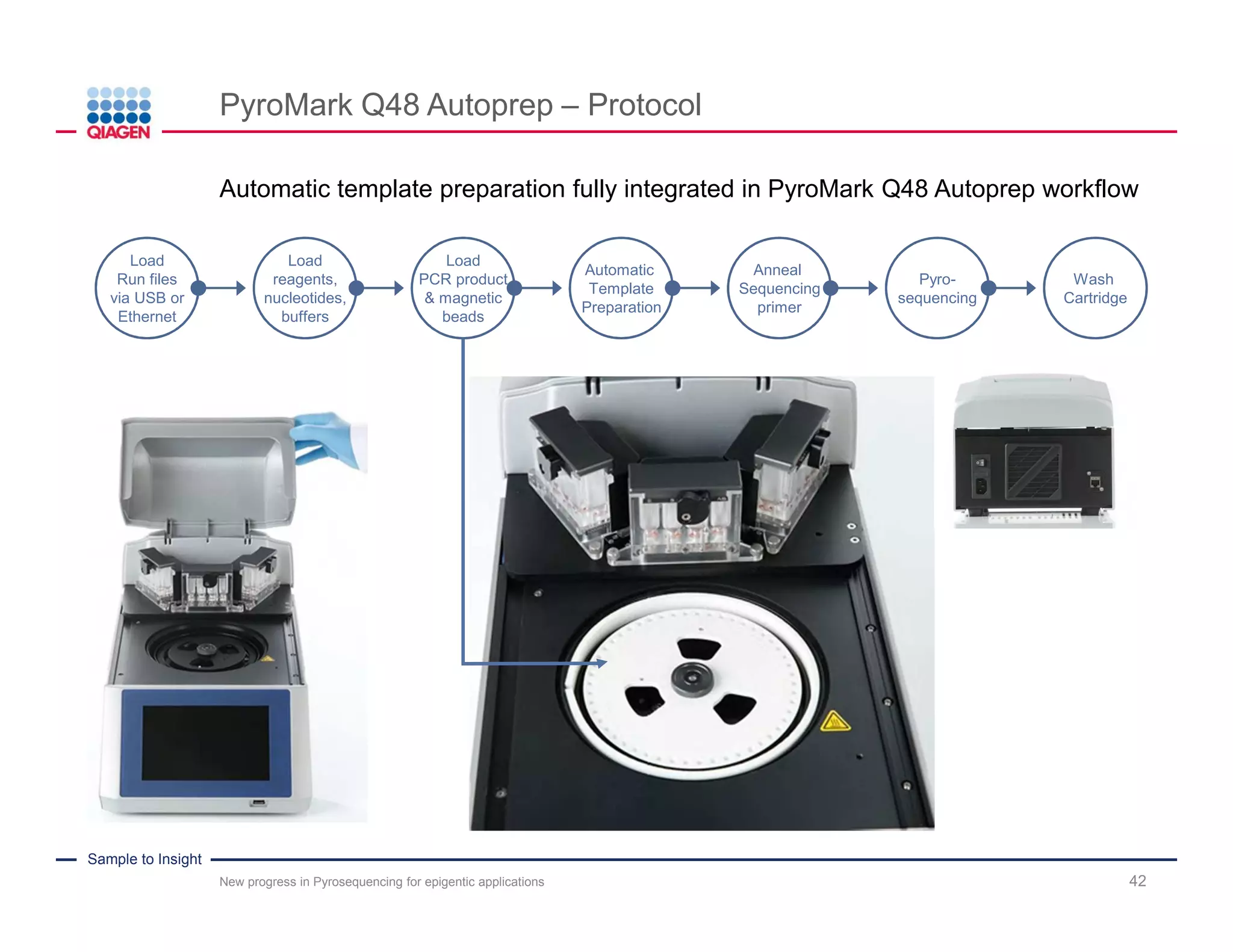
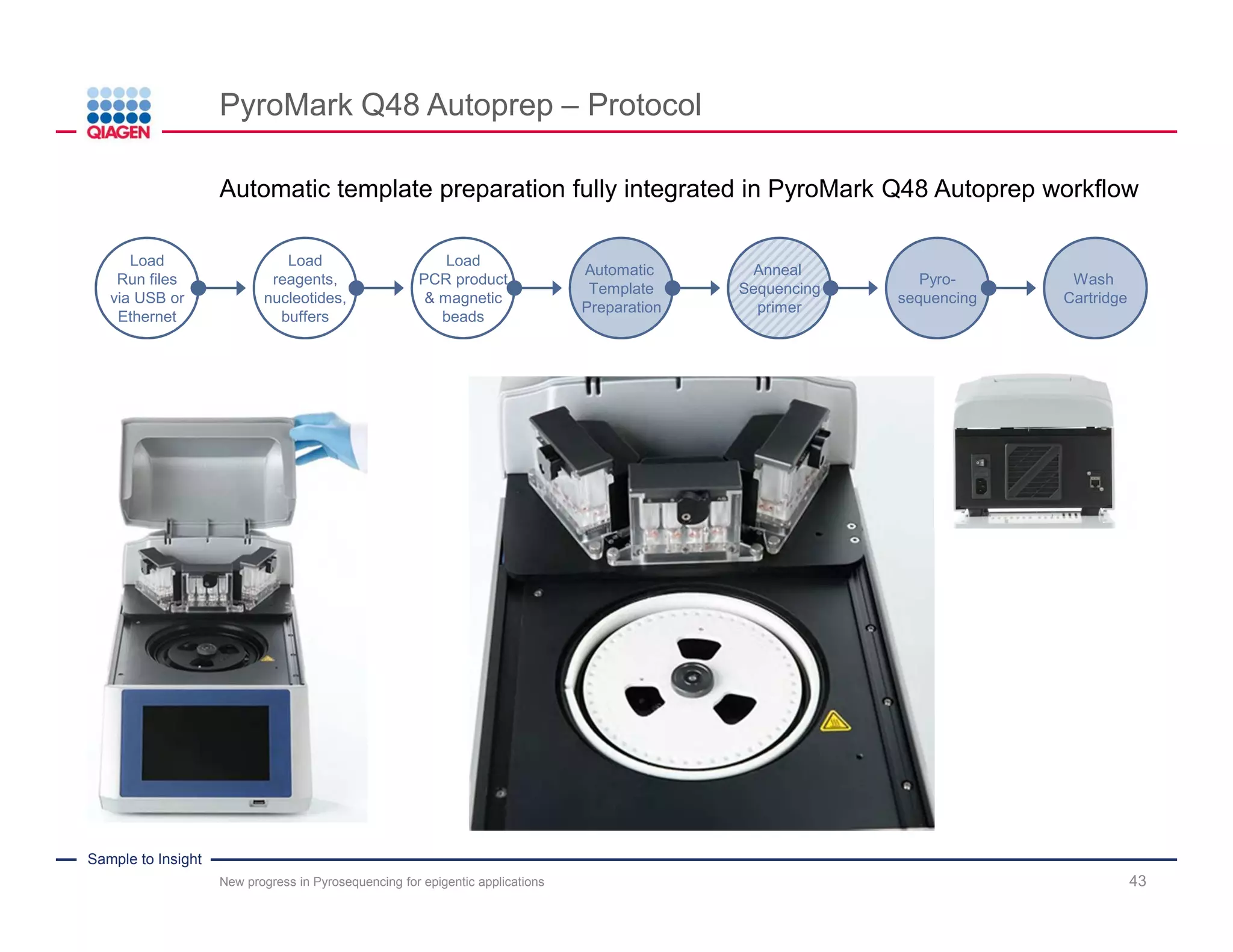
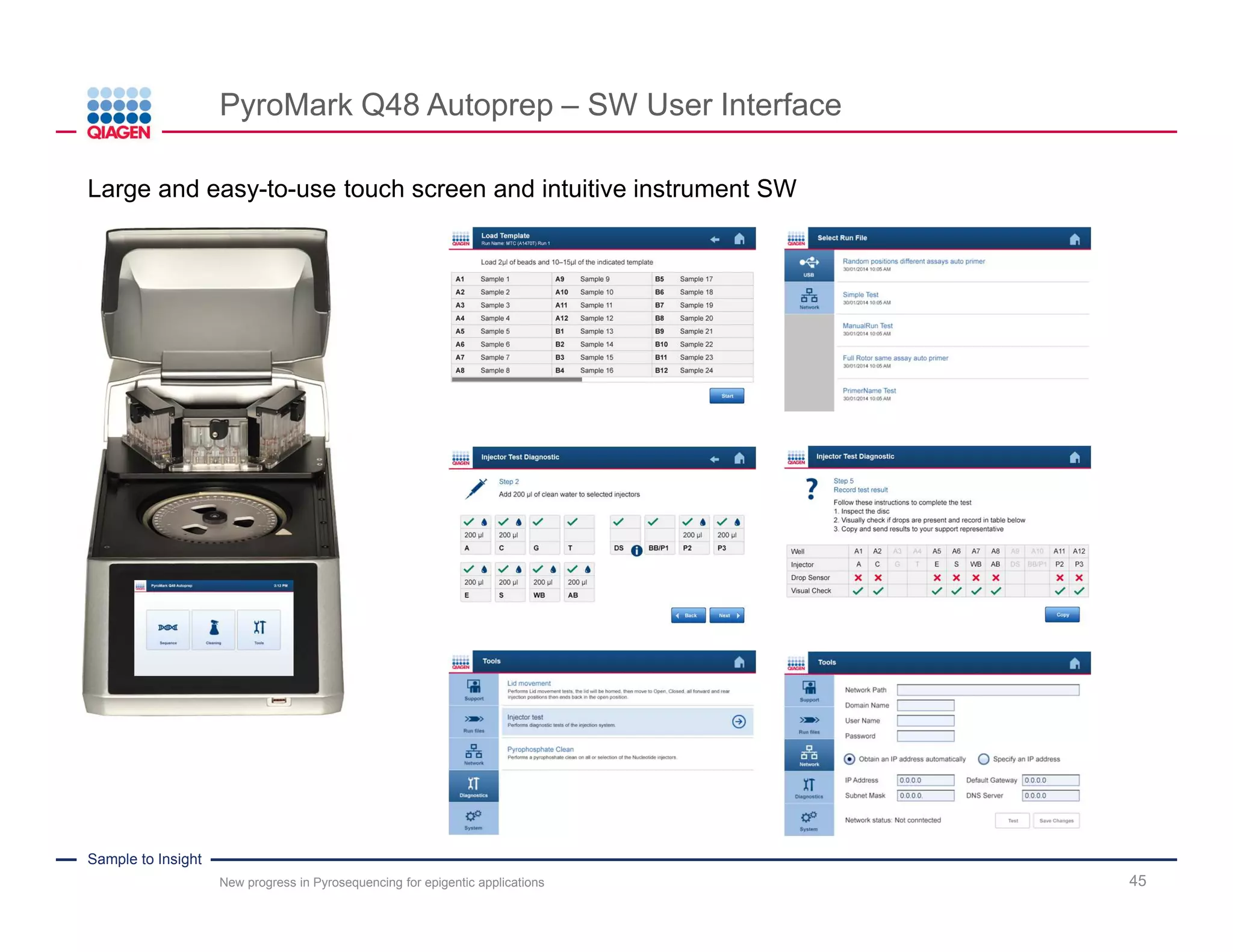
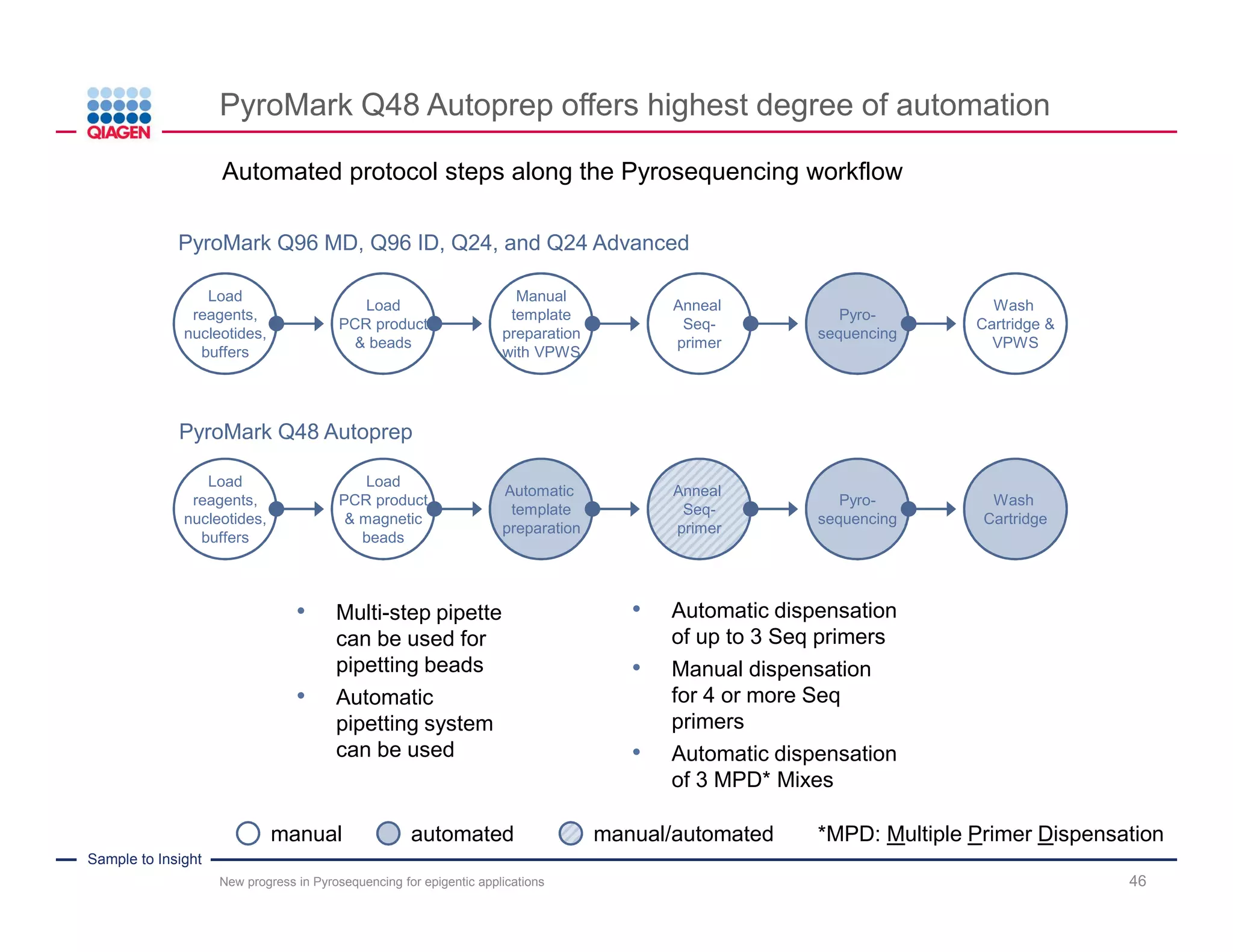
The document discusses advancements in pyrosequencing technology for automated DNA methylation analysis, highlighting challenges such as insufficient read lengths and incomplete bisulfite conversion. It introduces the PyroMark Q48 Autoprep and emphasizes its potential in overcoming these bottlenecks for epigenetic research. Additionally, it outlines the workflow for sample preparation and data analysis critical for reliable quantification of DNA methylation.