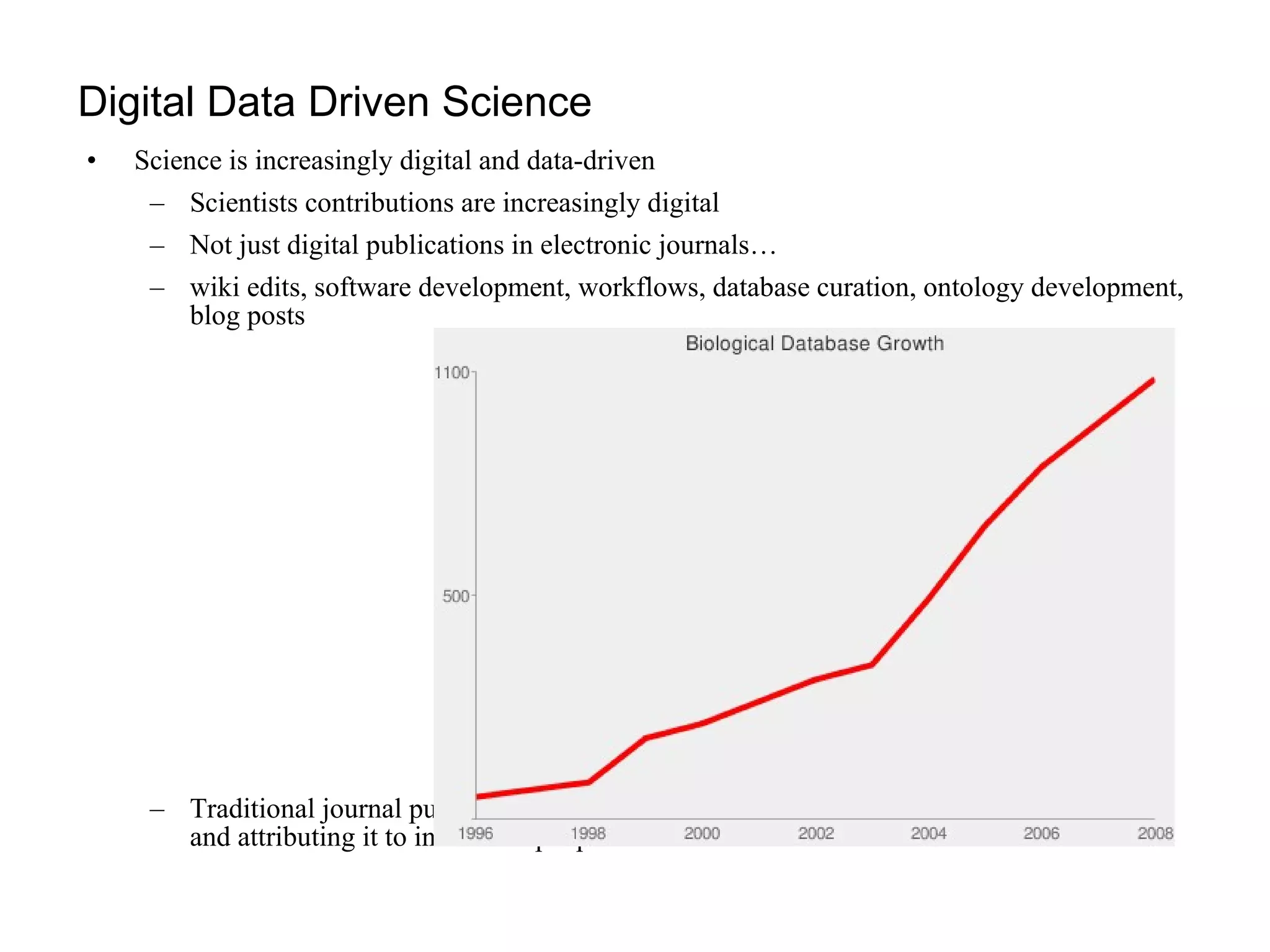
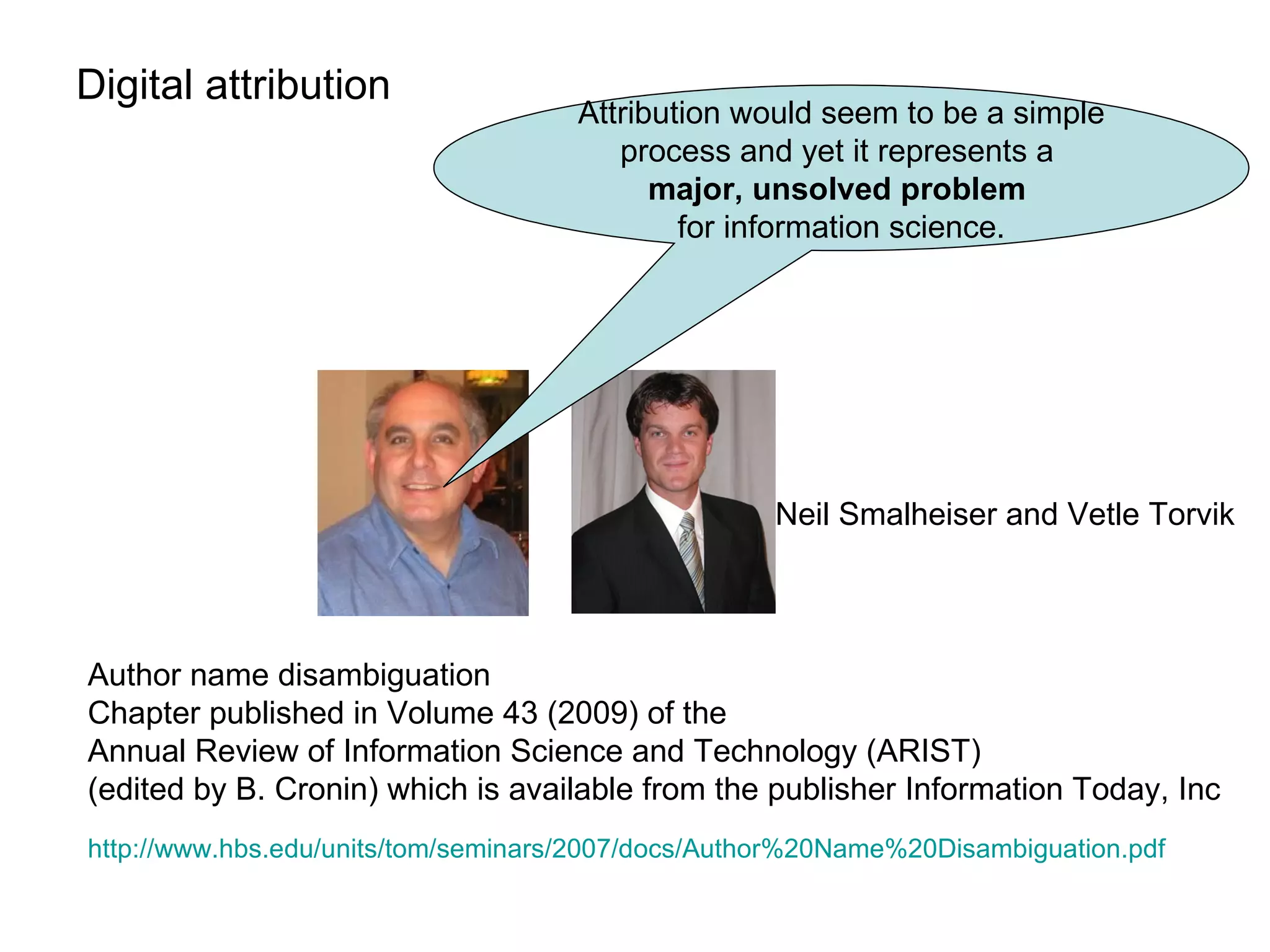
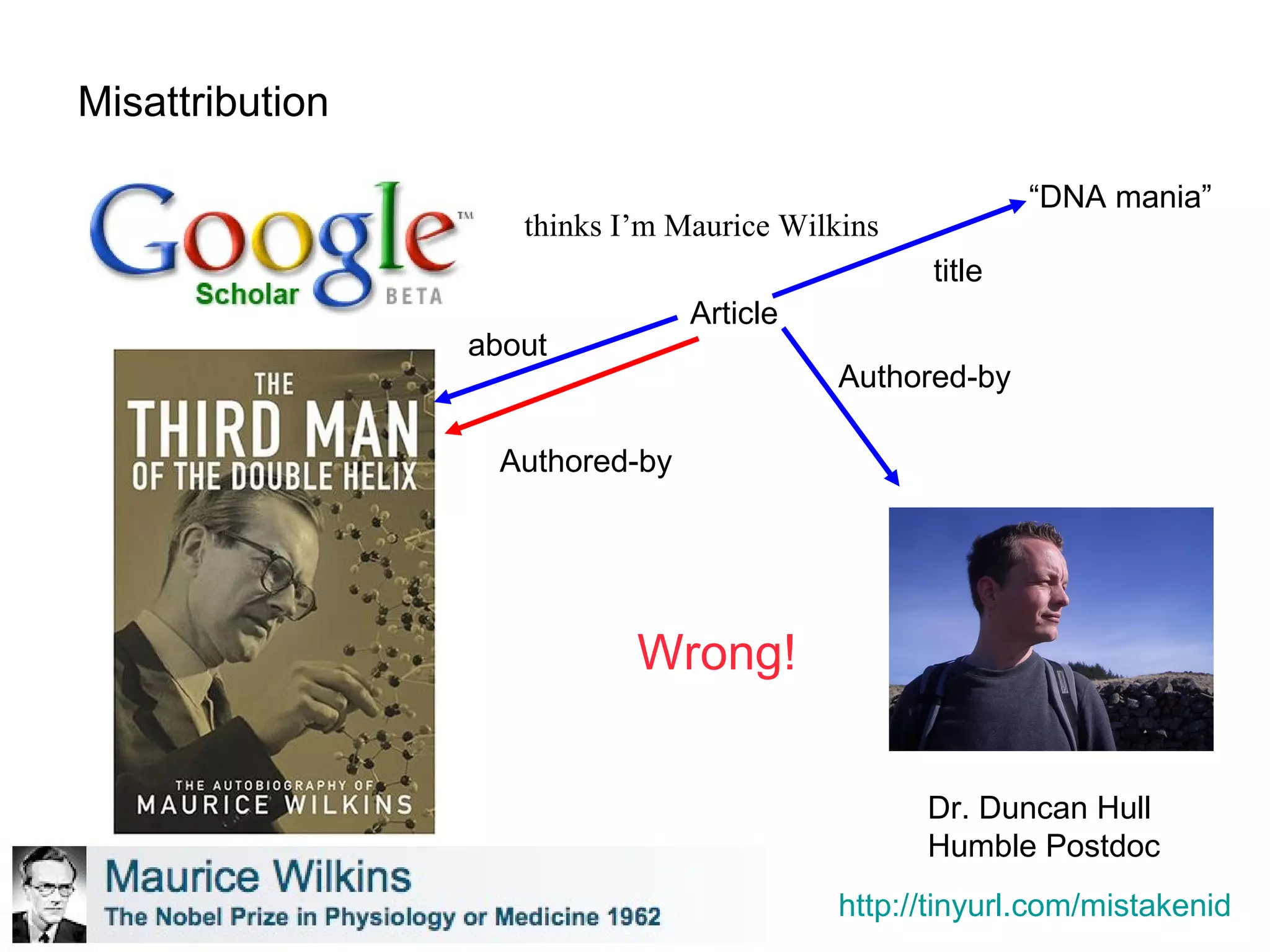
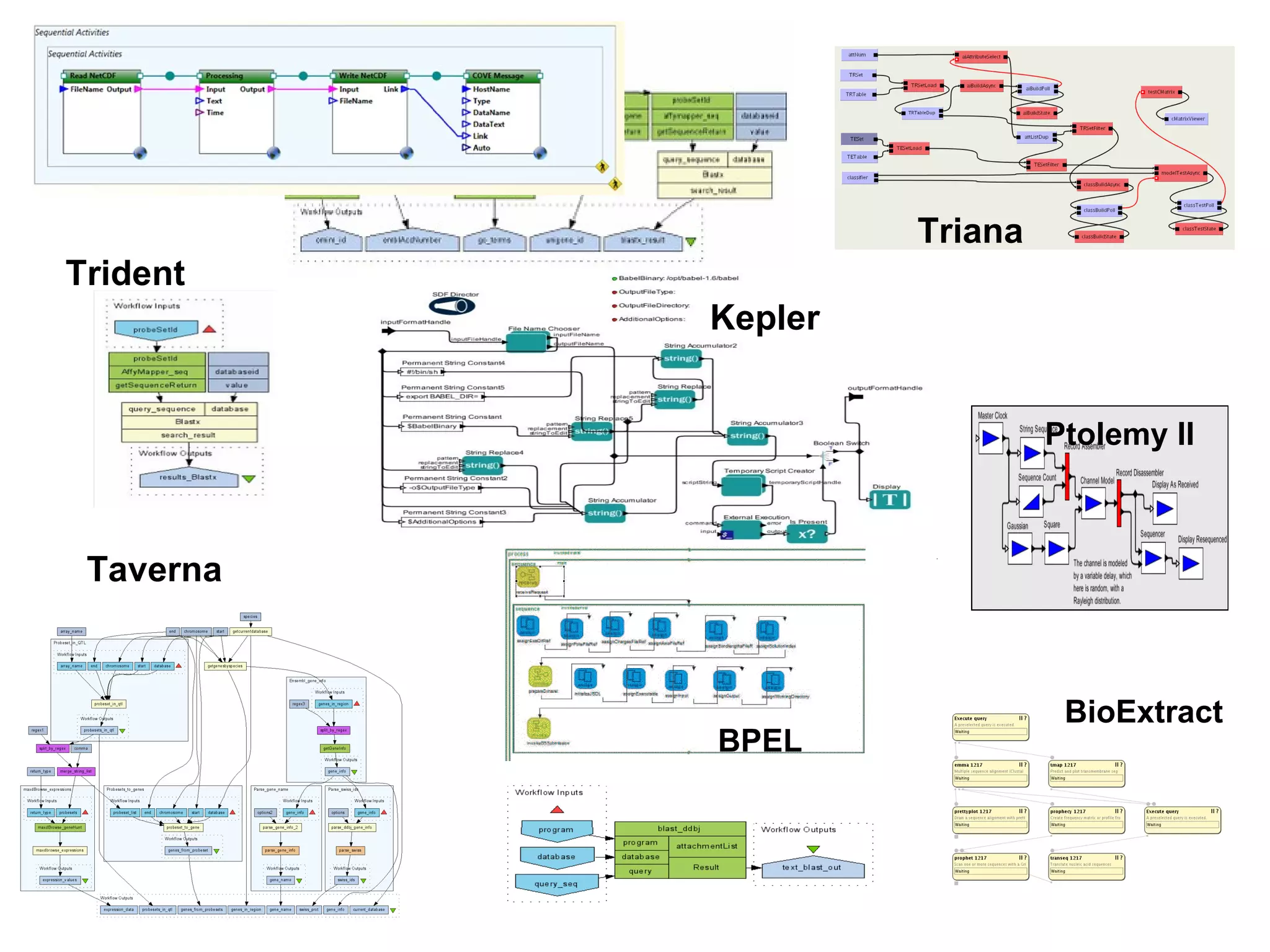
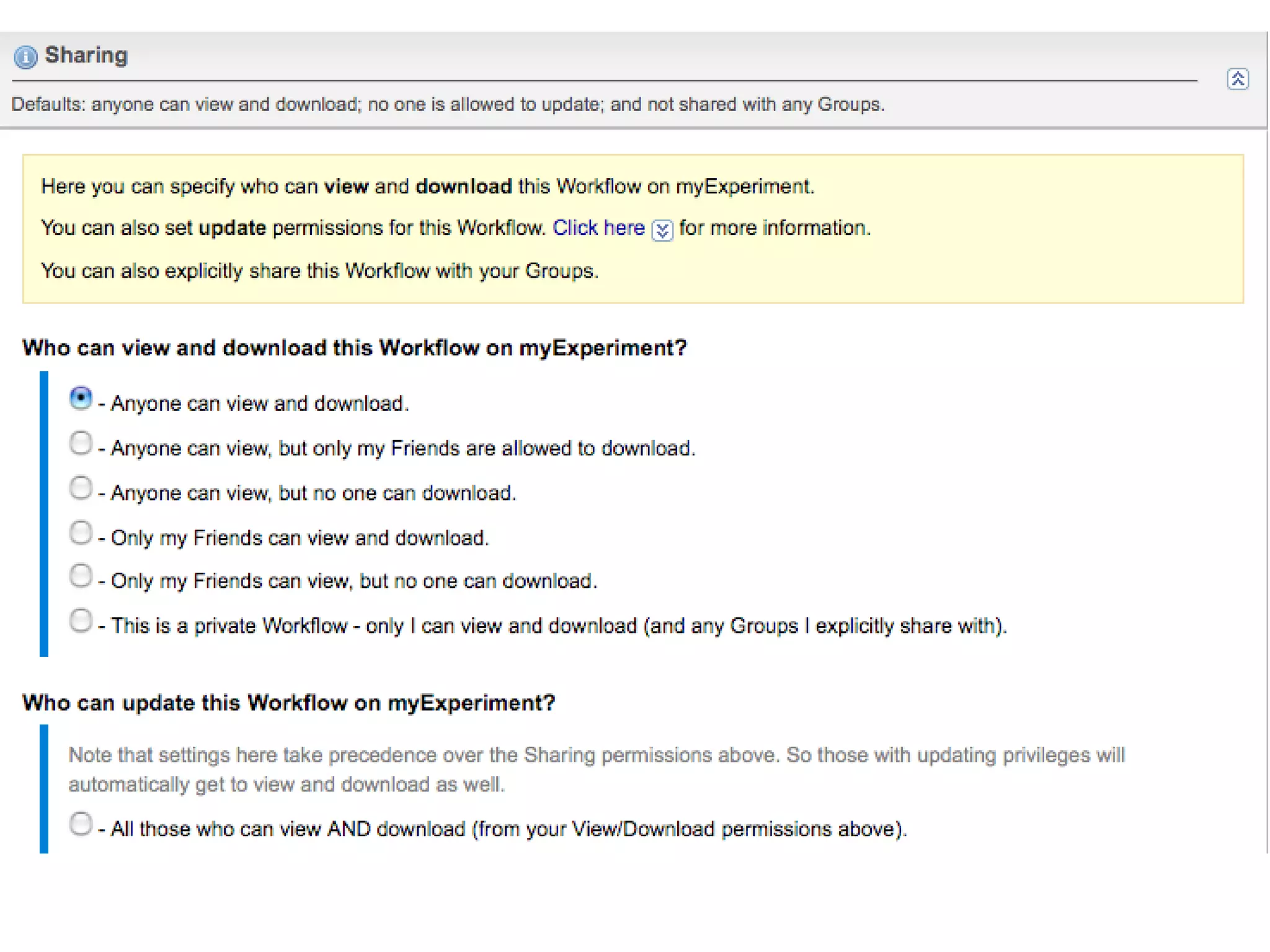
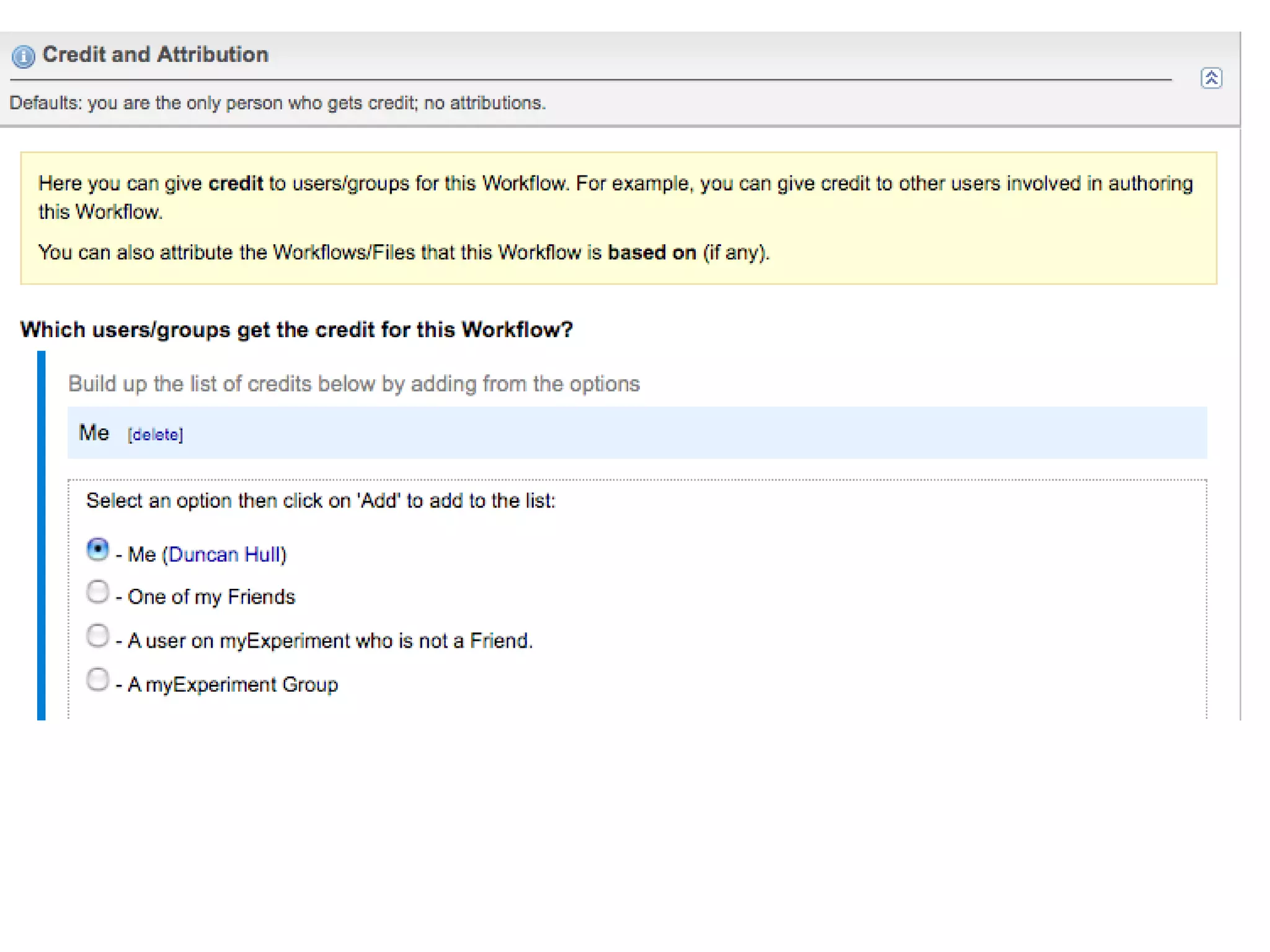
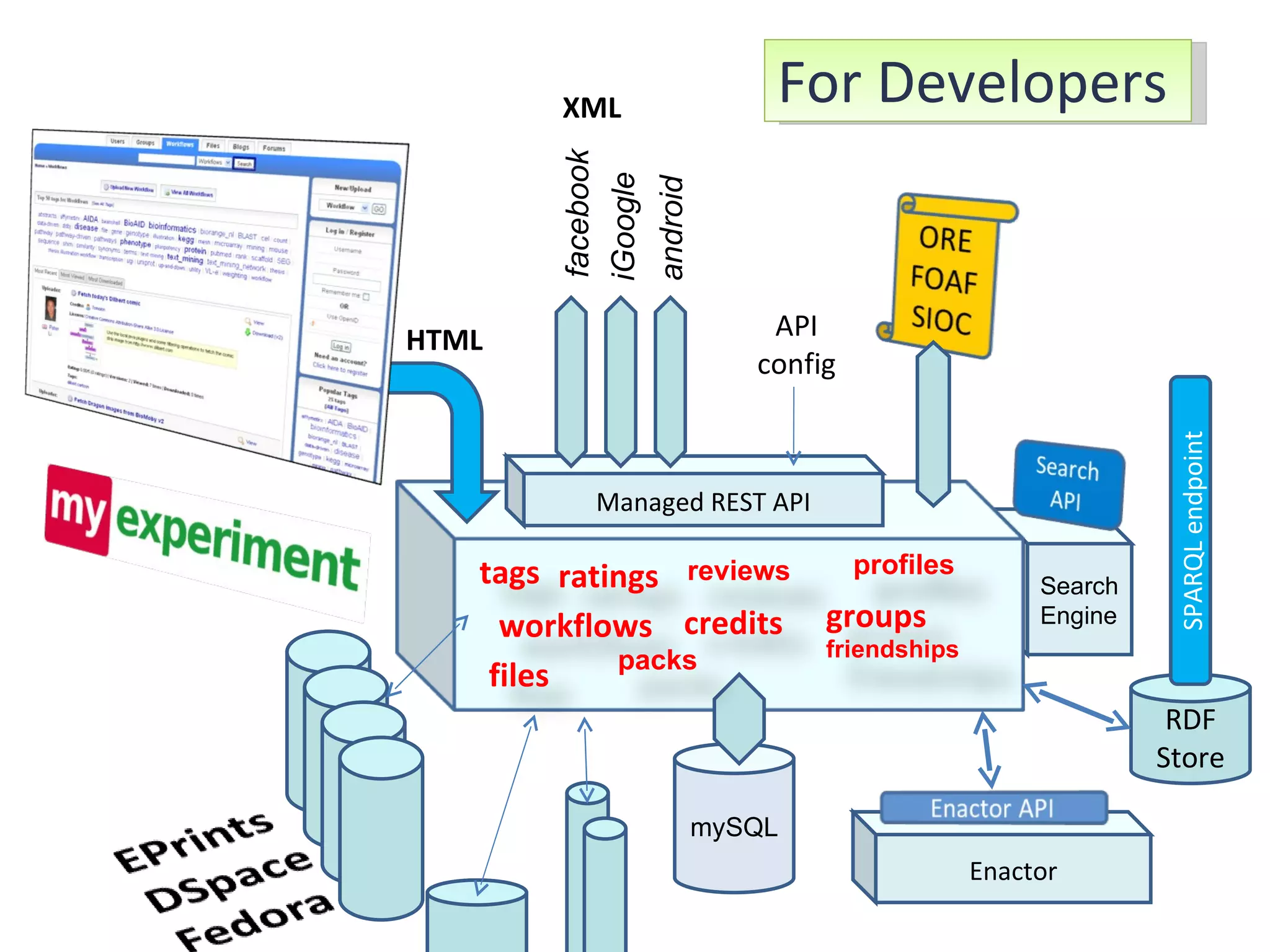
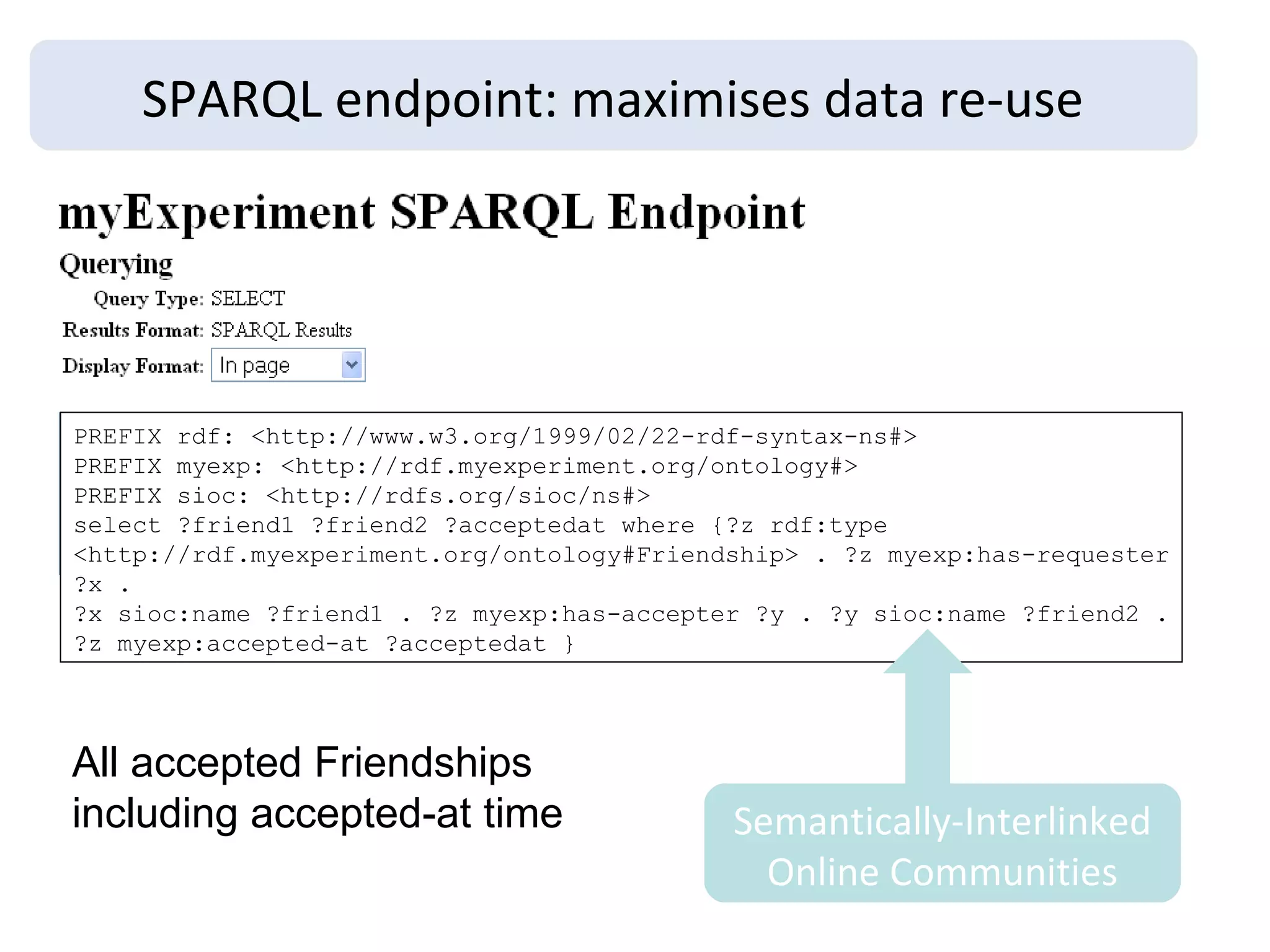
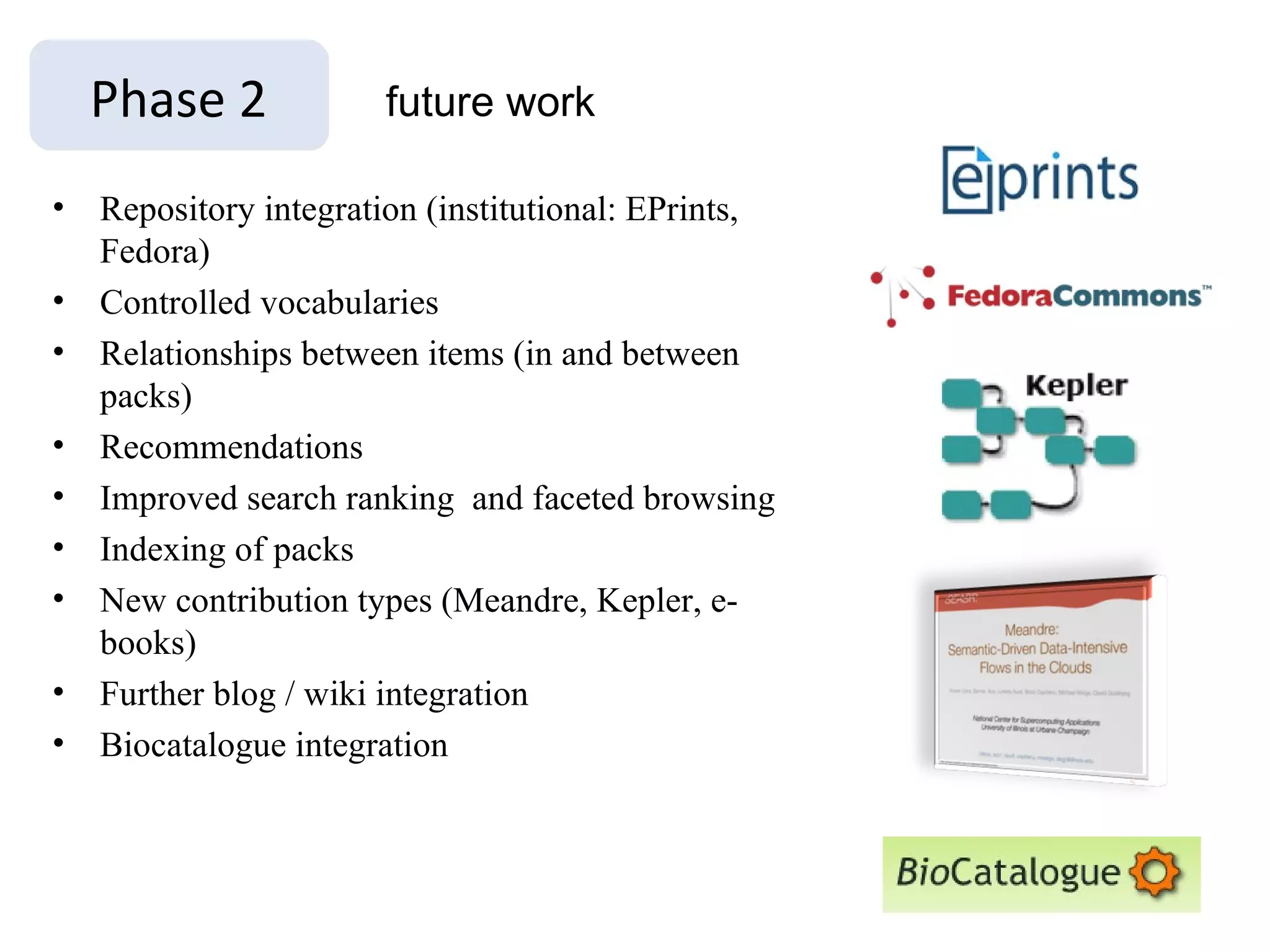
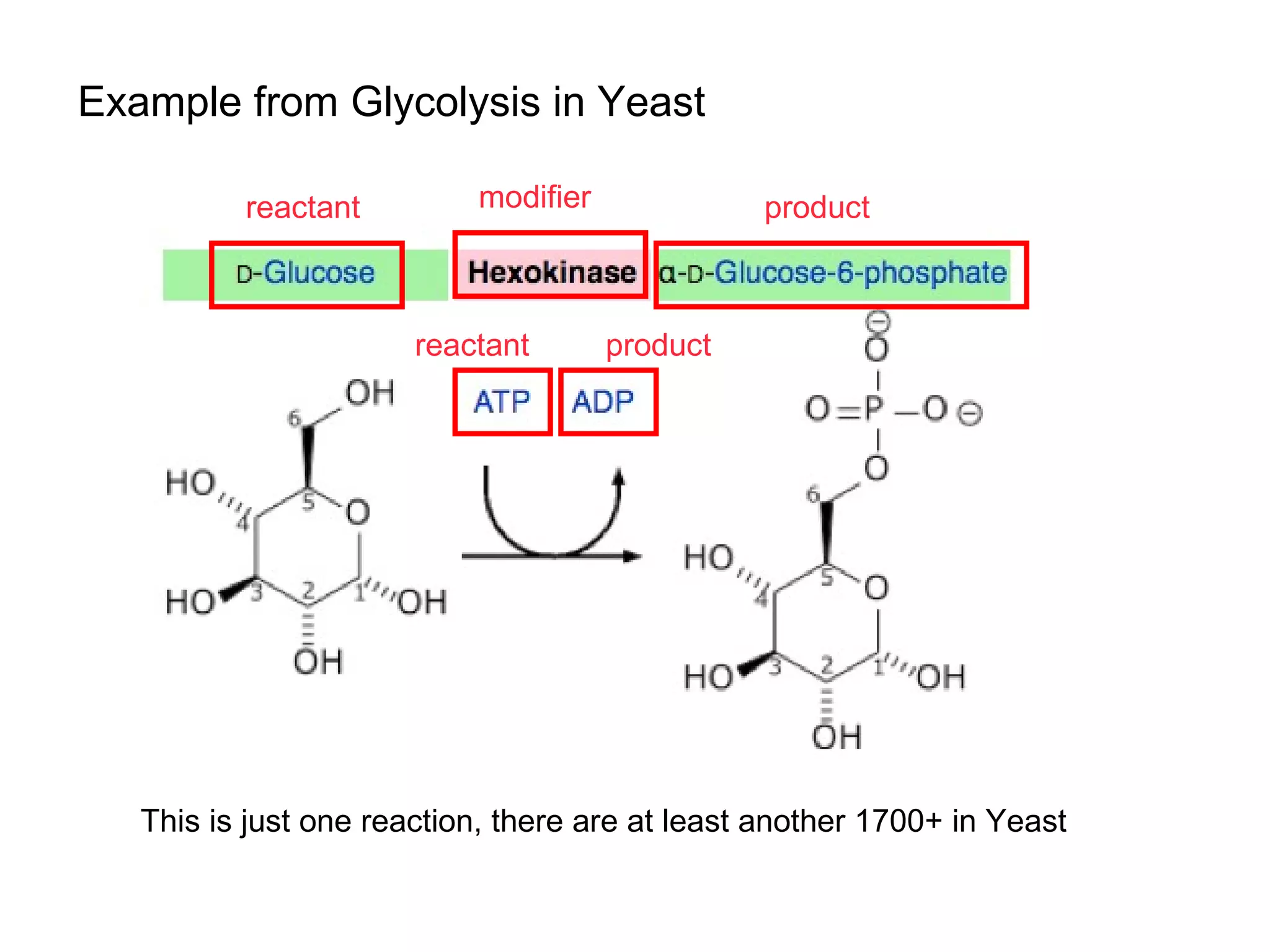
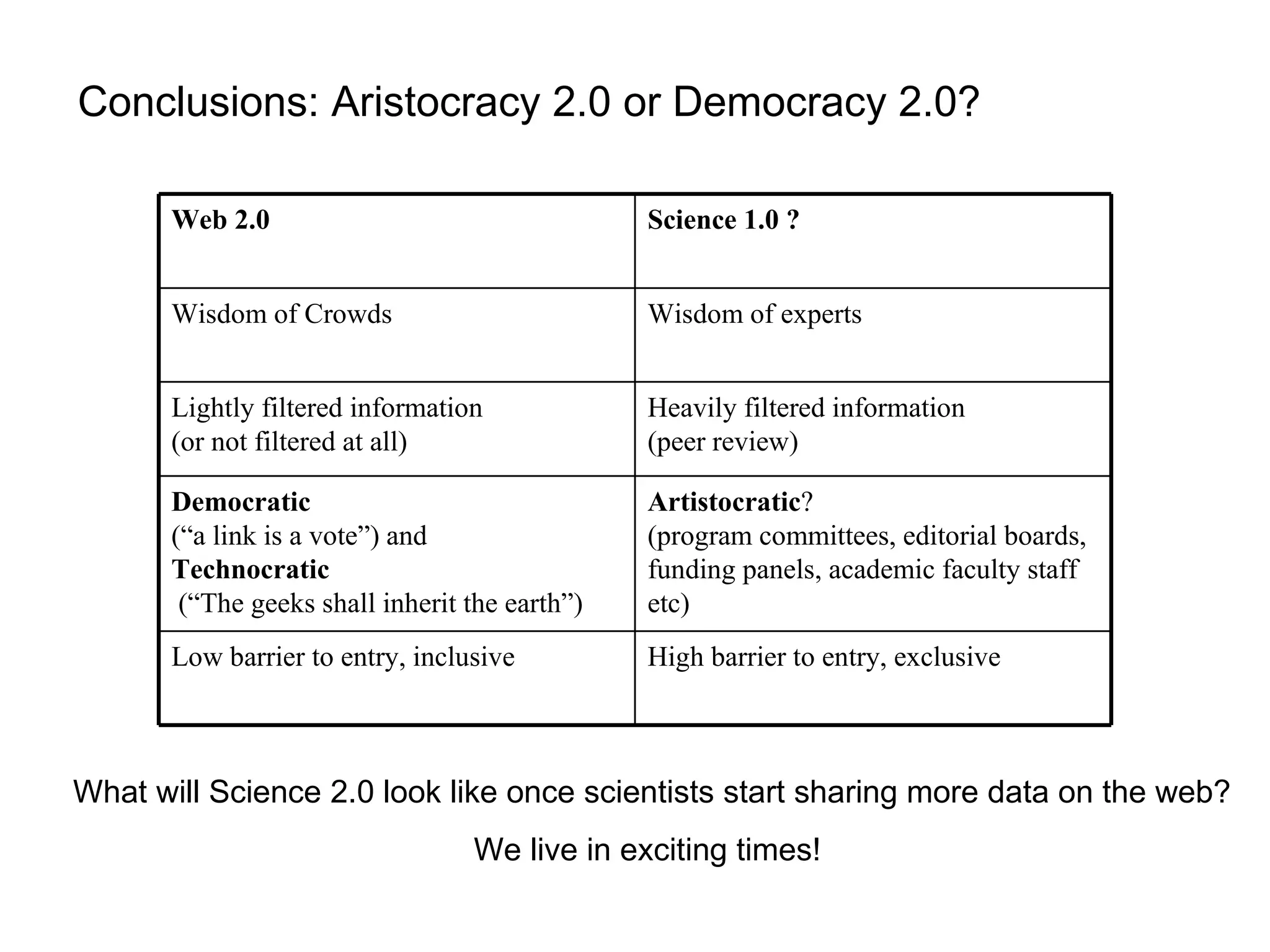
The document discusses the challenges and solutions related to managing digital identities in bioinformatics, particularly through the online platform MyExperiment, which fosters data sharing among scientists. It highlights issues like the inadequacies of traditional publishing in handling digital data, the complexity of attribution and authentication, and the importance of collaborative tools in scientific research. The insights presented emphasize the need for refined digital identities and improved data sharing workflows to enhance the efficiency of scientific communication.






![Burying or Destroying Data and Metadata? Publishing can be inadequte, difficult to mine Barend Mons Wikiproteins Why bury it [data] first and then mine it again? Which gene did you mean? http://pubmed.gov/15941477 BMC Bioinformatics. 2005 Jun 7;6:142. In other cases important data and metadata gets destroyed completely (author, title, gene, protein, chemical names etc) Make digital libraries difficult to use Defrosting the Digital Library Hull, Pettifer and Kell http://www.pubmed.gov/18974831 PLoS Computational Biology 2008 Oct;4(10):e1000204](https://image.slidesharecdn.com/the-real-nettab-090612105541-phpapp02/75/myExperiment-Nettab-8-2048.jpg)

![Digital Identity is currently a mess (part 1) One person, can be identified by many different URIs People who know Paolo can tell the difference People who don’t (and software) face a significant challenge to disambiguate Digital Identity is a second-class citizen on the Web (see http://www.flickr.com/photos/dullhunk/3618998907/ for web e.g.) http://www.nettab.org/promano/ (nettab organiser) mailto:paolo.rmano@istge.it http://www.paoloromano.it/ en. wikipedia . org/wiki/Paolo_Romano (sculptor) it. wikipedia .org/wiki/Paolo_Romano (actor) www.linkedin.com/in/paoloromano http://pubmed.gov?Term=Paolo+Romano[author] myspace .com/paoloromano (musician) www.paoloromano.net/ (politician and friend of Berlusconi) citeulike . org/tag/paolo-romano ... uni-trier .de/~ley/db/indices/a-tree/r/Romano_0001:Paolo.html Will the real Paolo Romano please stand up? URI’s are used for identifying people on the web](https://image.slidesharecdn.com/the-real-nettab-090612105541-phpapp02/75/myExperiment-Nettab-10-2048.jpg)
![Digital identity is currently a mess (part 2) On three levels, the three A ’s: Authentication : is Paolo is who he says he is? Or a fake? Authorisation : is Paolo authorised to view/operate-on workflow? Attribution : Paolo AuthorOf Nettab-Workflow or Paolo Reused Workshop-Workflow Currently done through combination of username-and-password http://tinyurl.com/too-many-passwords Paolo Romano Simon Willison (The Guardian) The average user has [at least] 18 user accounts and 3.49 passwords”](https://image.slidesharecdn.com/the-real-nettab-090612105541-phpapp02/75/myExperiment-Nettab-13-2048.jpg)




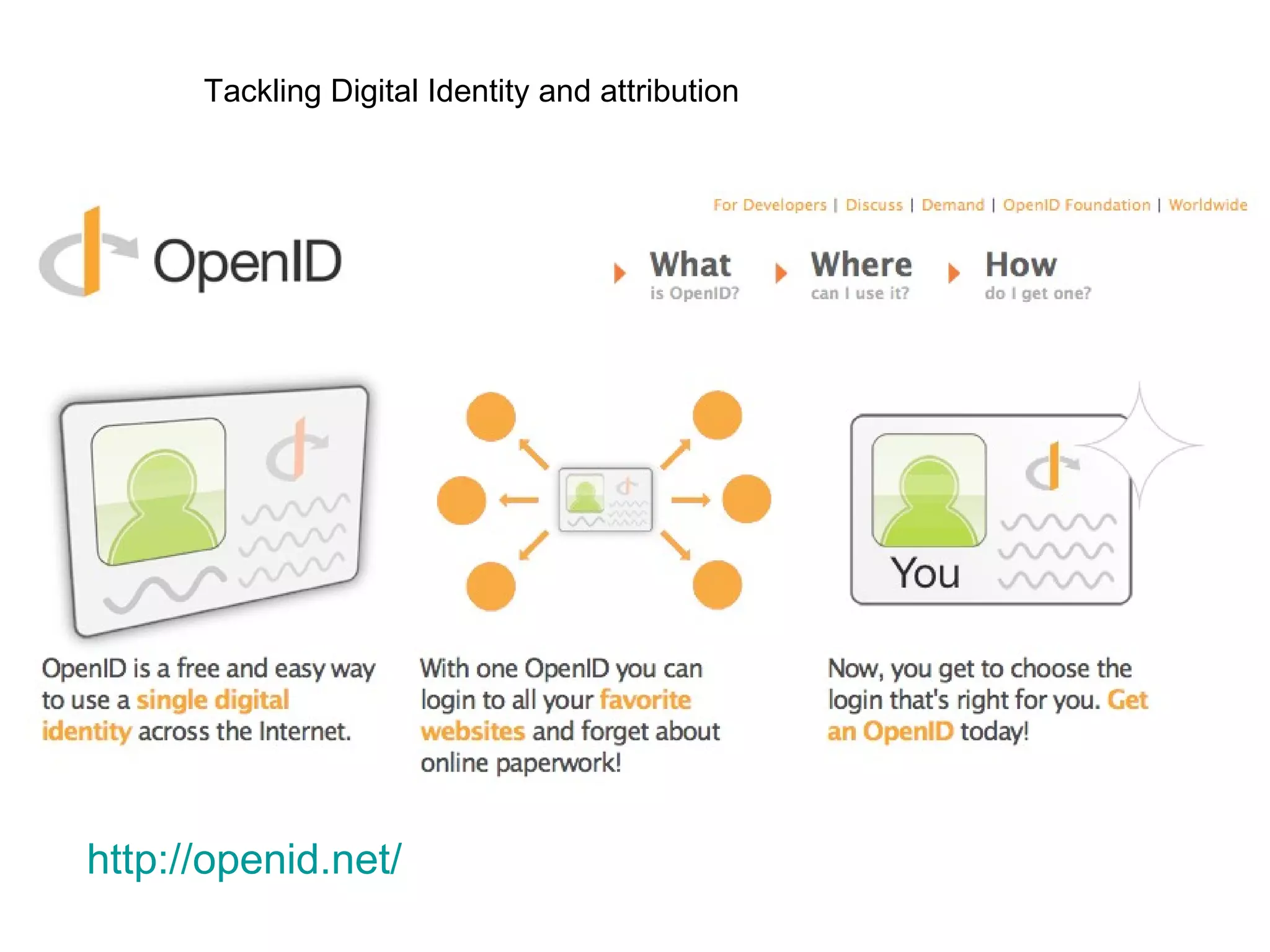
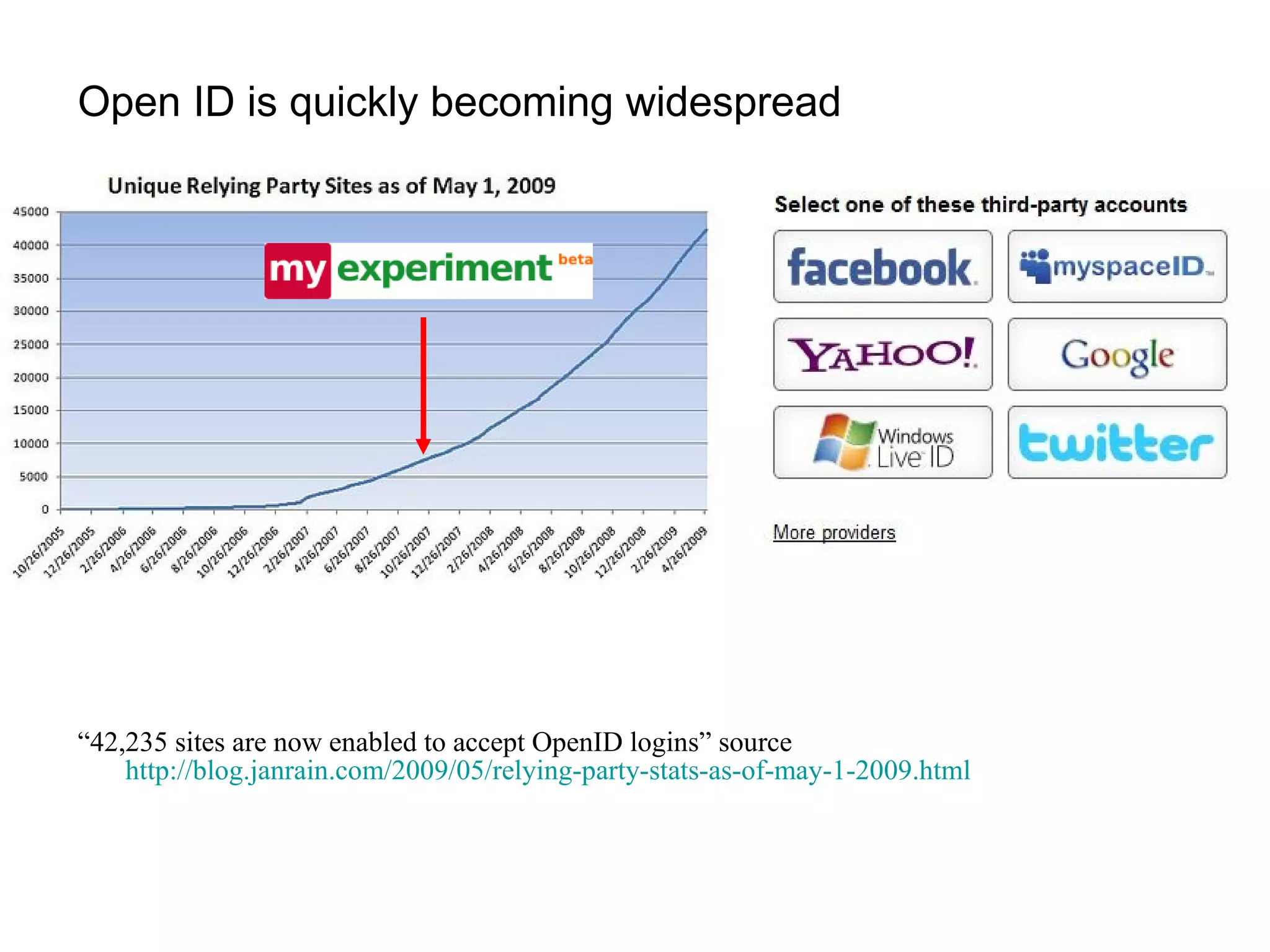
![But you can’t force OpenID on people…(yet) http://romano.myopenid.com/ [email_address] nettab OR Password handled by third party OpenID provider + 84% 16%](https://image.slidesharecdn.com/the-real-nettab-090612105541-phpapp02/75/myExperiment-Nettab-21-2048.jpg)
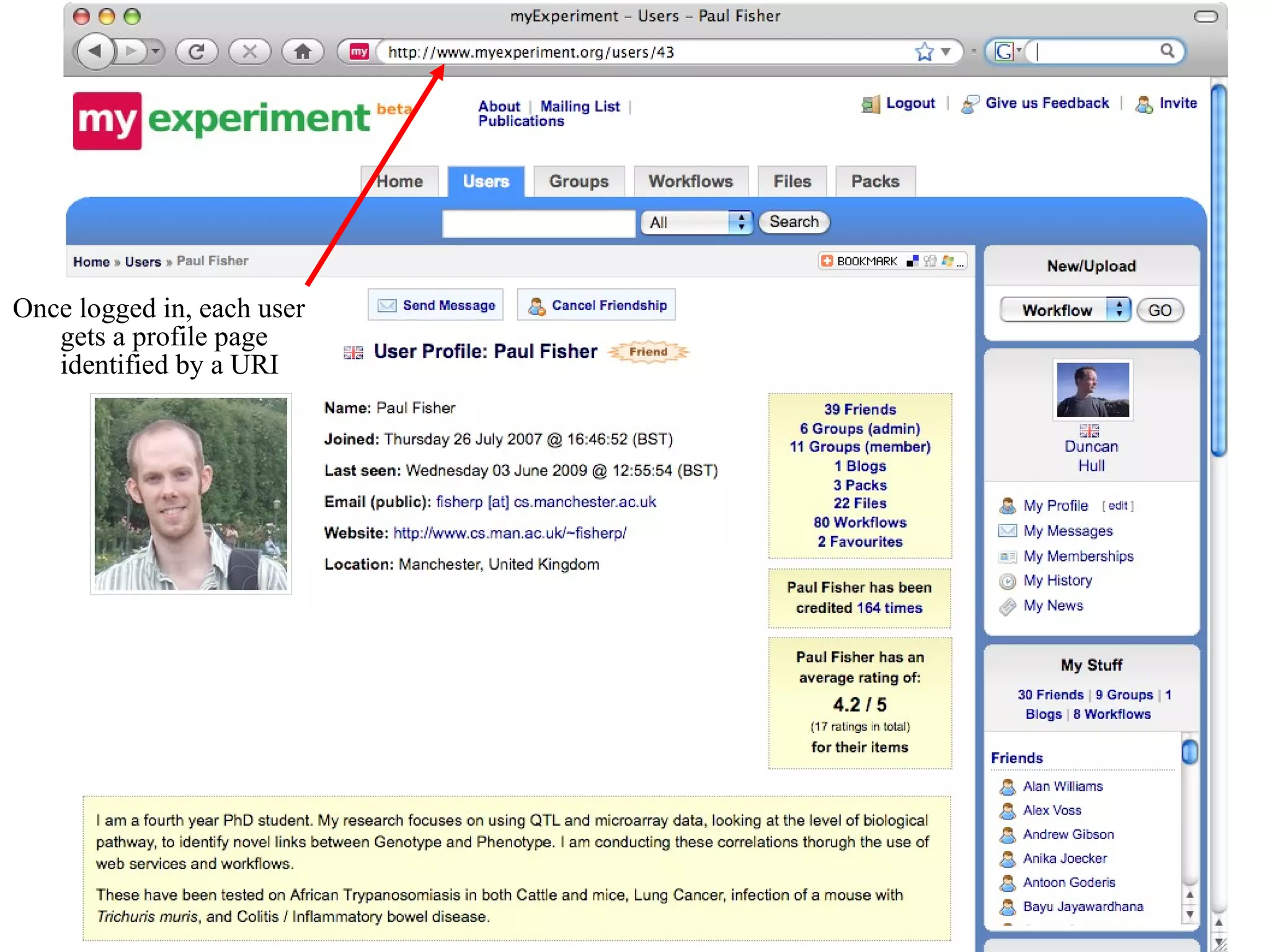
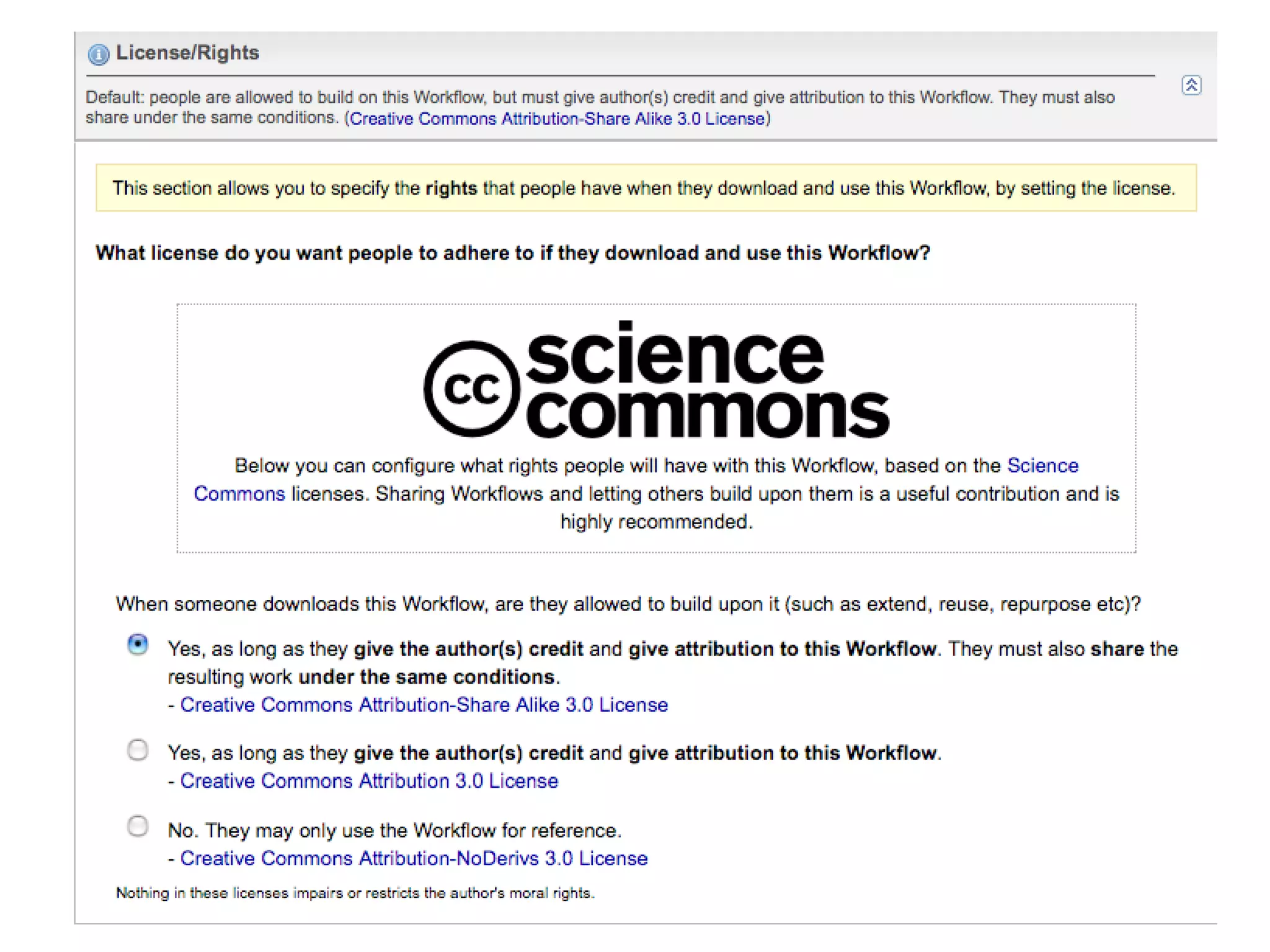



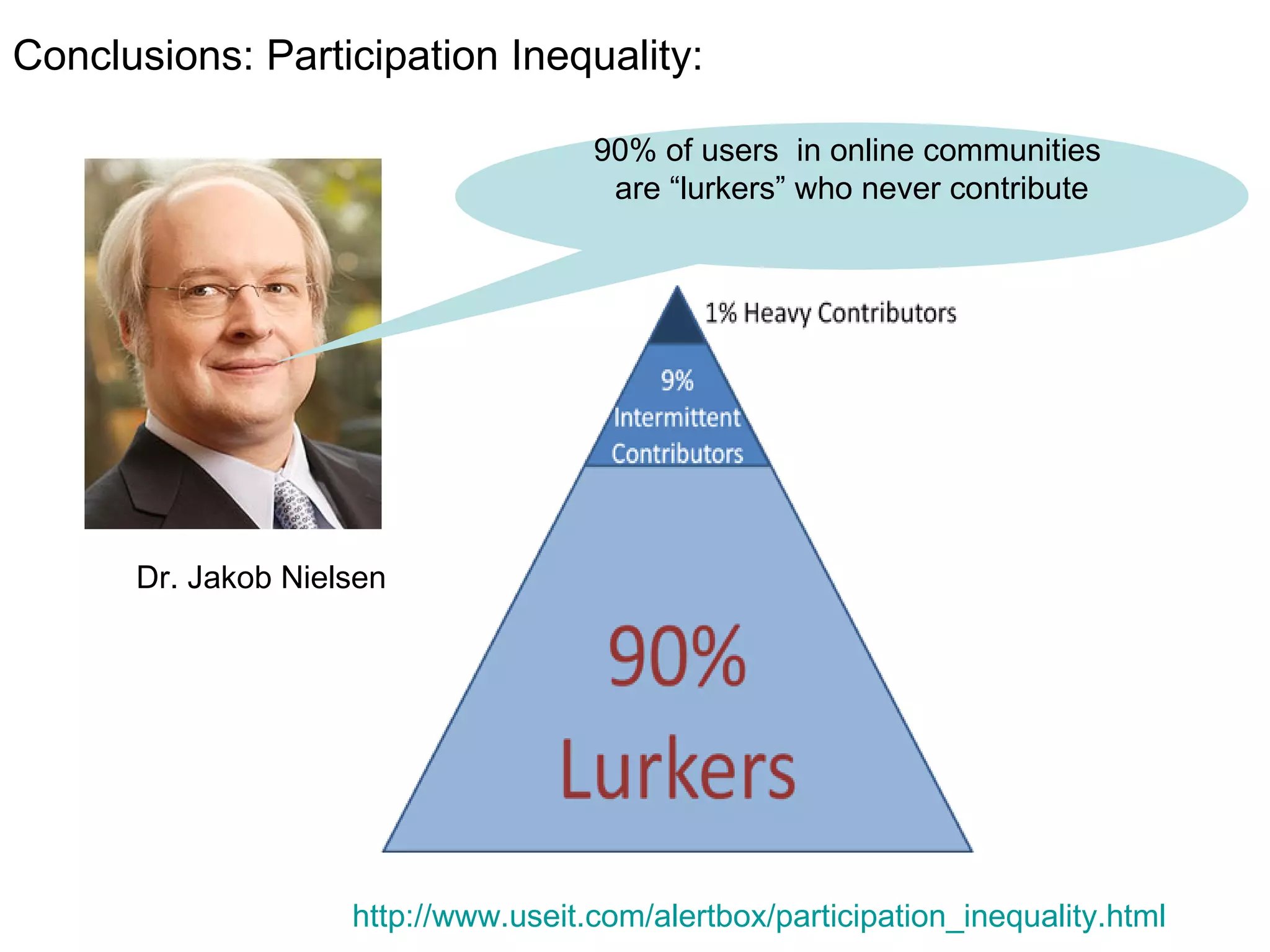
![We need you! It’s all about collaboration Sign up for an account at http://www.myexperiment.org Please get in touch if you’d like to join in Mailing list [email_address] Questions? … and now for a live demonstration](https://image.slidesharecdn.com/the-real-nettab-090612105541-phpapp02/75/myExperiment-Nettab-37-2048.jpg)
