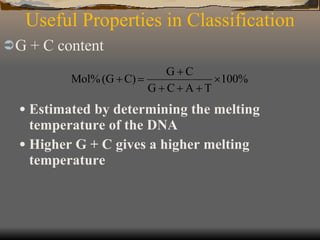
The document discusses microbial taxonomy and classification systems. It describes the levels of taxonomy from domain to species. Key points include that taxonomy aims to classify organisms based on evolutionary relationships, there are three domains of life, and Bergey's Manual of Systematic Bacteriology is the standard reference for prokaryotic taxonomy. The domains are Bacteria, Archaea, and Eukarya, with Bacteria further divided into 23 phyla including Proteobacteria, Firmicutes, and Actinobacteria.