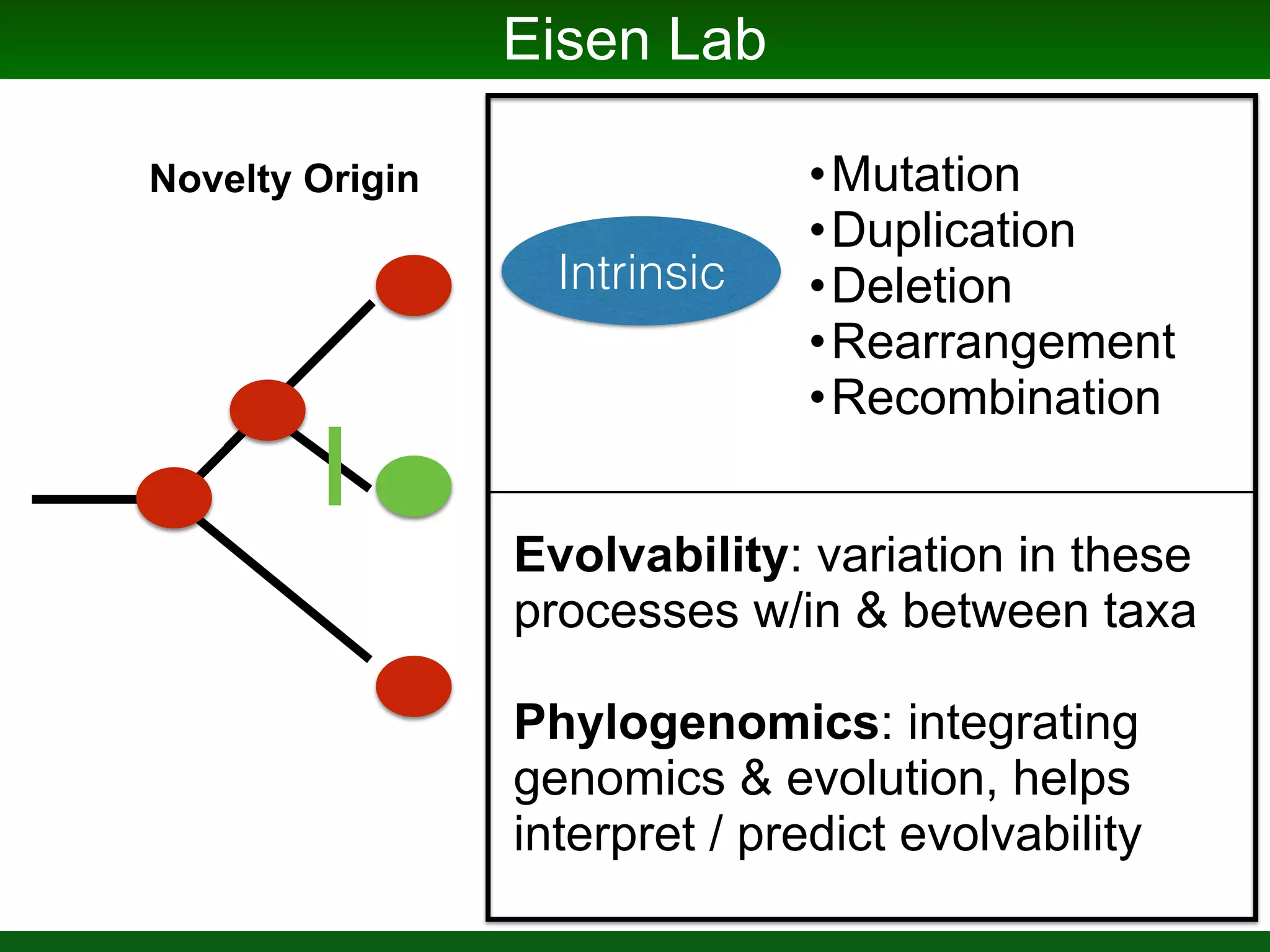
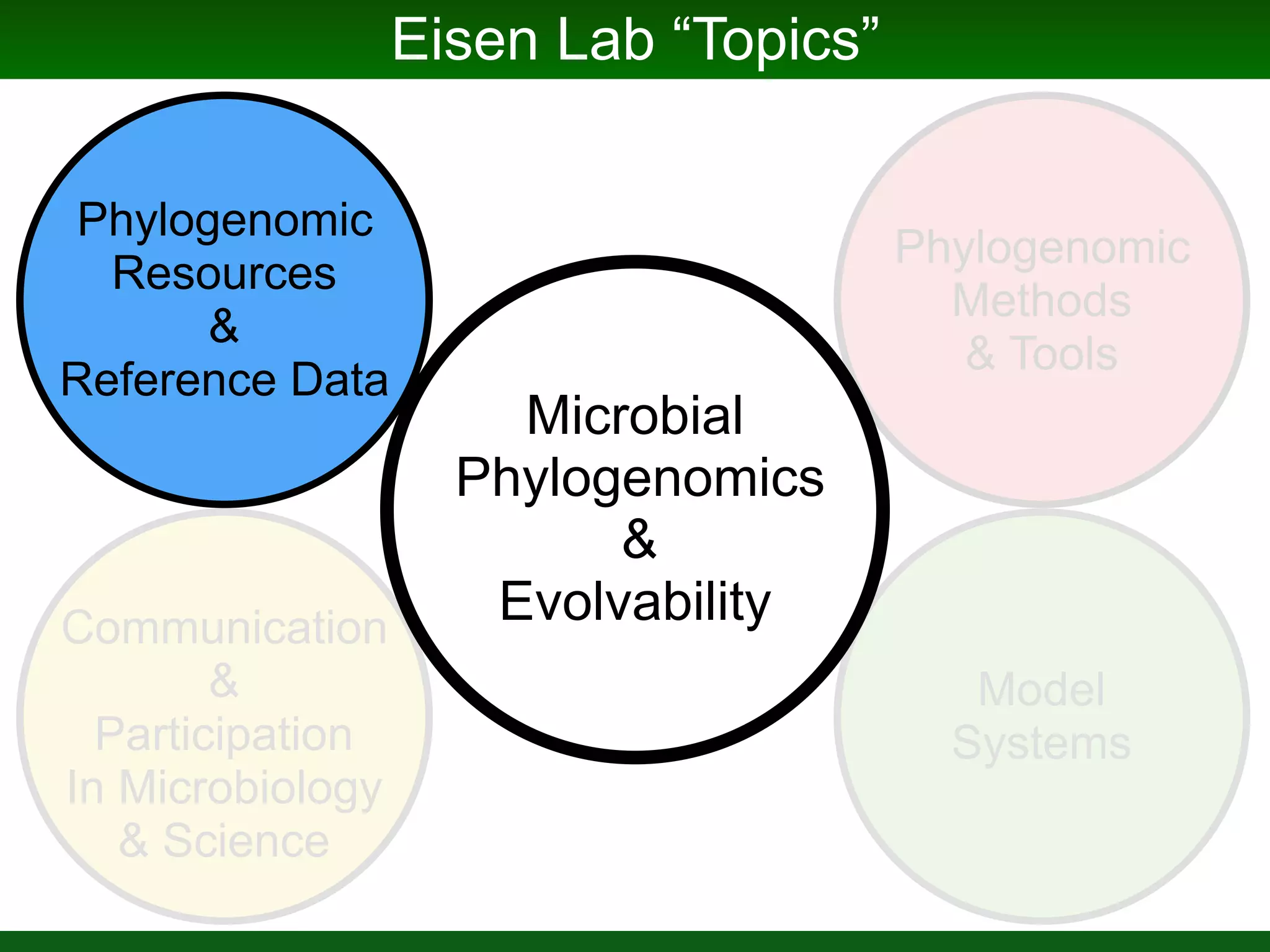
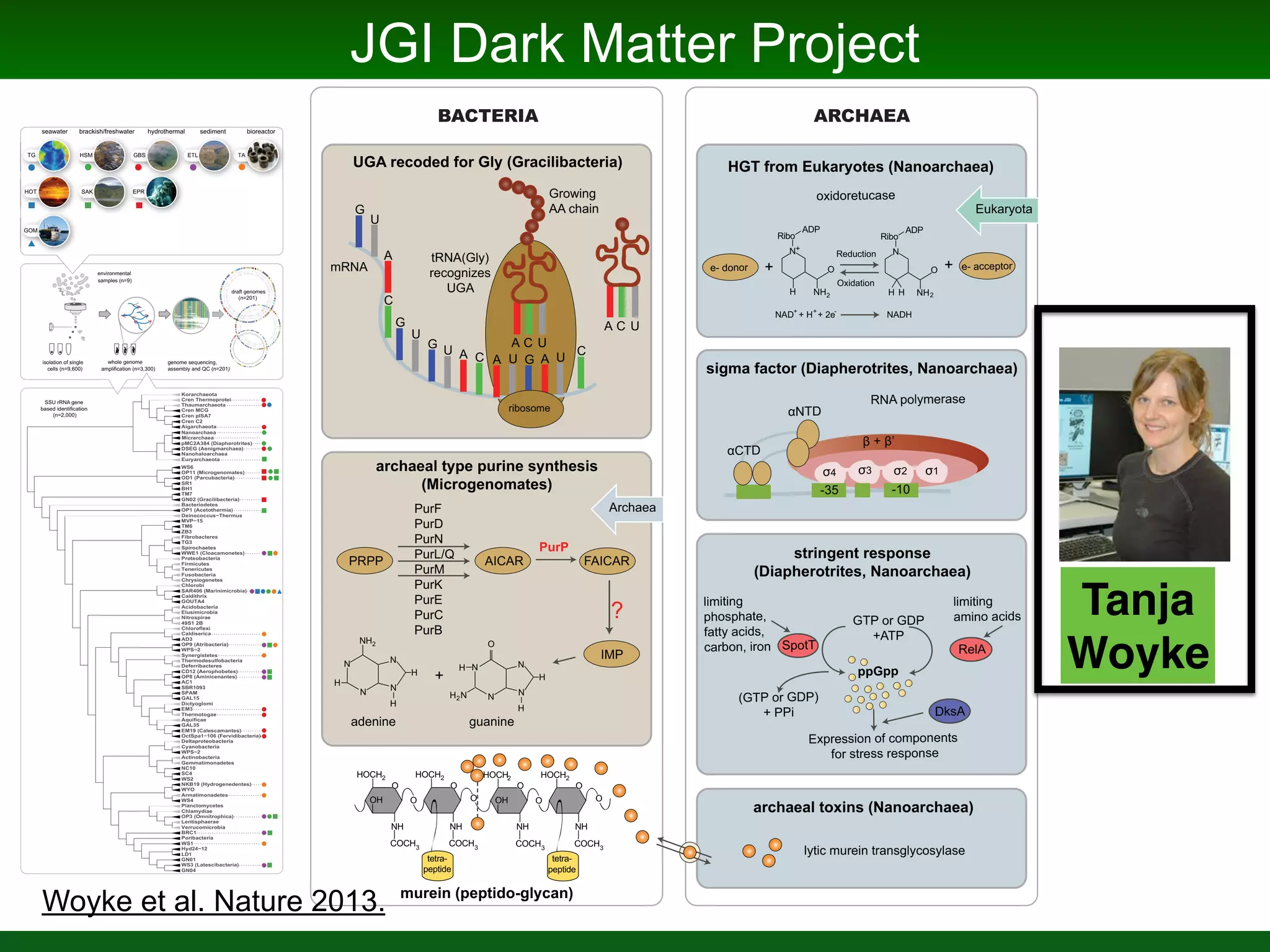
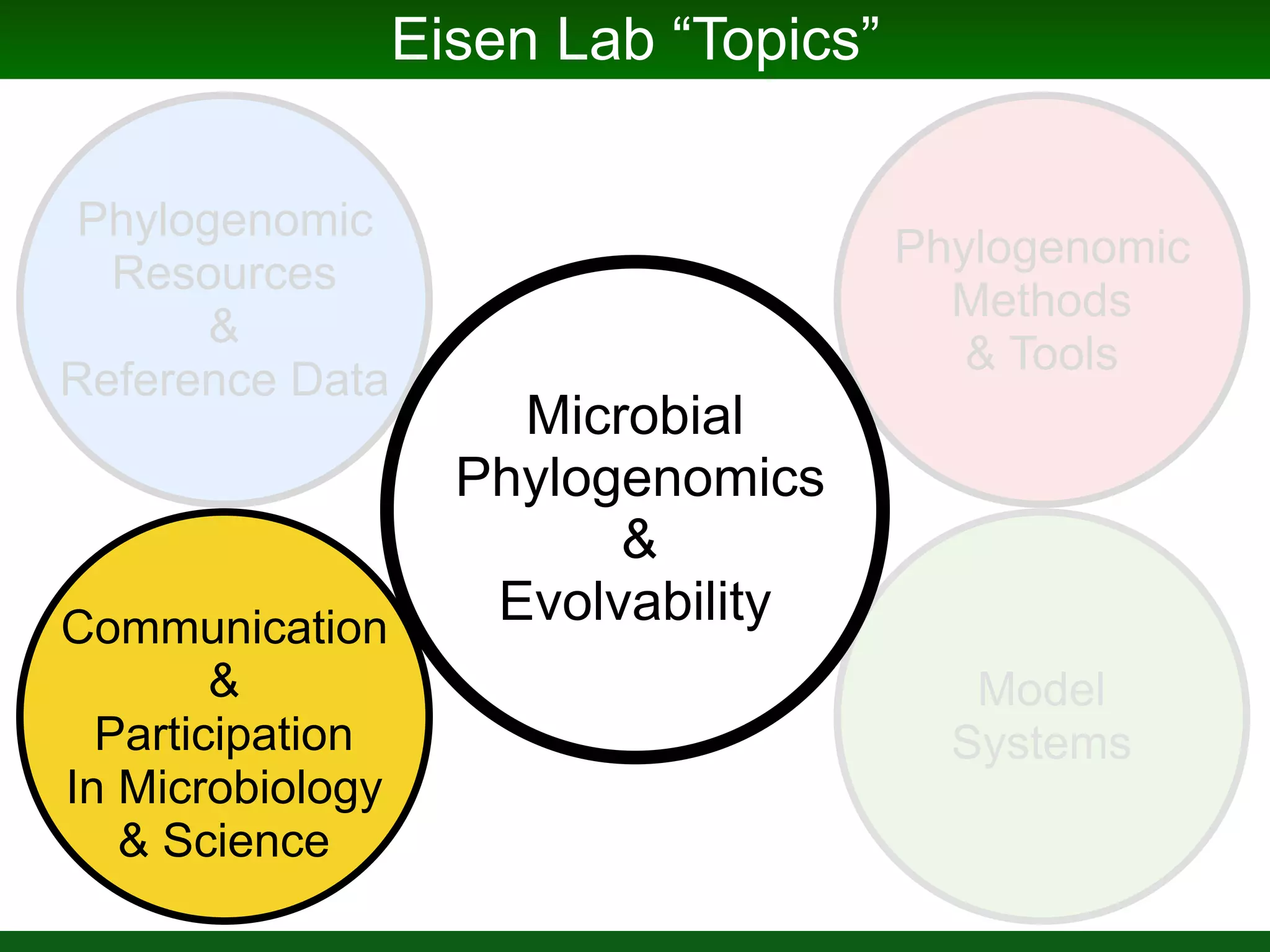
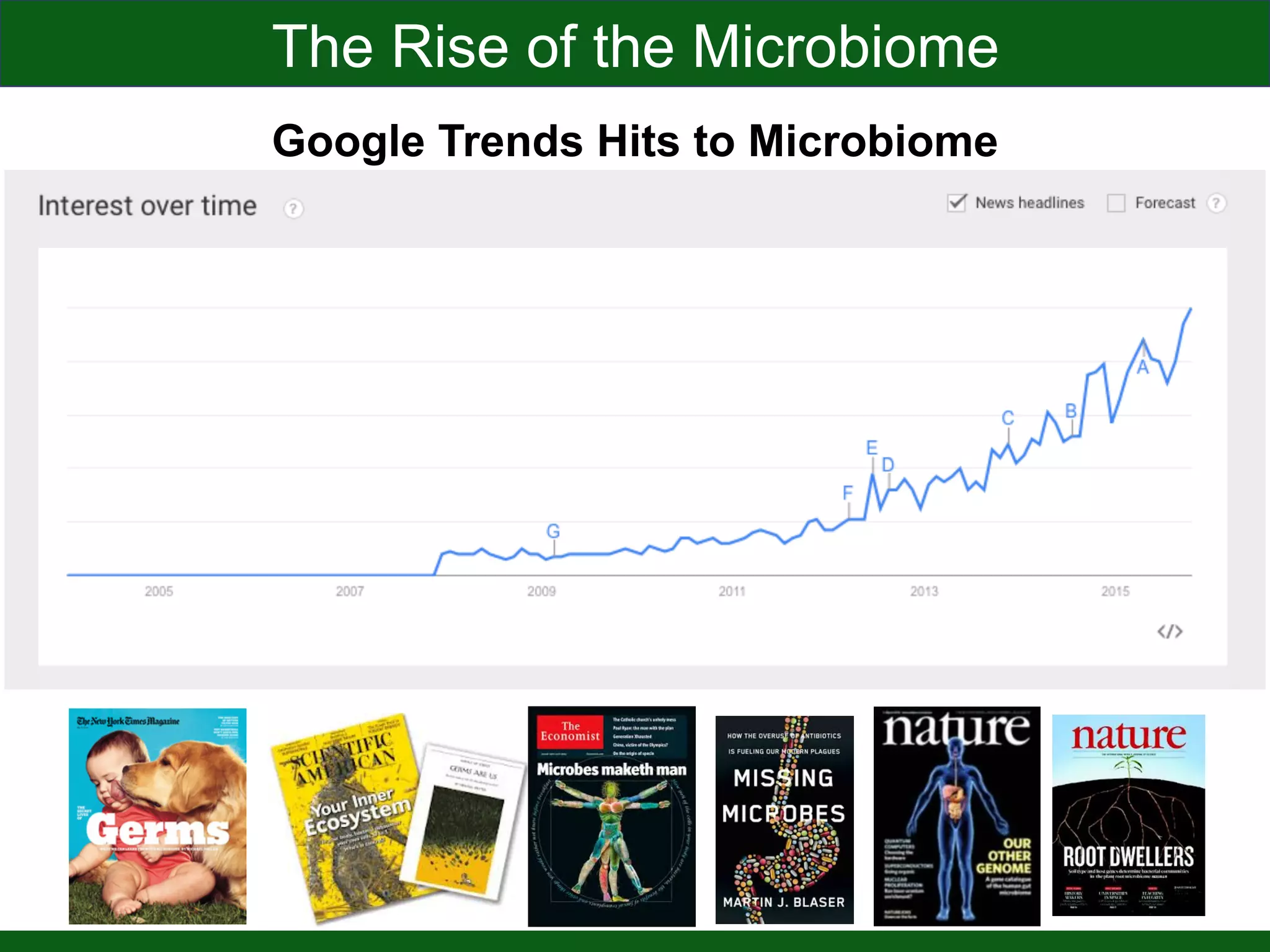
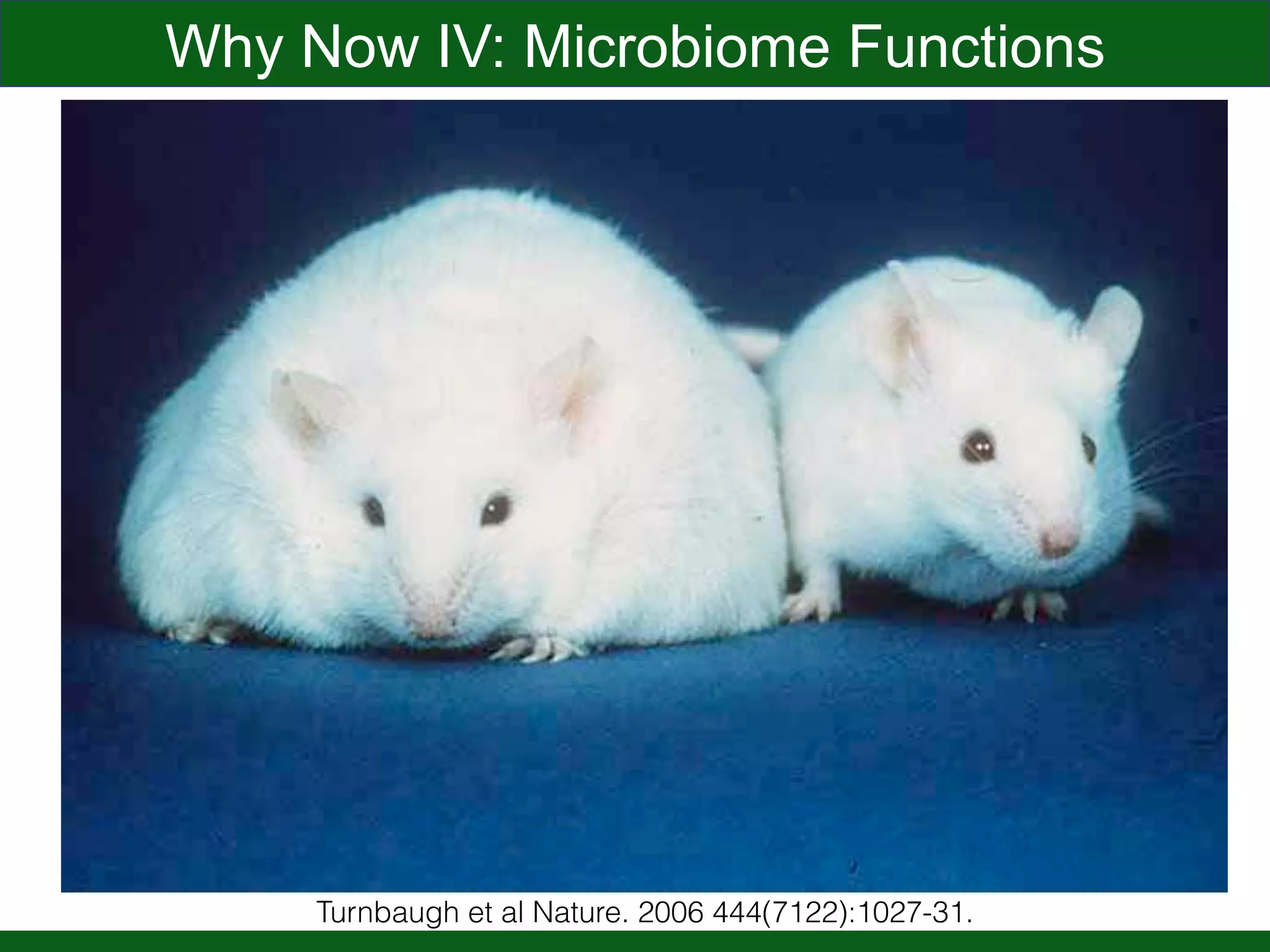
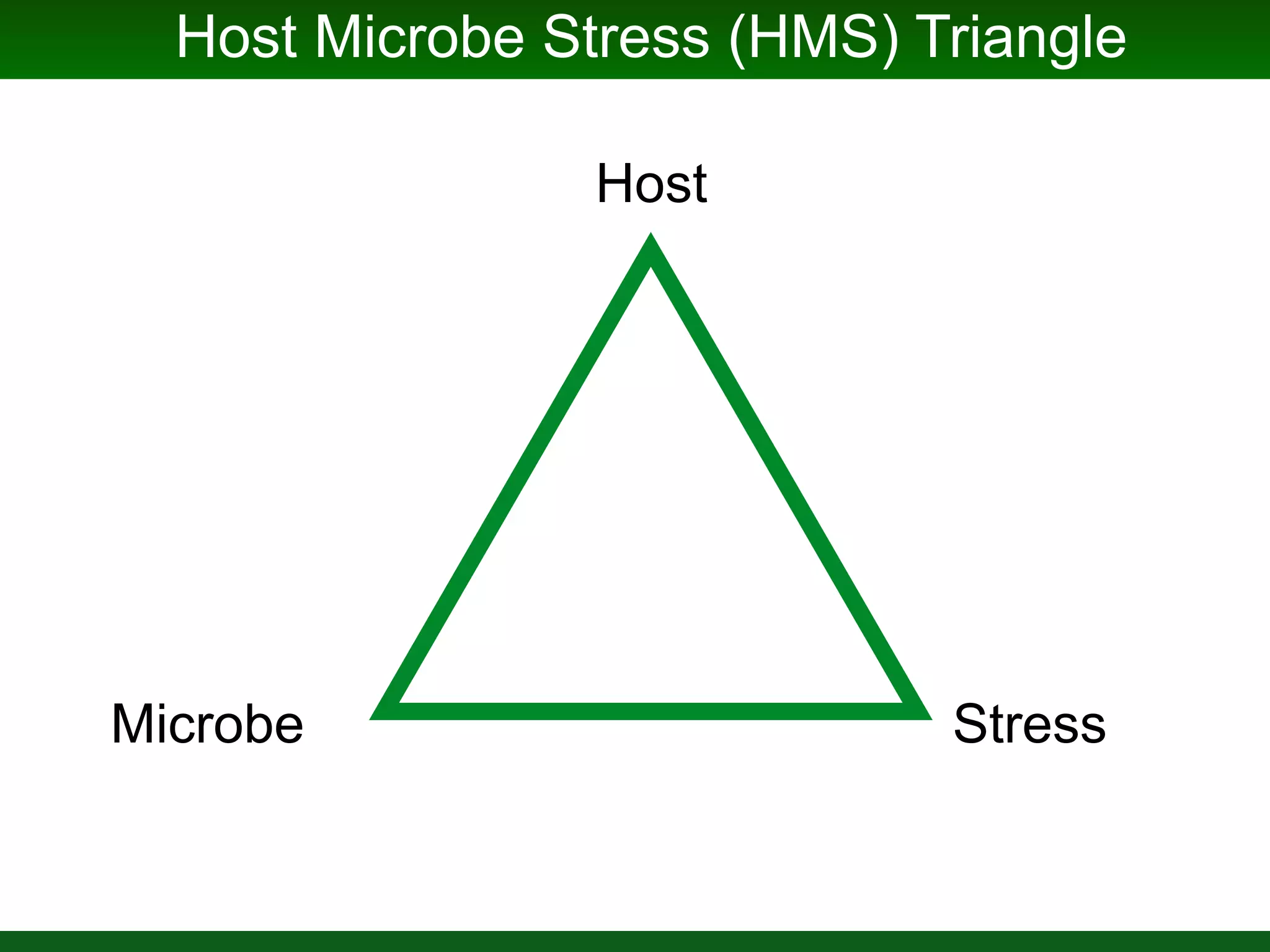
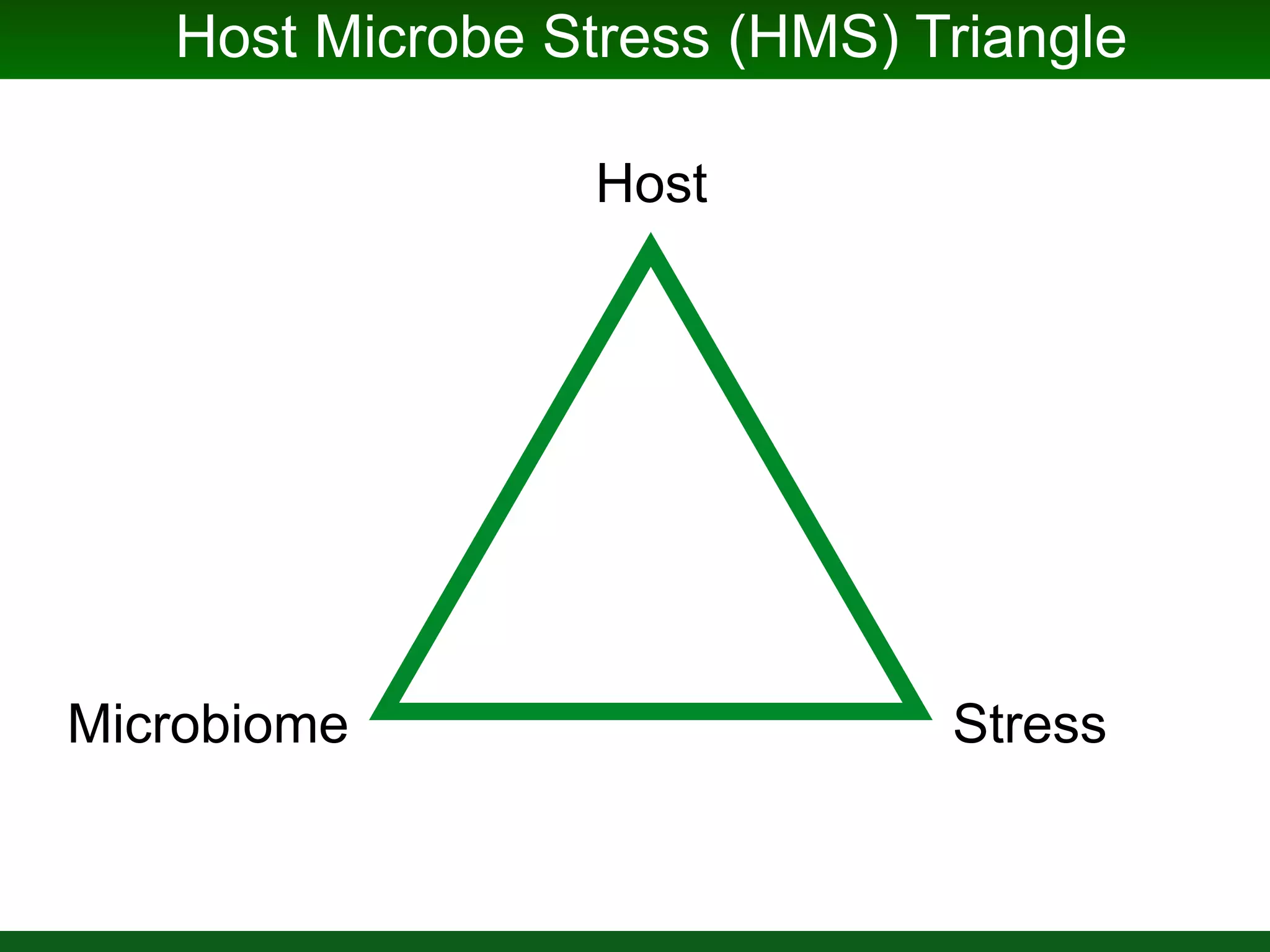
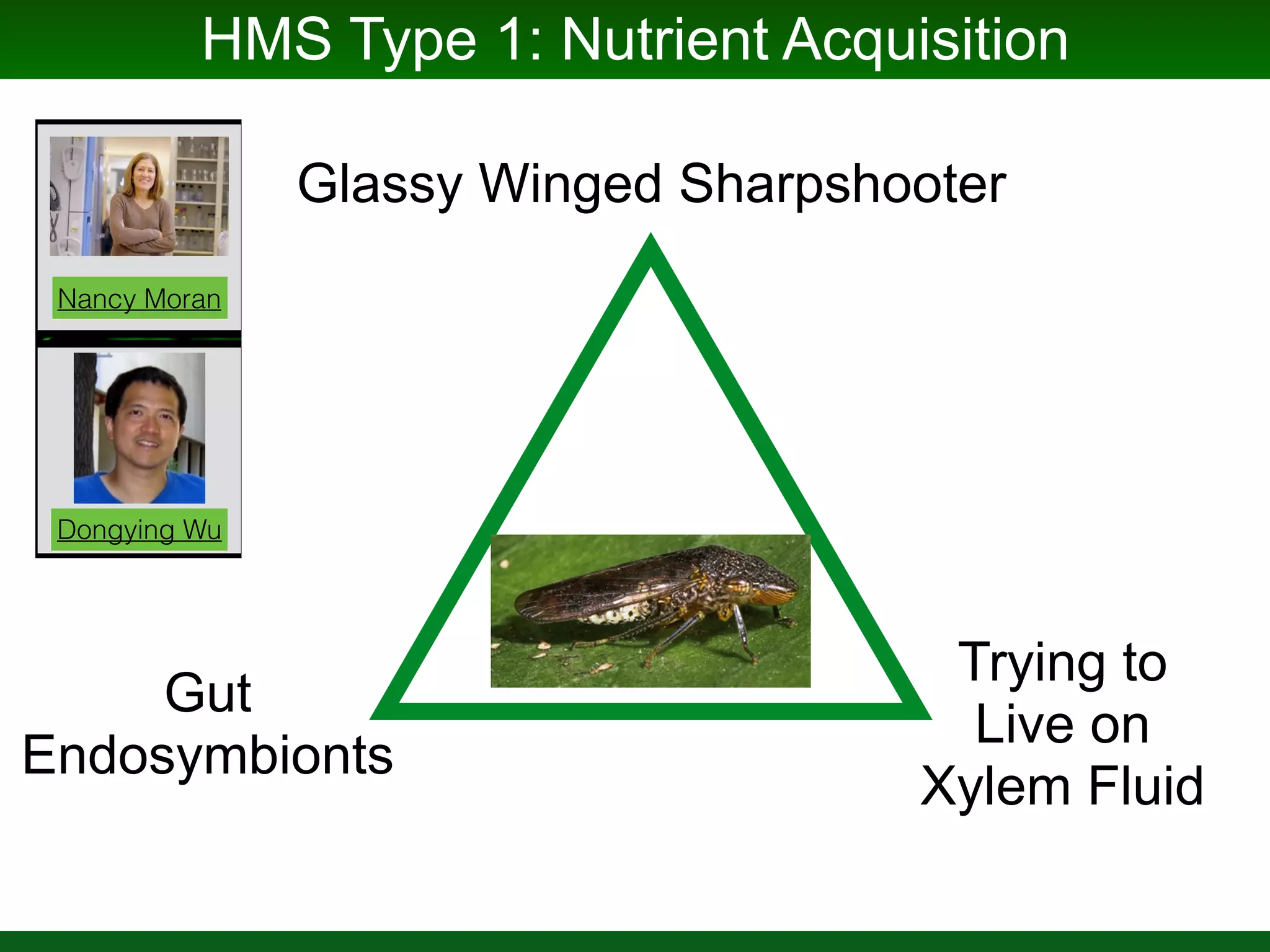
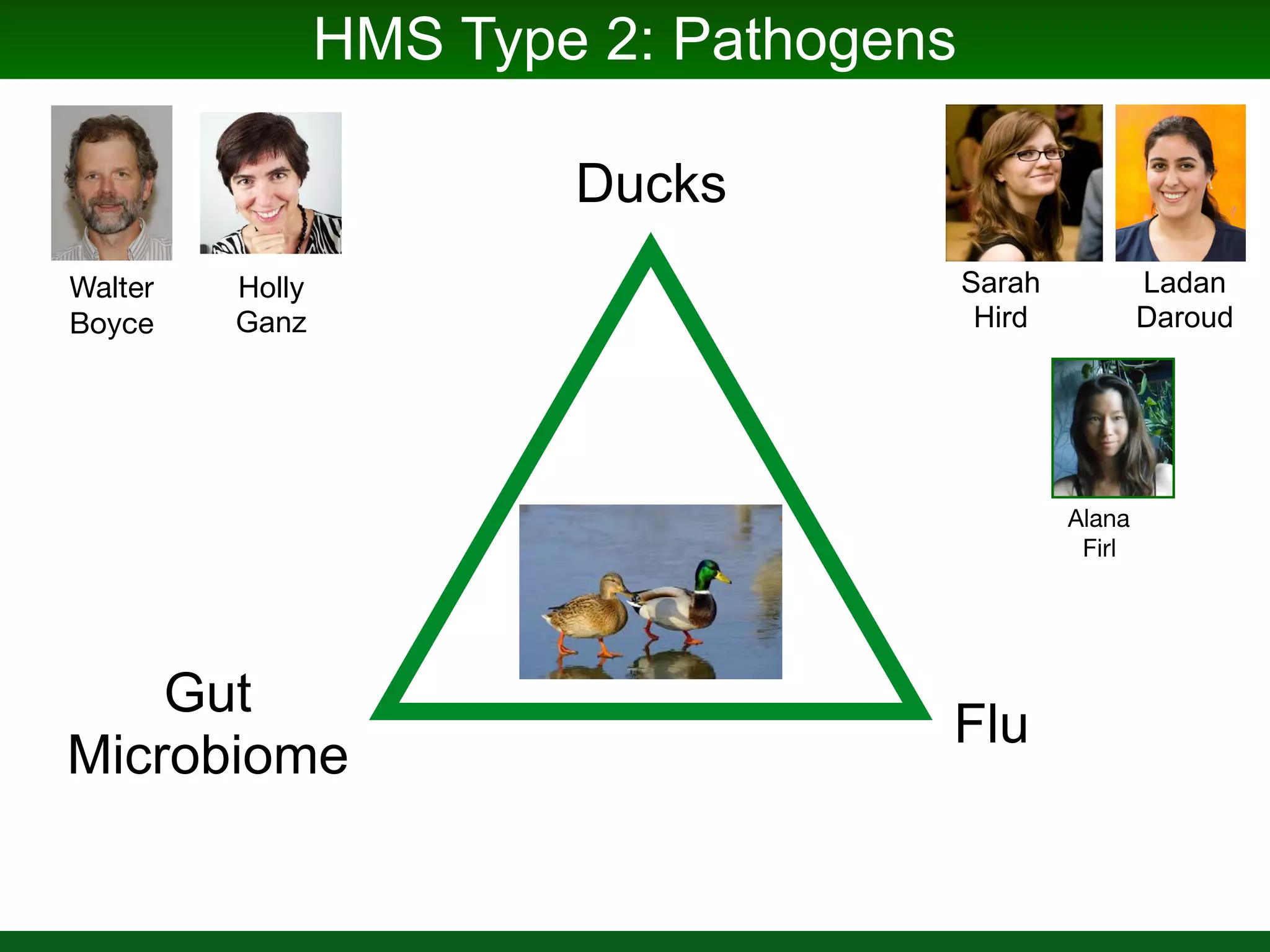
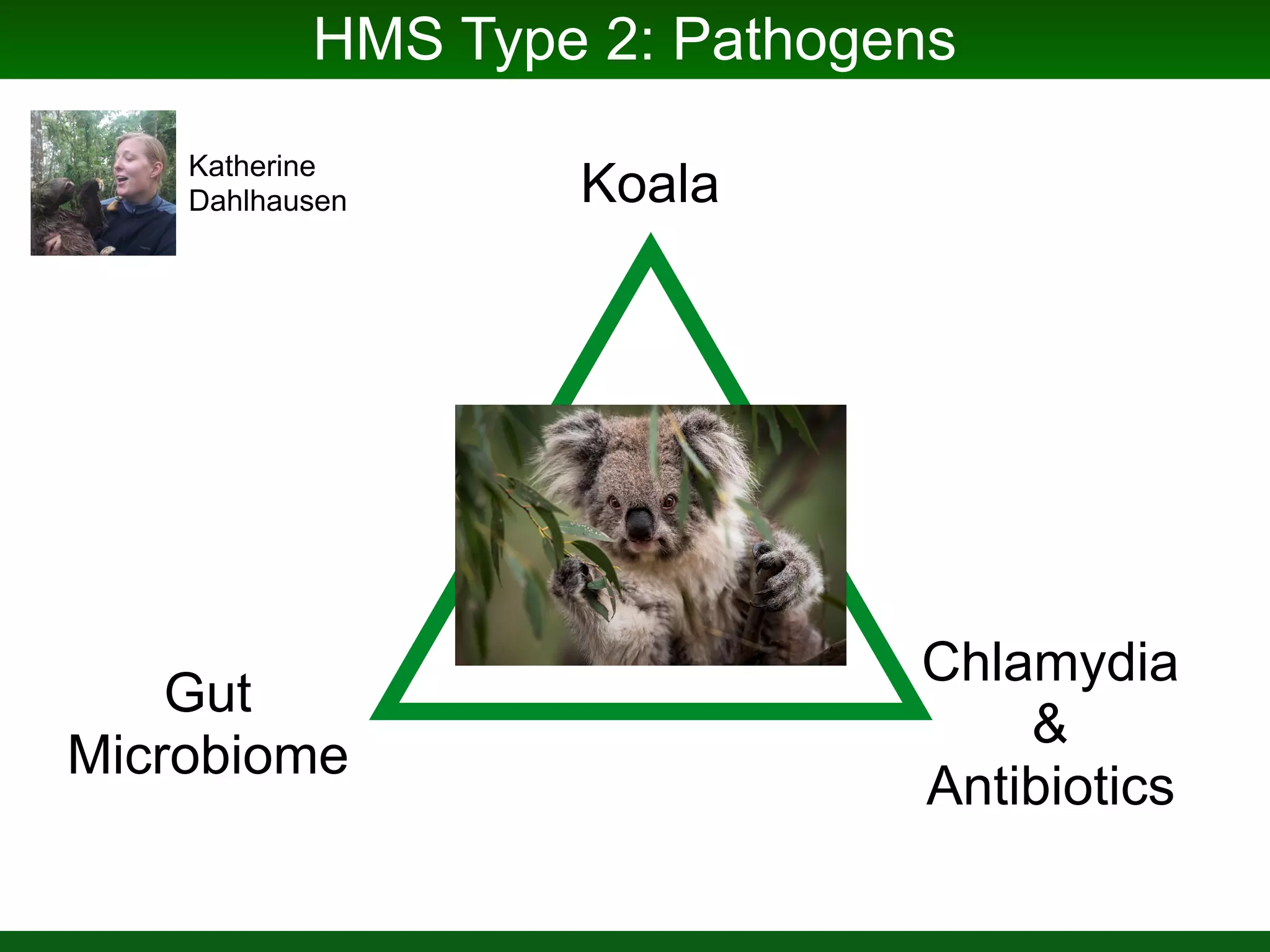
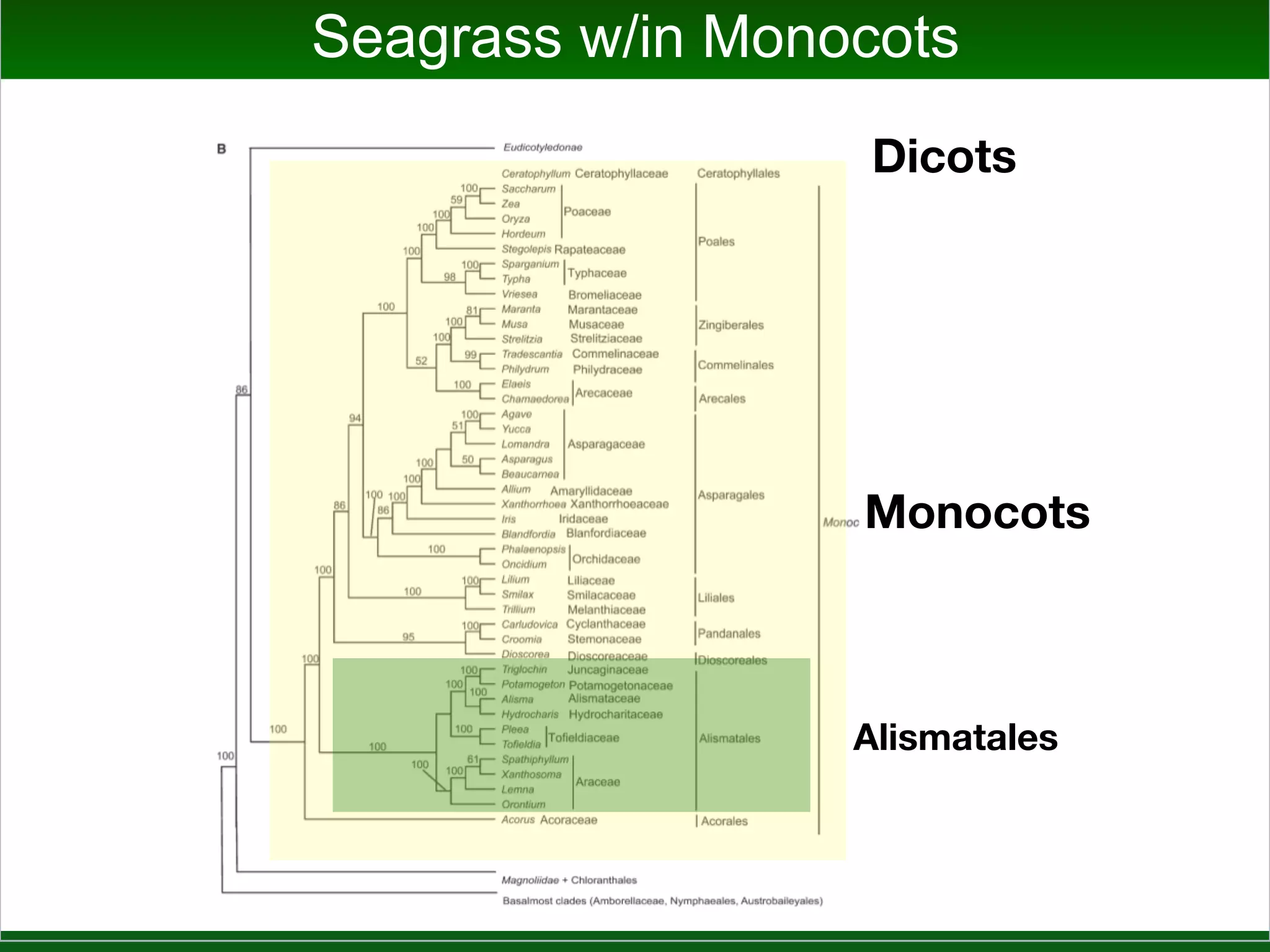
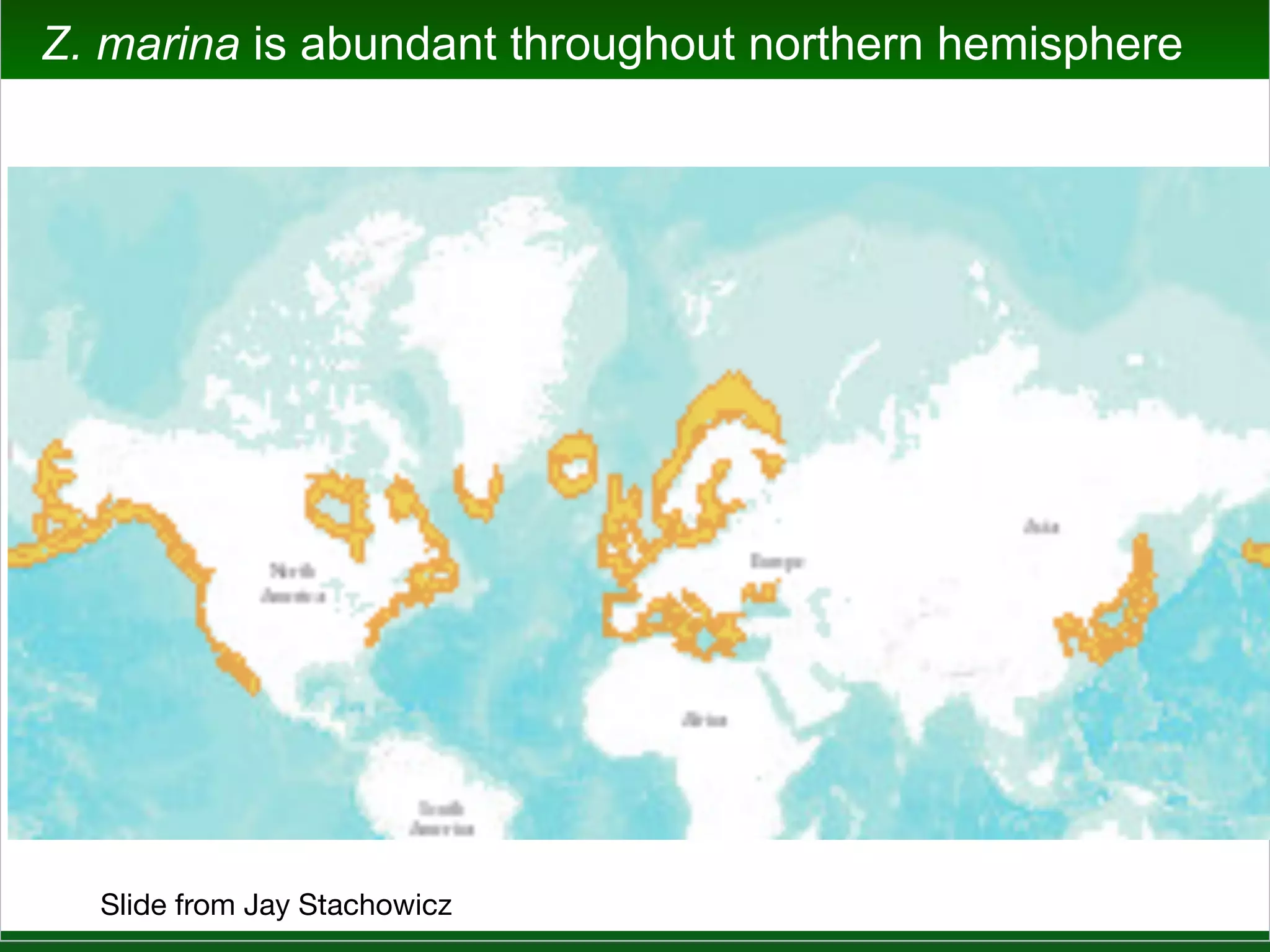
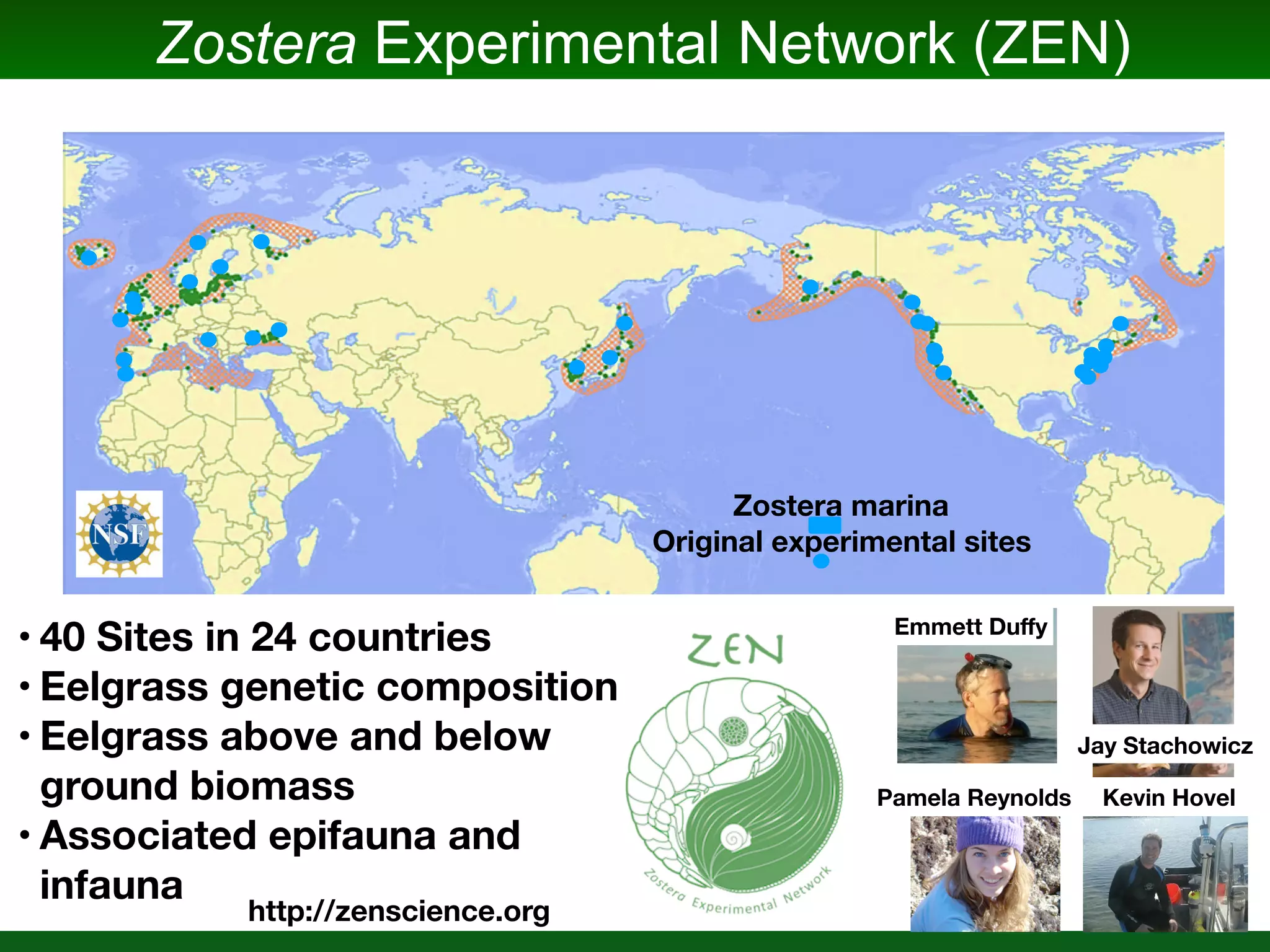
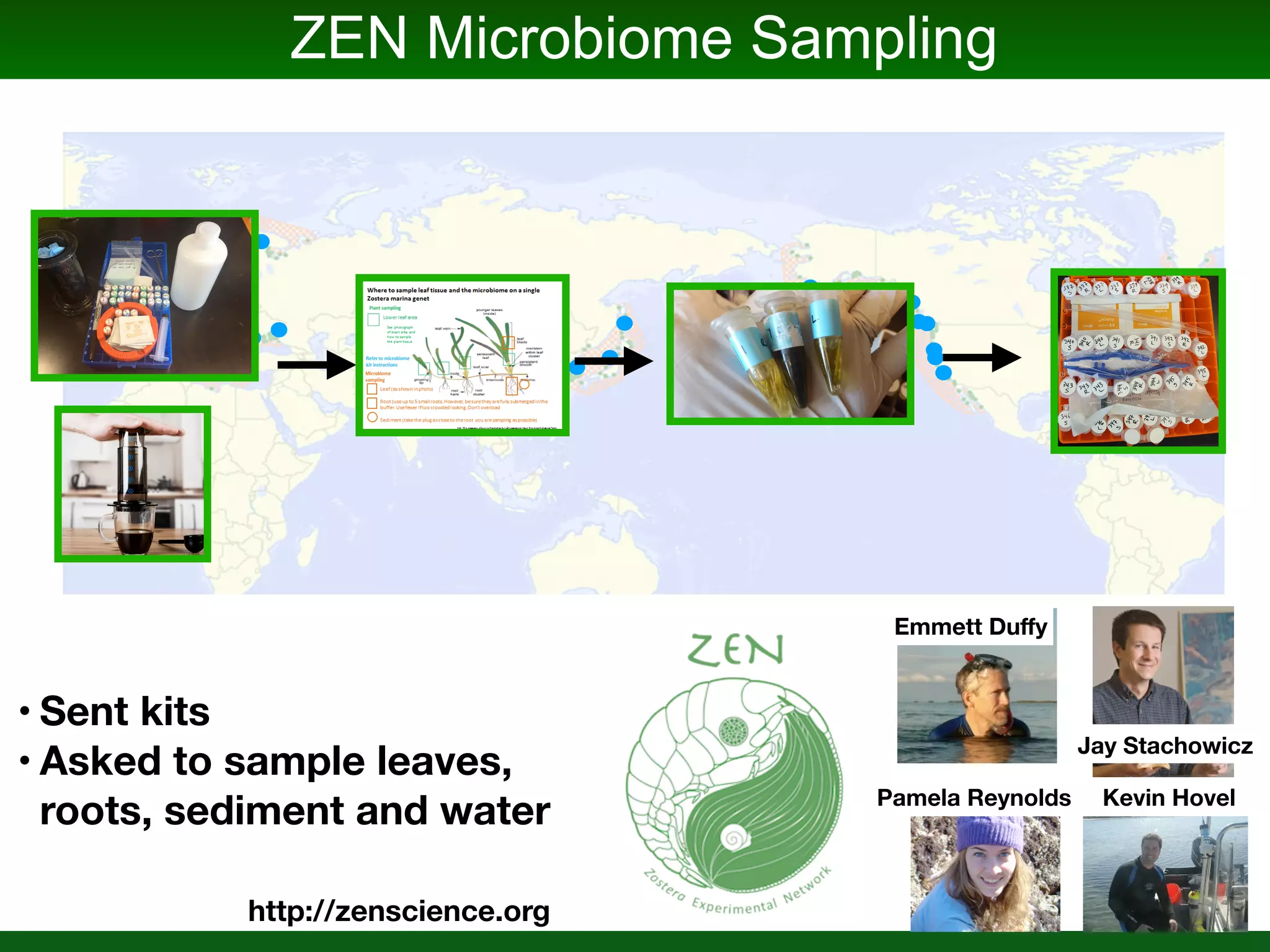
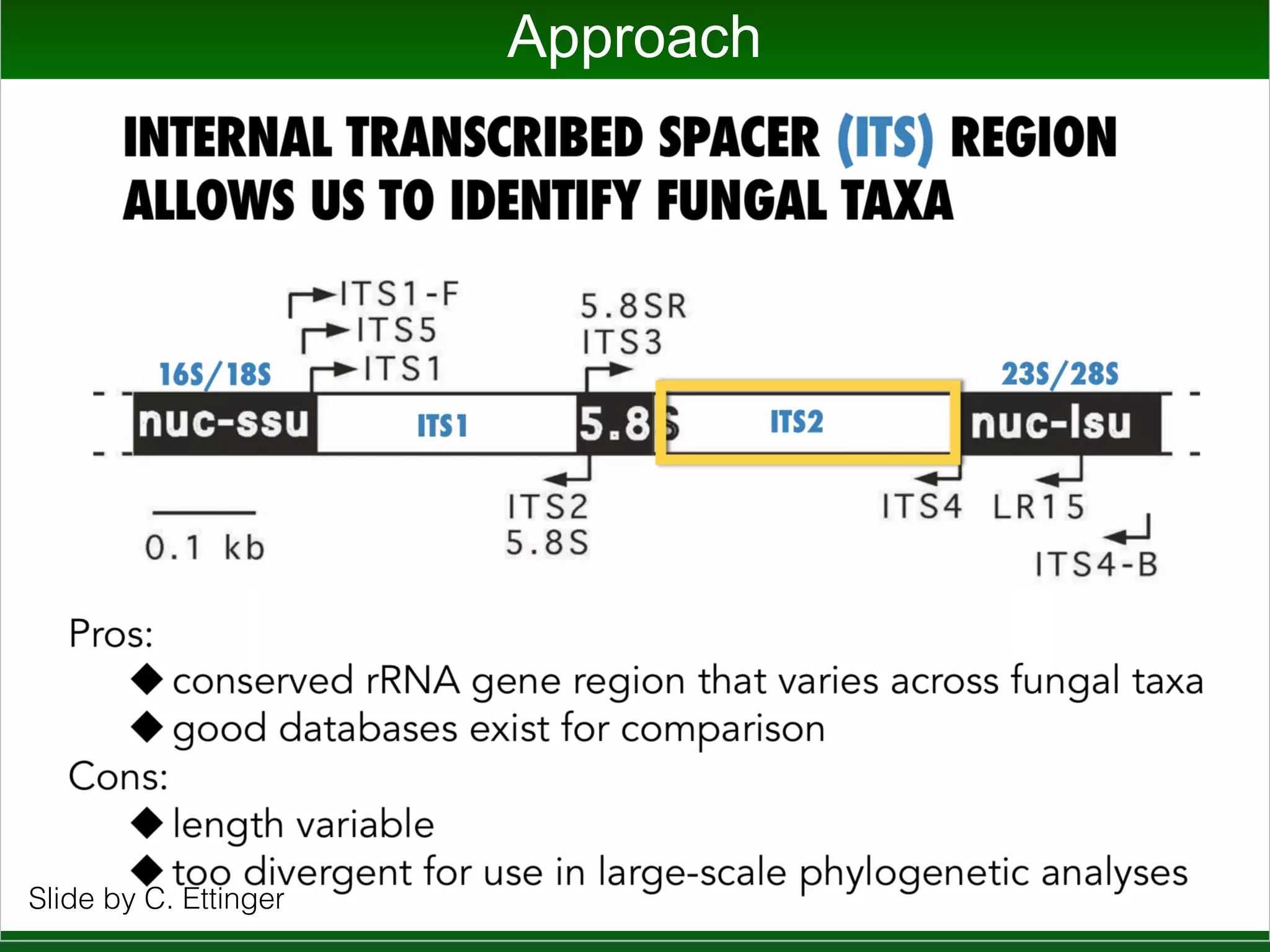
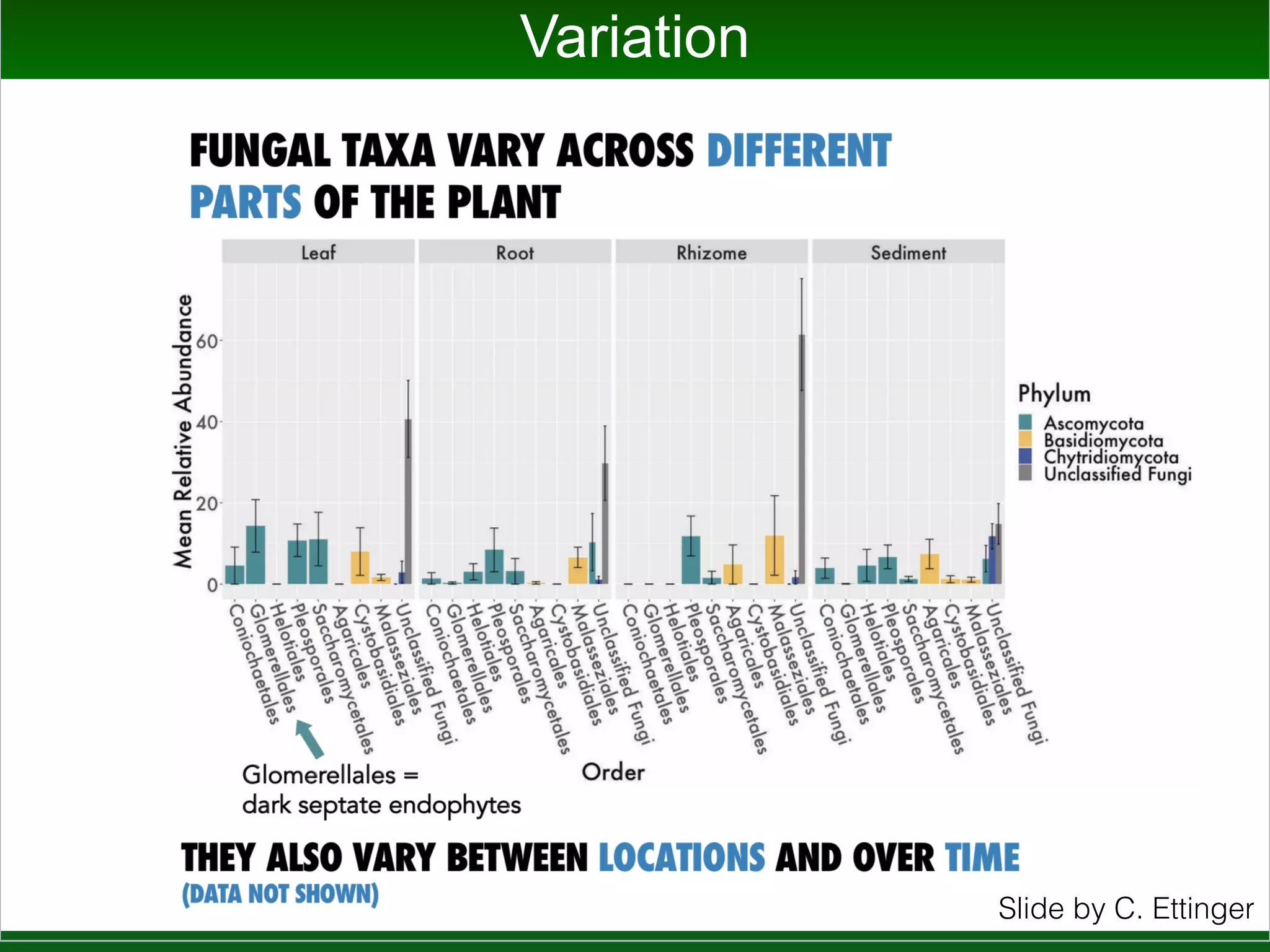
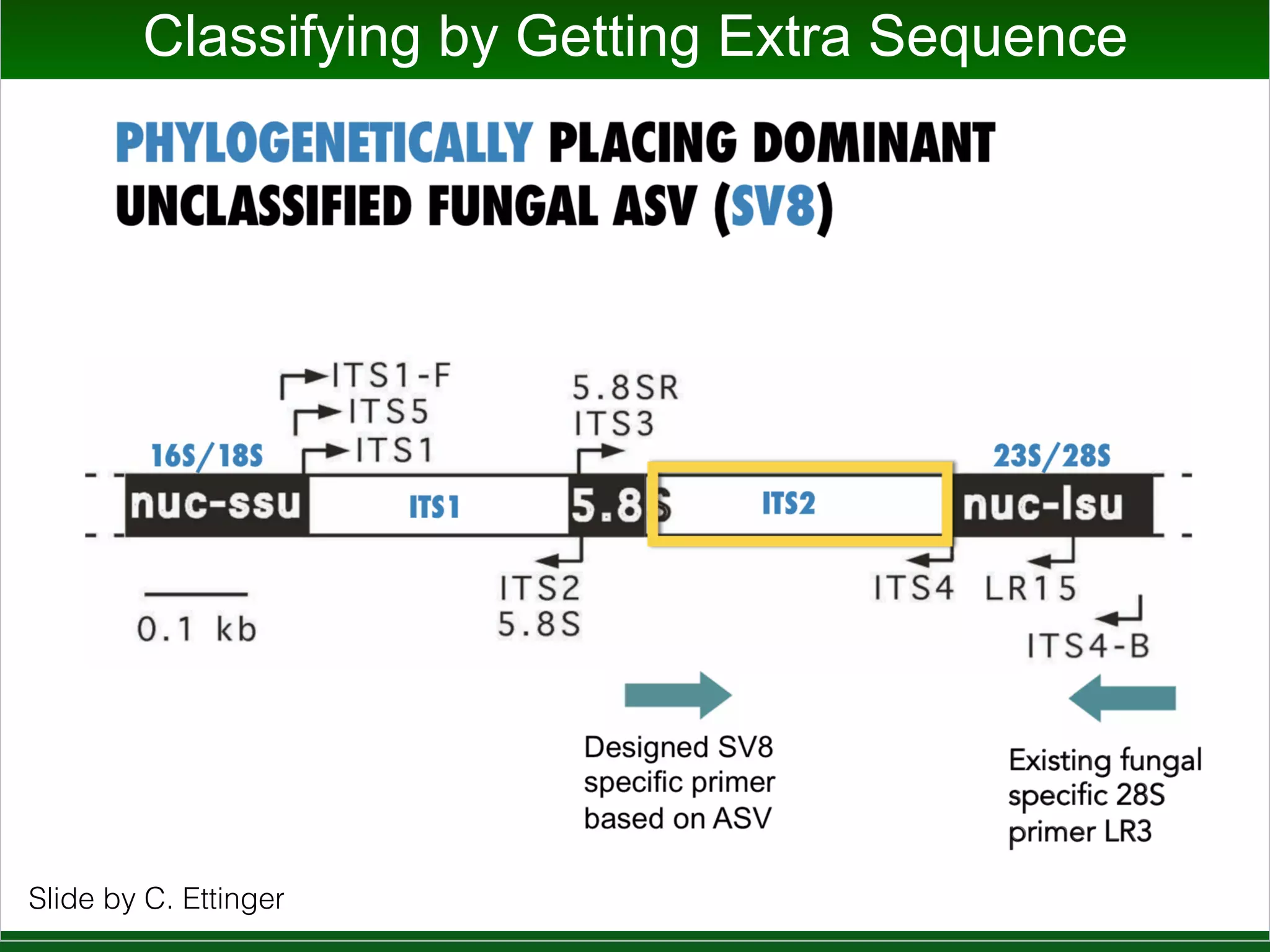
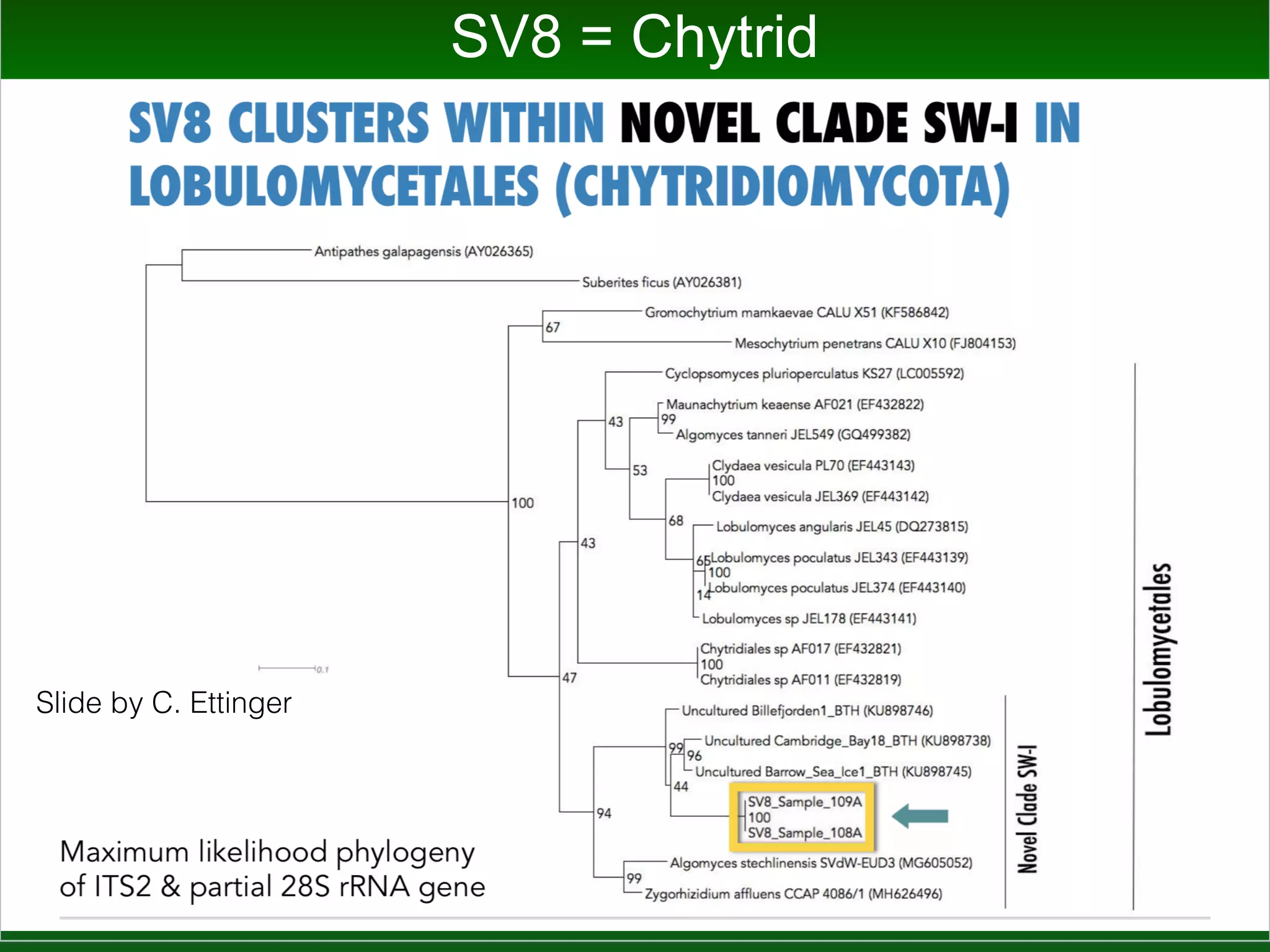
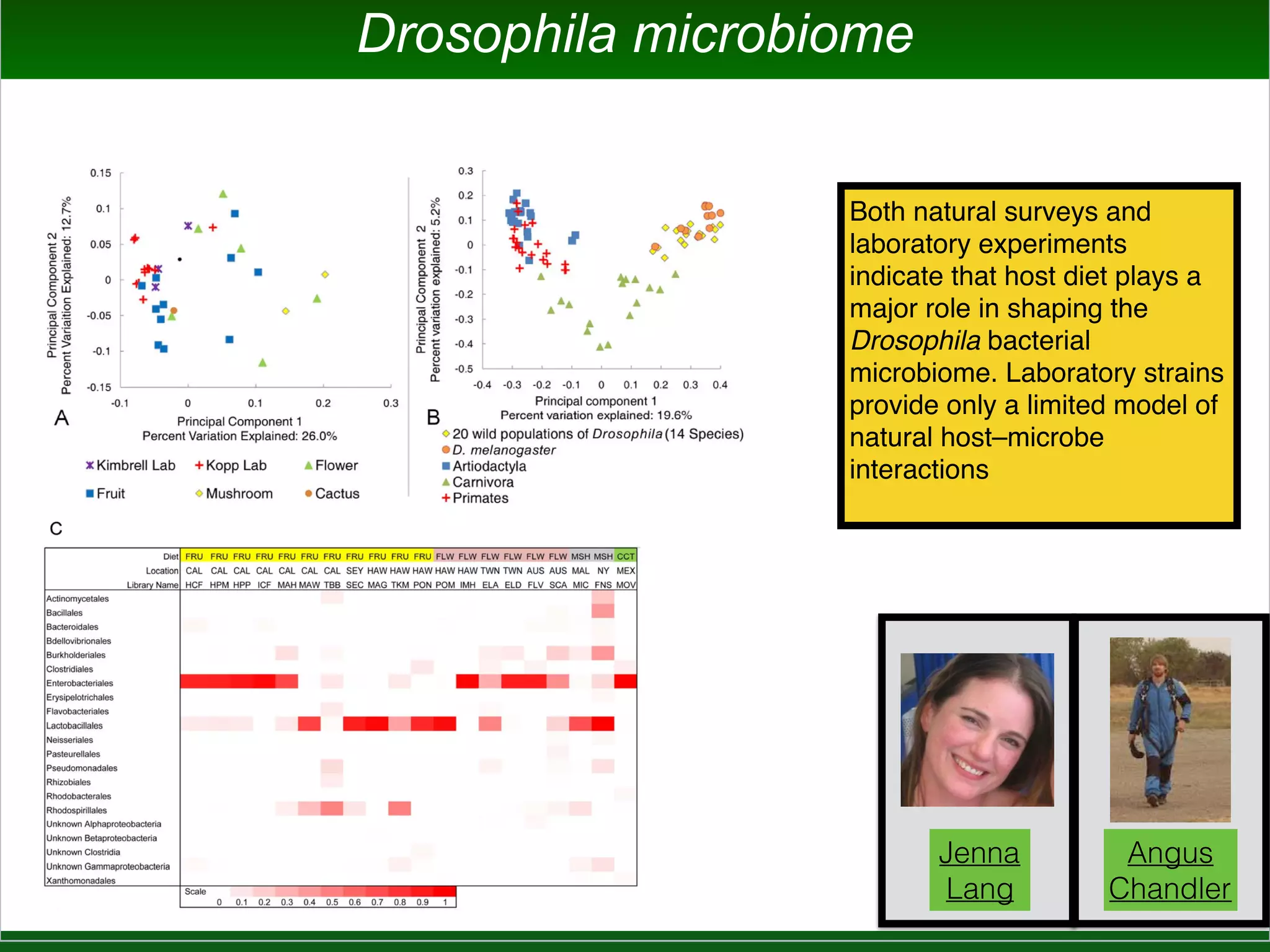
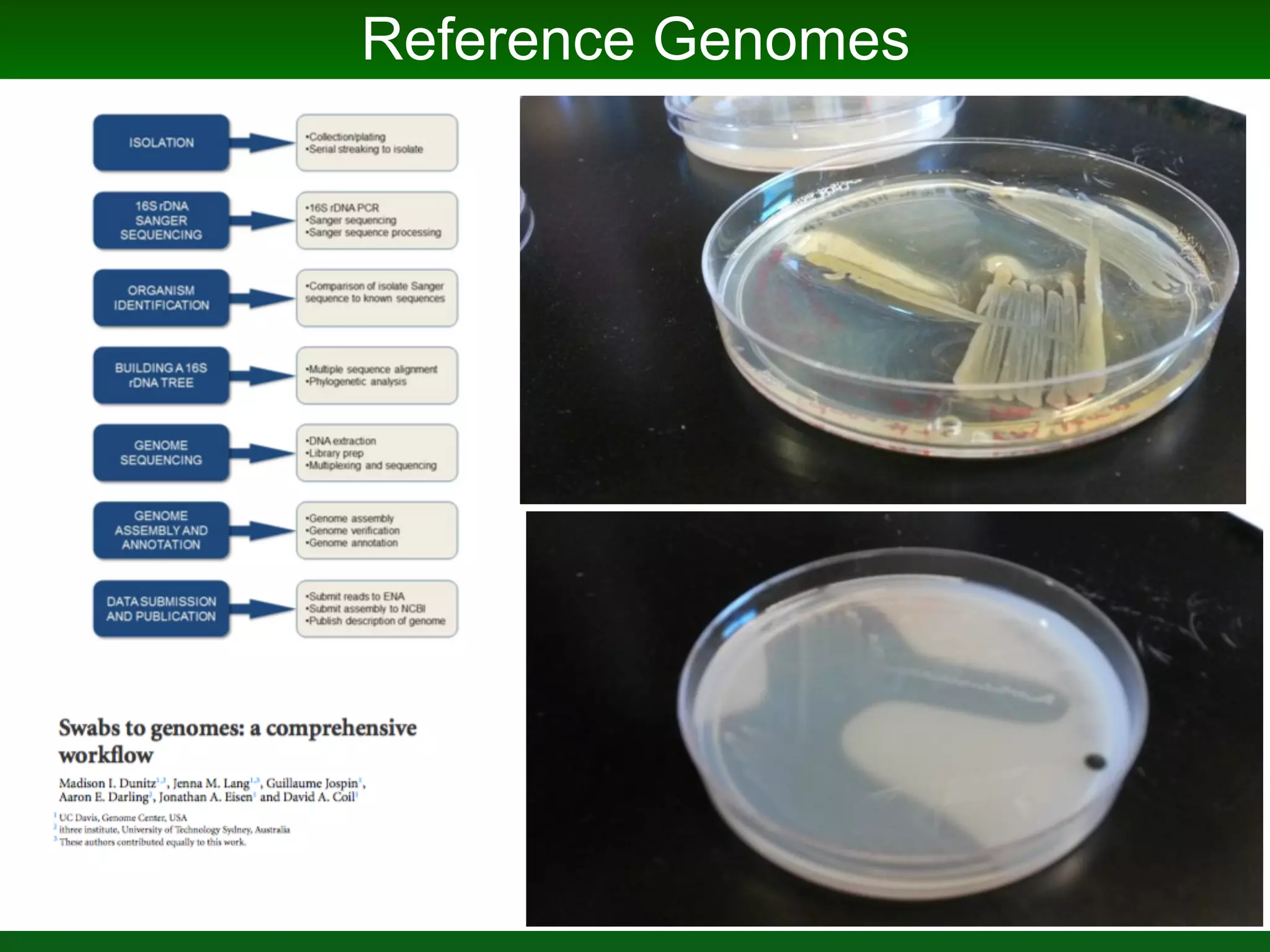
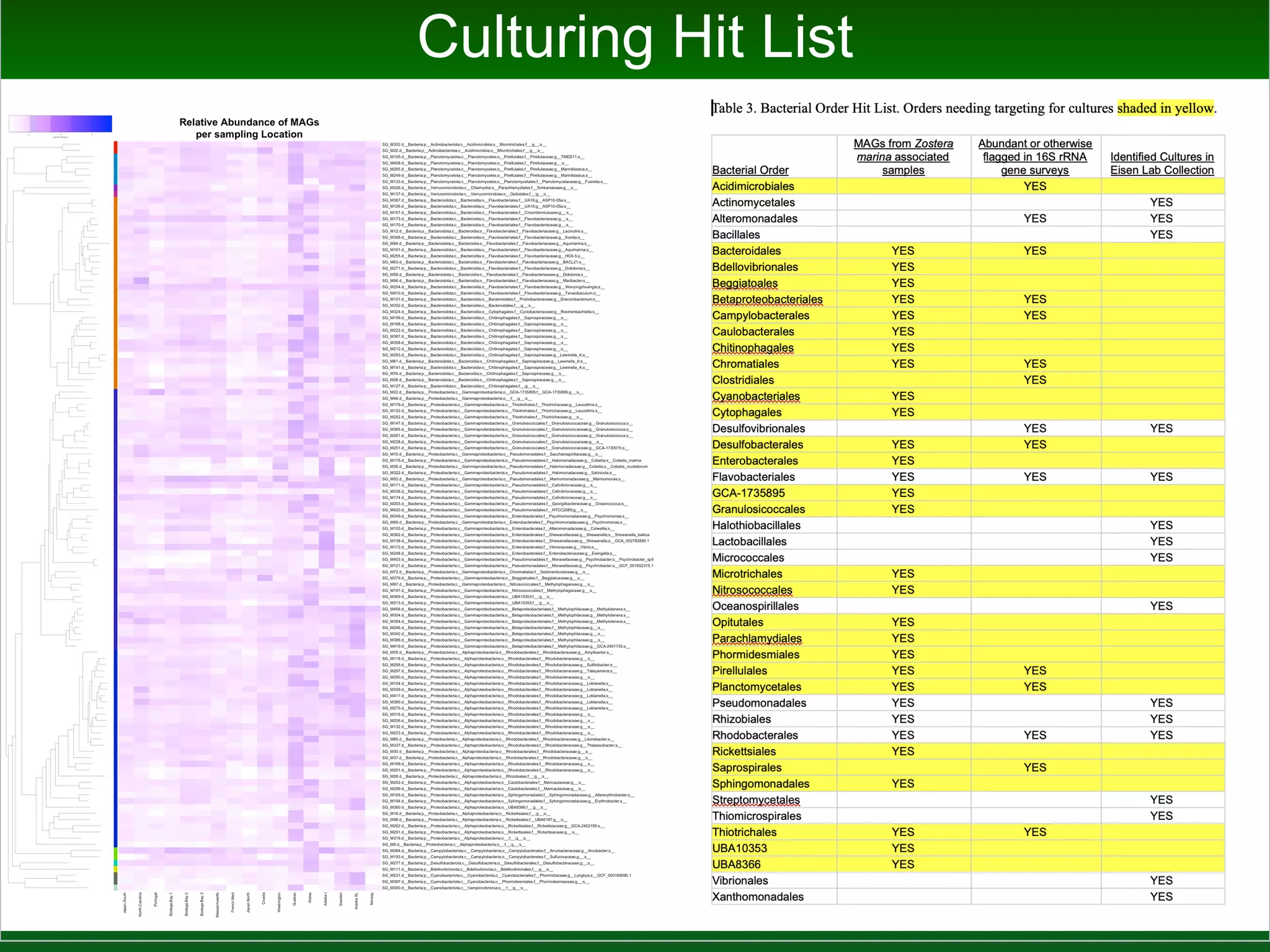
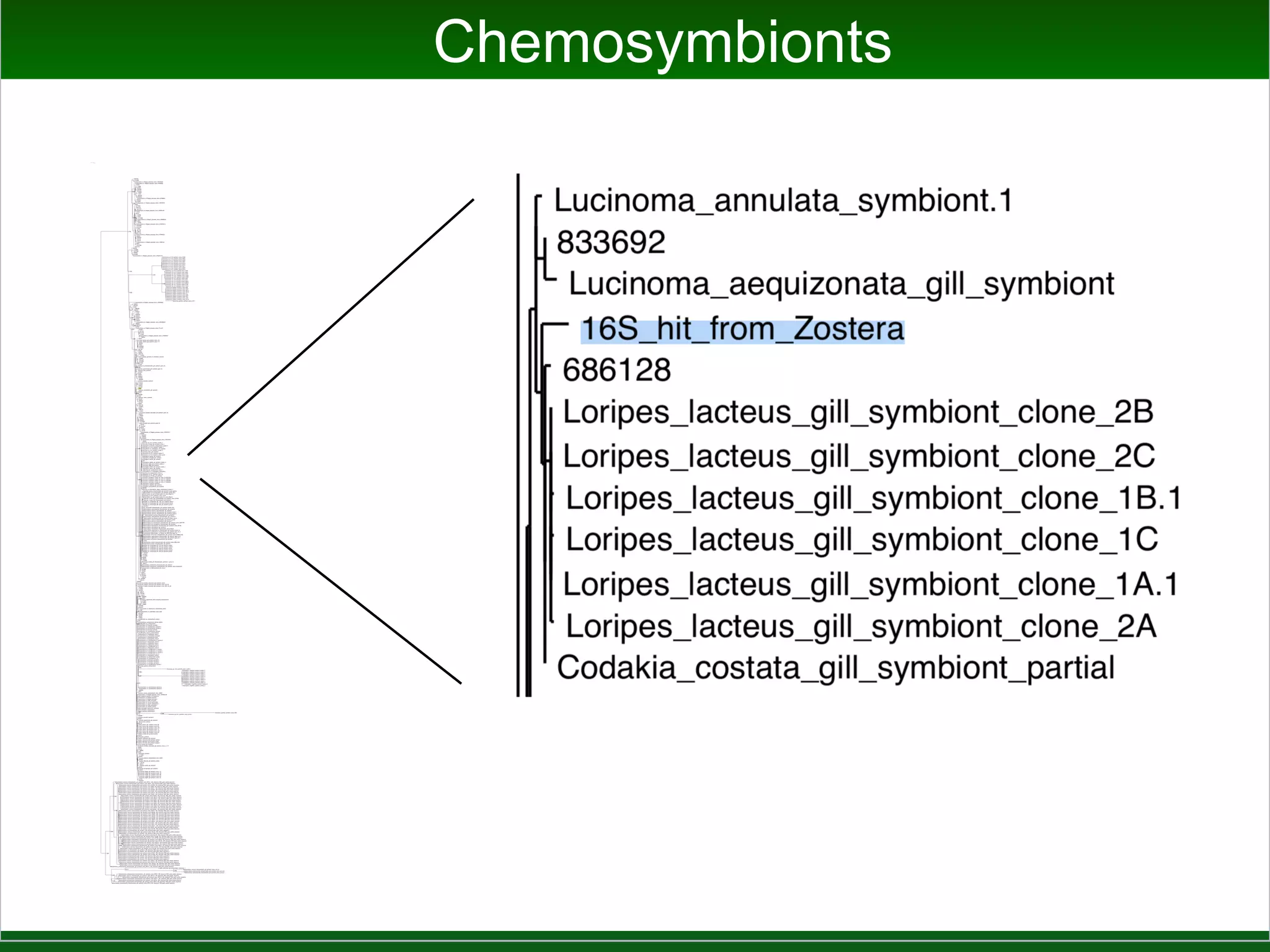
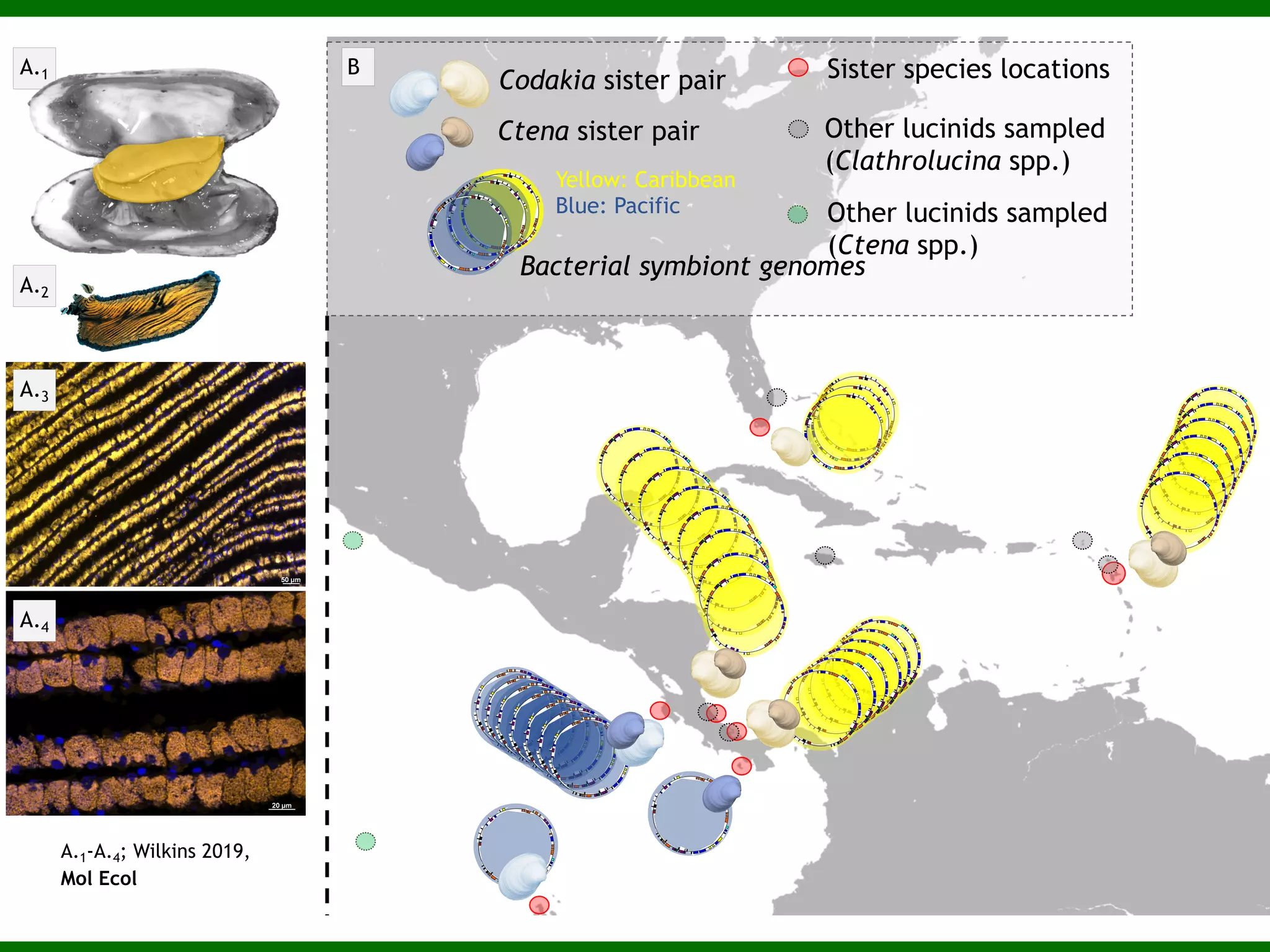
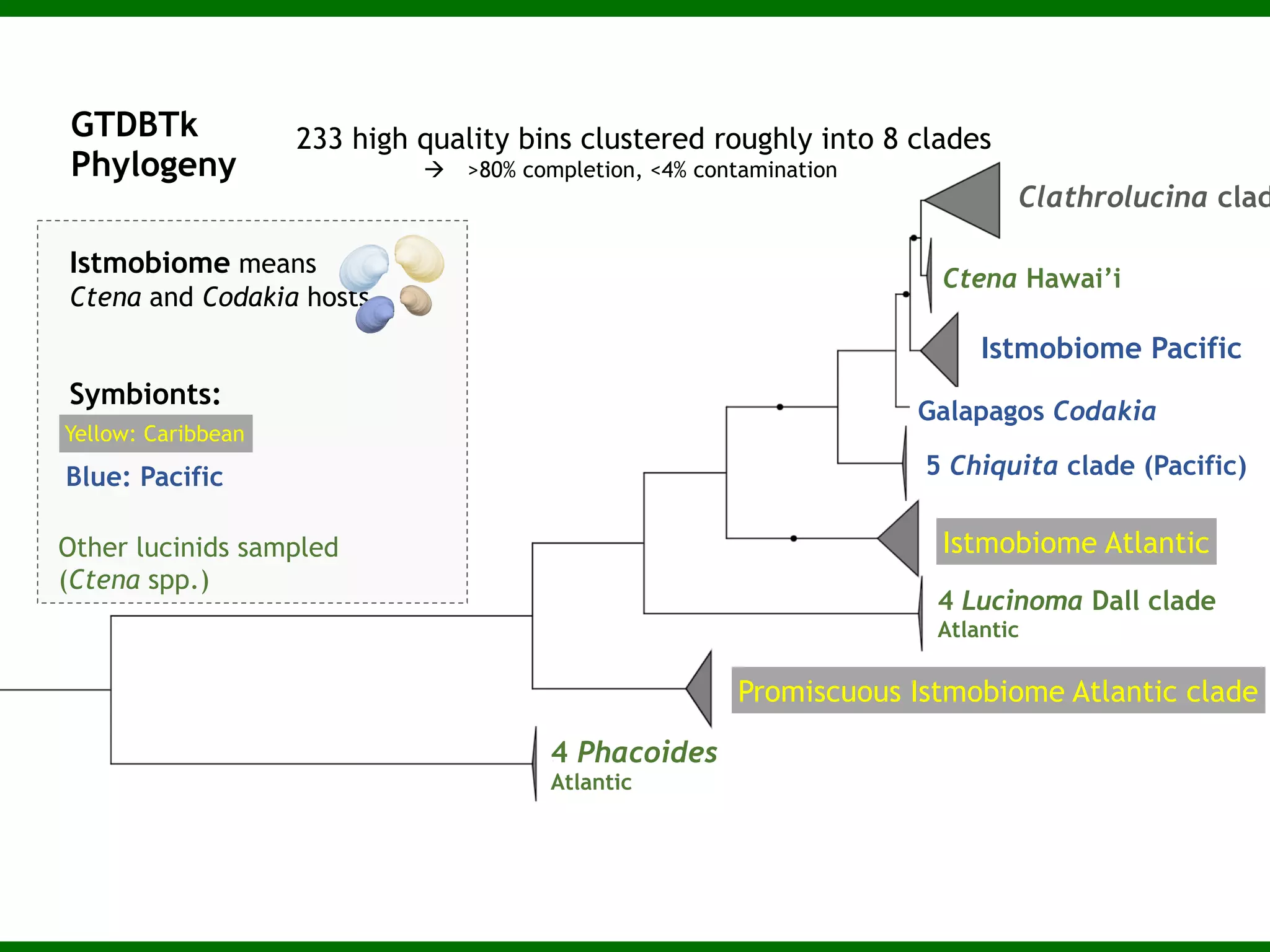
This document summarizes a talk given by Jonathan Eisen on marine host-microbiome interactions. It discusses various topics researched in Eisen's lab, including phylogenomic methods and tools, microbial phylogenomics and evolvability, reference data resources, communication in science, and model systems. Specific projects are mentioned, such as automated genome trees, phylogenetic marker genes, the GEBA project, and dark matter microbes. The document then introduces the concept of the host-microbiome stress triangle and gives examples of stress types including nutrient acquisition, pathogens, and environmental change. It concludes by discussing a potential project on seagrass microbiomes in collaboration with Jay Stachowicz's lab.