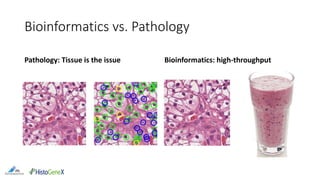
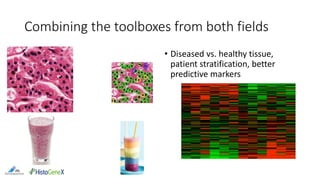
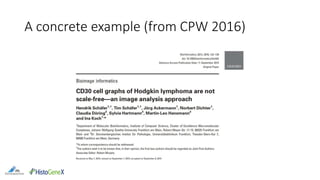
The document discusses the integration of digital pathology with bioinformatics to form computational pathology, highlighting its significance as an omics data layer. It reviews the historical development of digital pathology and emphasizes the necessity of collaboration between the two fields to improve patient diagnosis and treatment. The author presents various suggestions for bridging gaps in the technology and fostering usability and replicability in research efforts.