This document discusses key concepts about enzyme structure and function including:
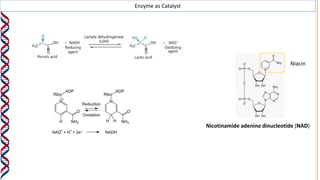
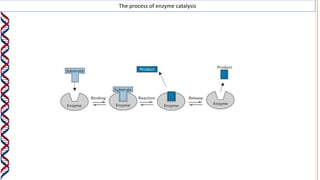
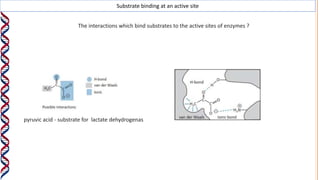
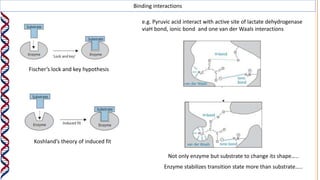
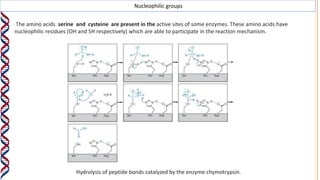
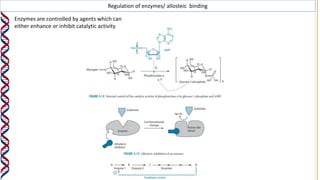
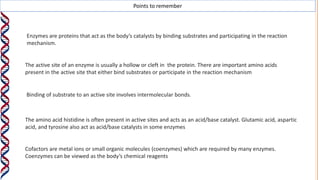
1) Enzymes catalyze reactions by providing a reaction surface that brings reactants together and positions them correctly at the active site. Amino acids at the active site can bind substrates or participate in reaction mechanisms.
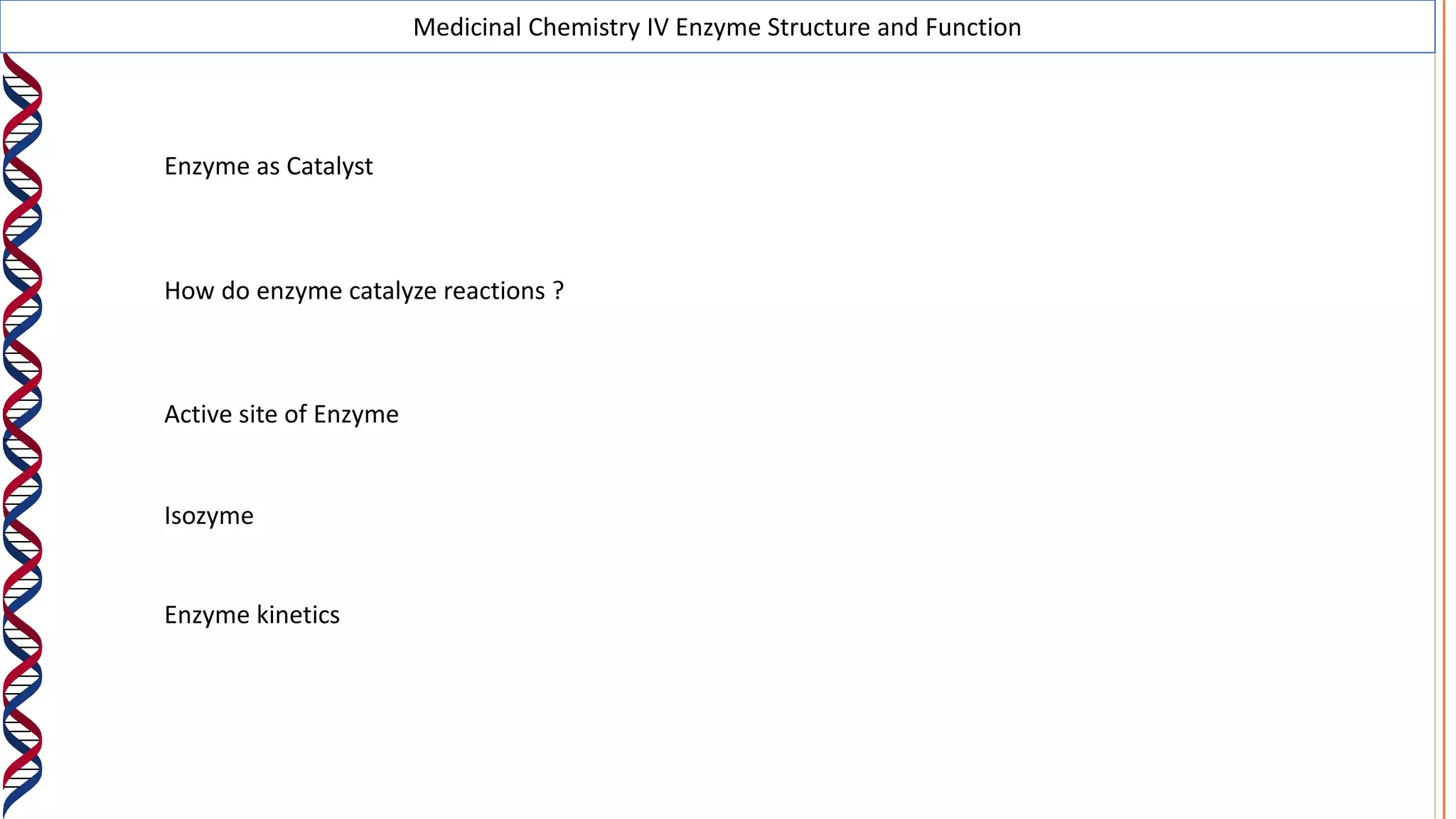
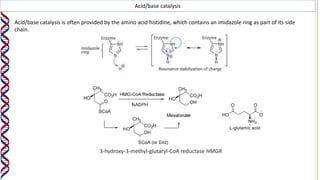
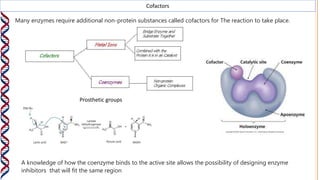
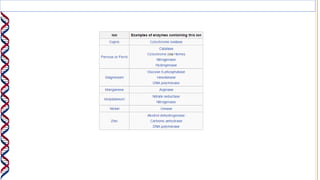
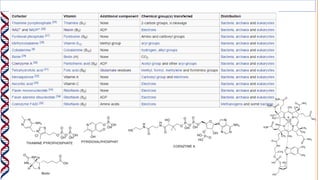
2) The active site contains catalytic amino acids like histidine that act as acid/base catalysts to weaken bonds in reactants. Some enzymes also require cofactors like metal ions or coenzymes.
3) Isozymes are variations in the combination of polypeptide subunits that make up enzyme complexes, resulting in different isozyme forms of the same enzyme in different tissues.