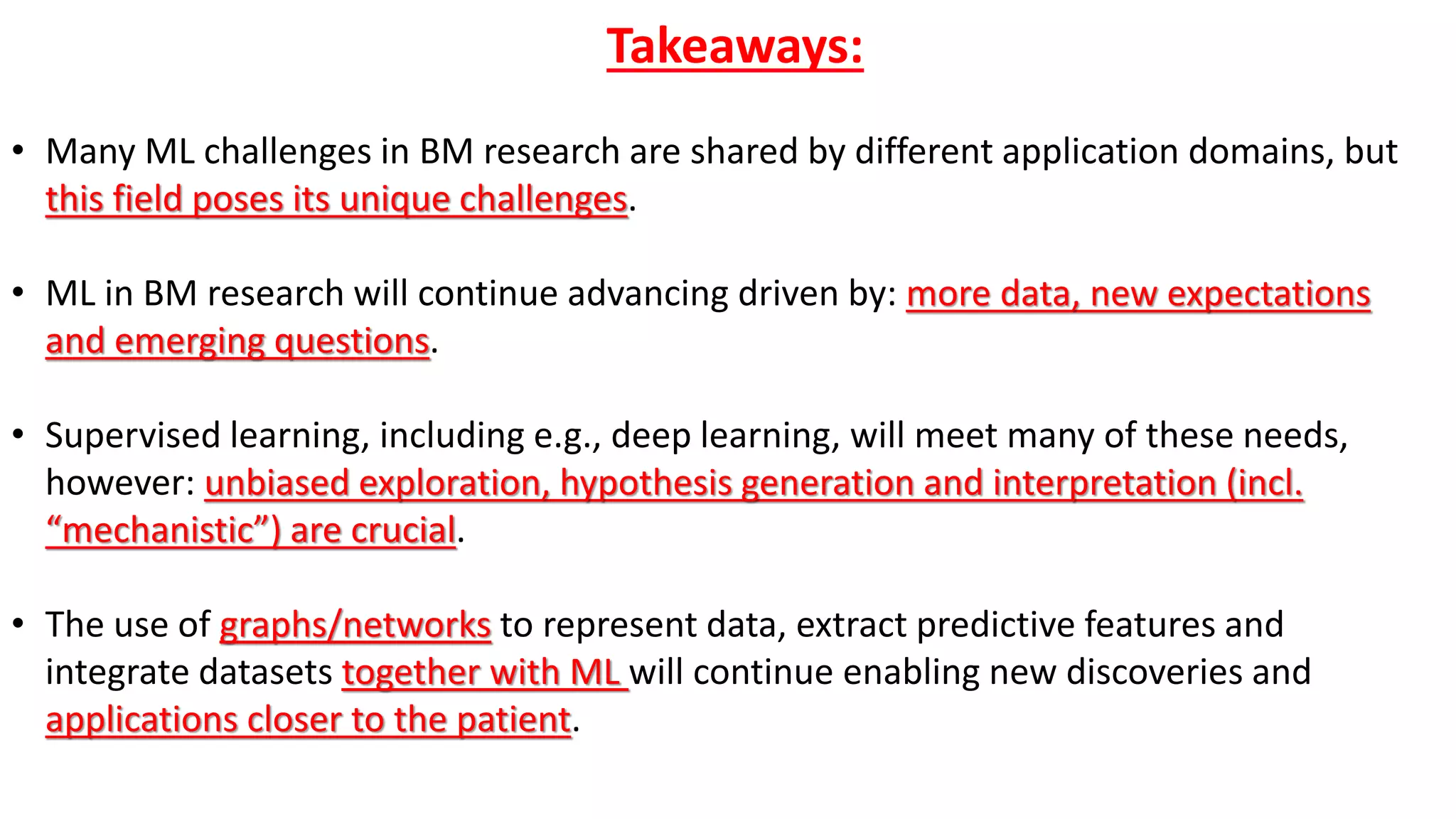
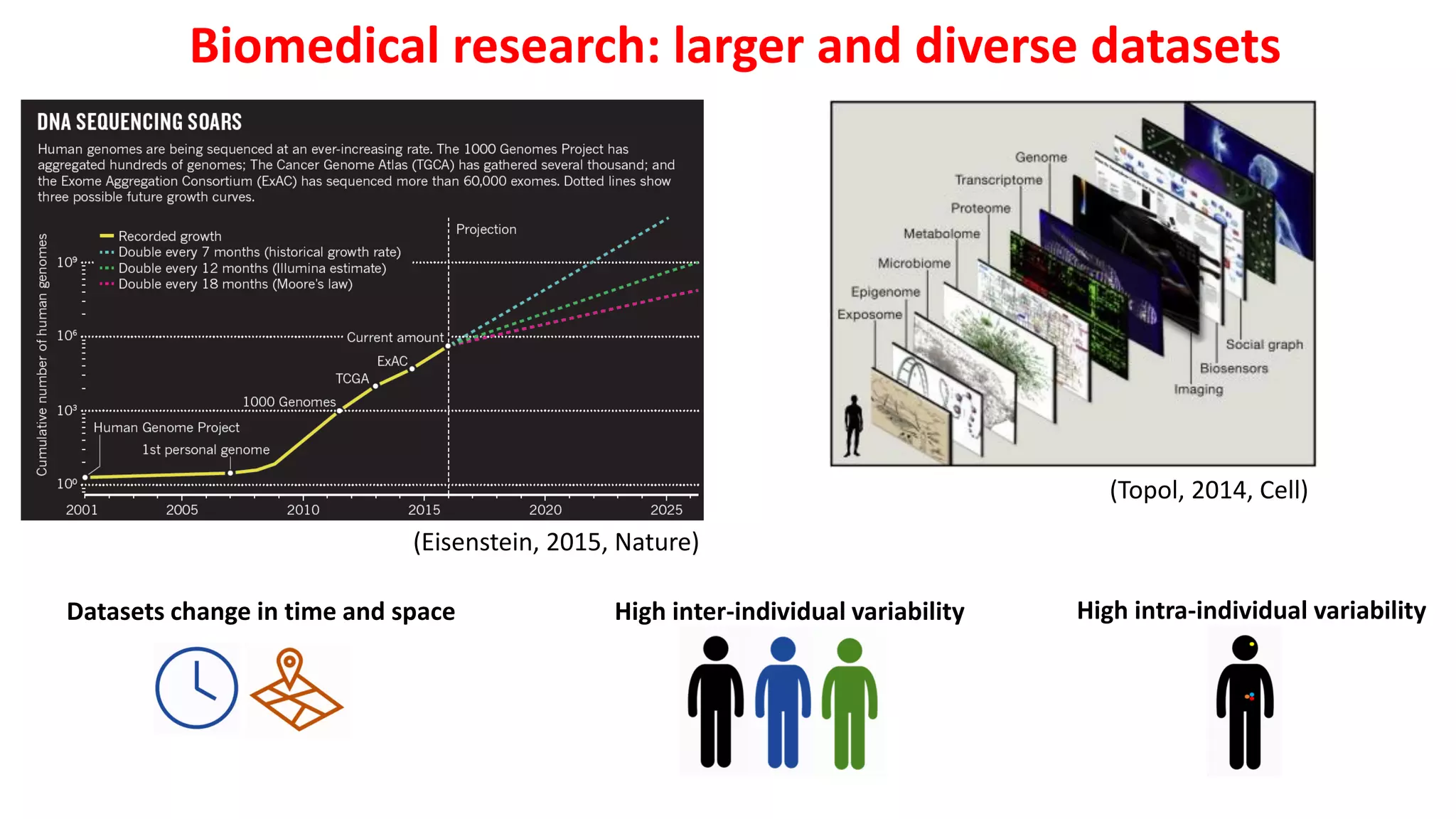
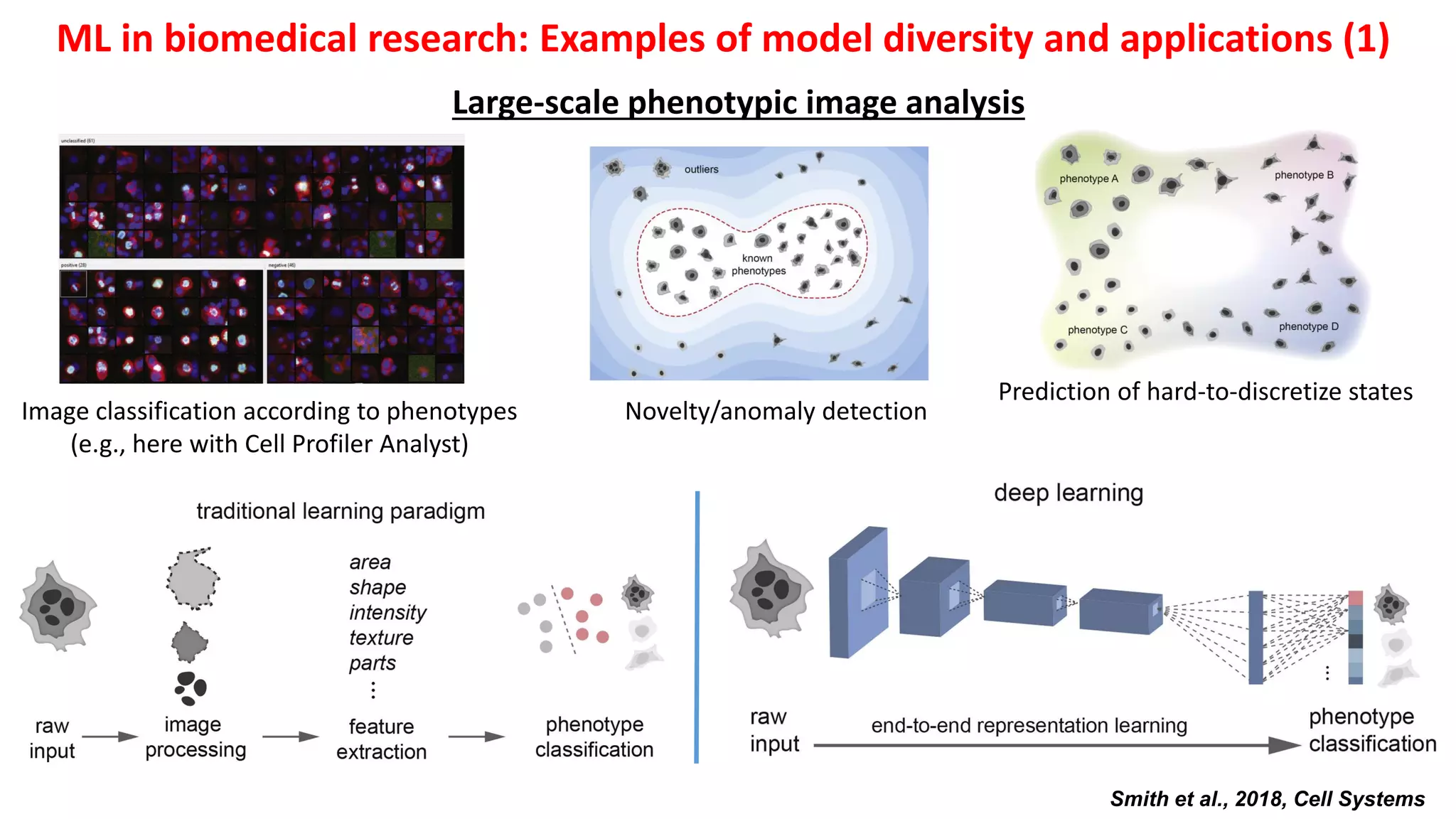
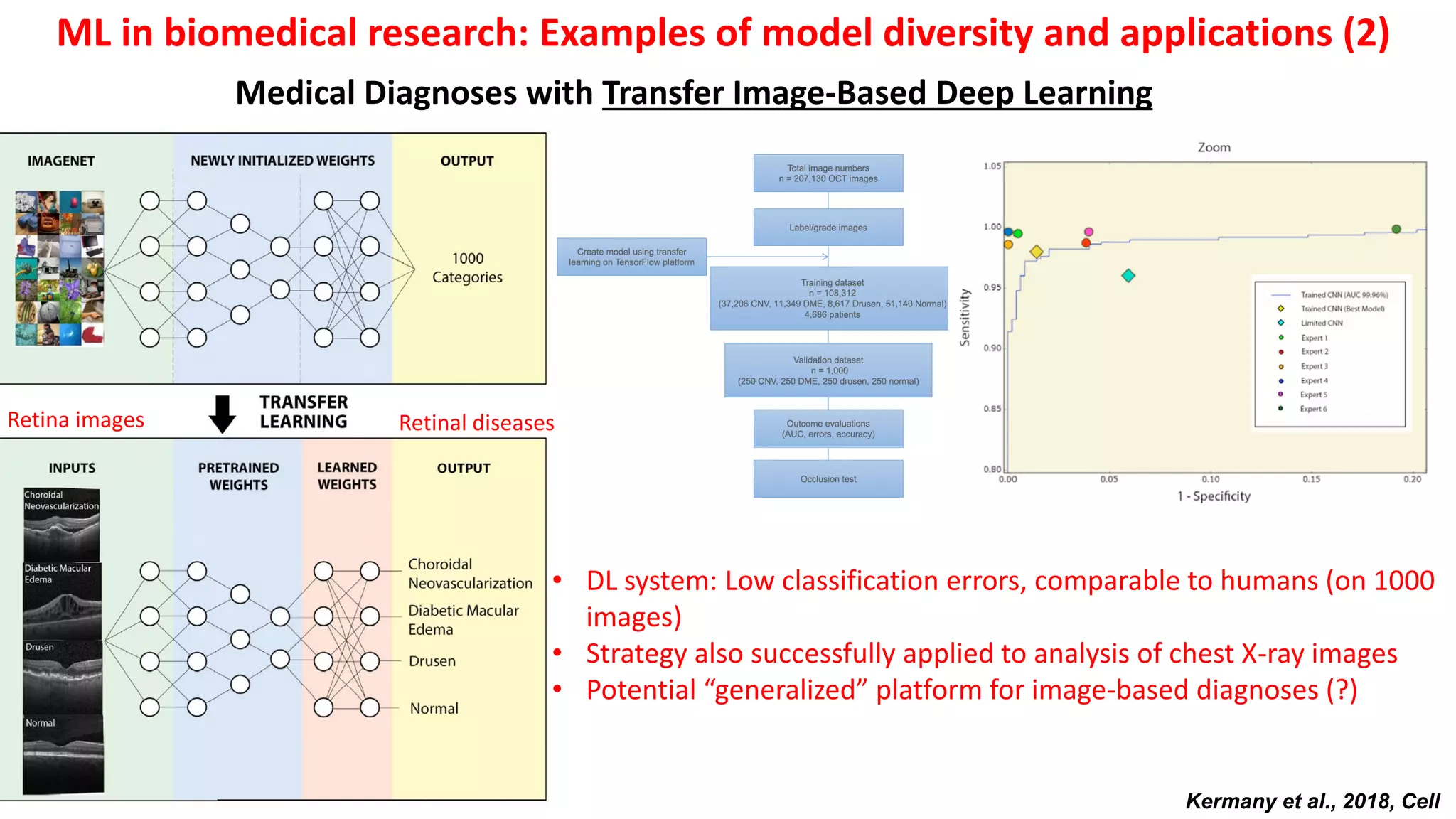
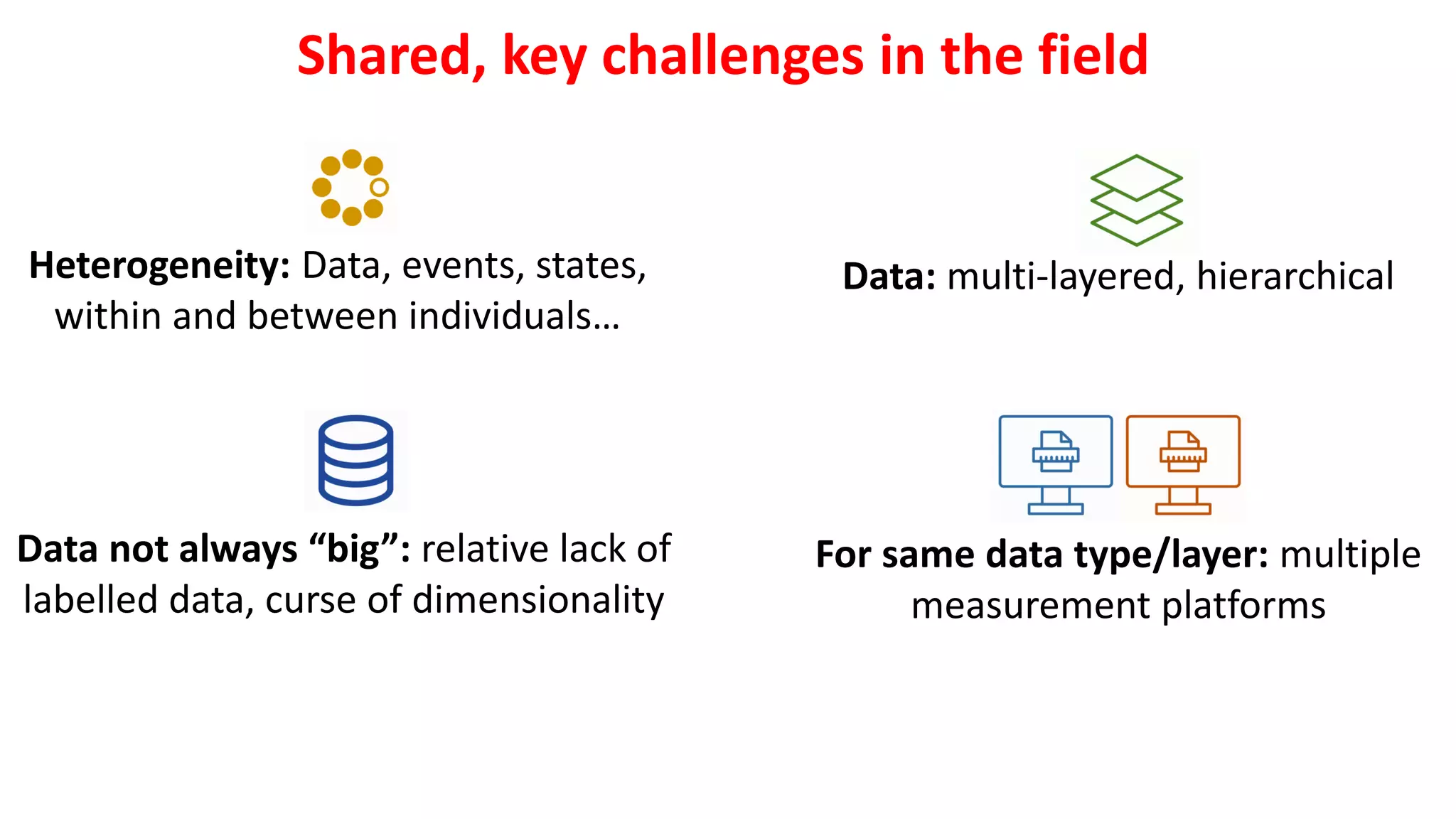
1. Machine learning faces challenges in biomedical research due to data heterogeneity, lack of labeled data, and complexity in biological patterns and networks.
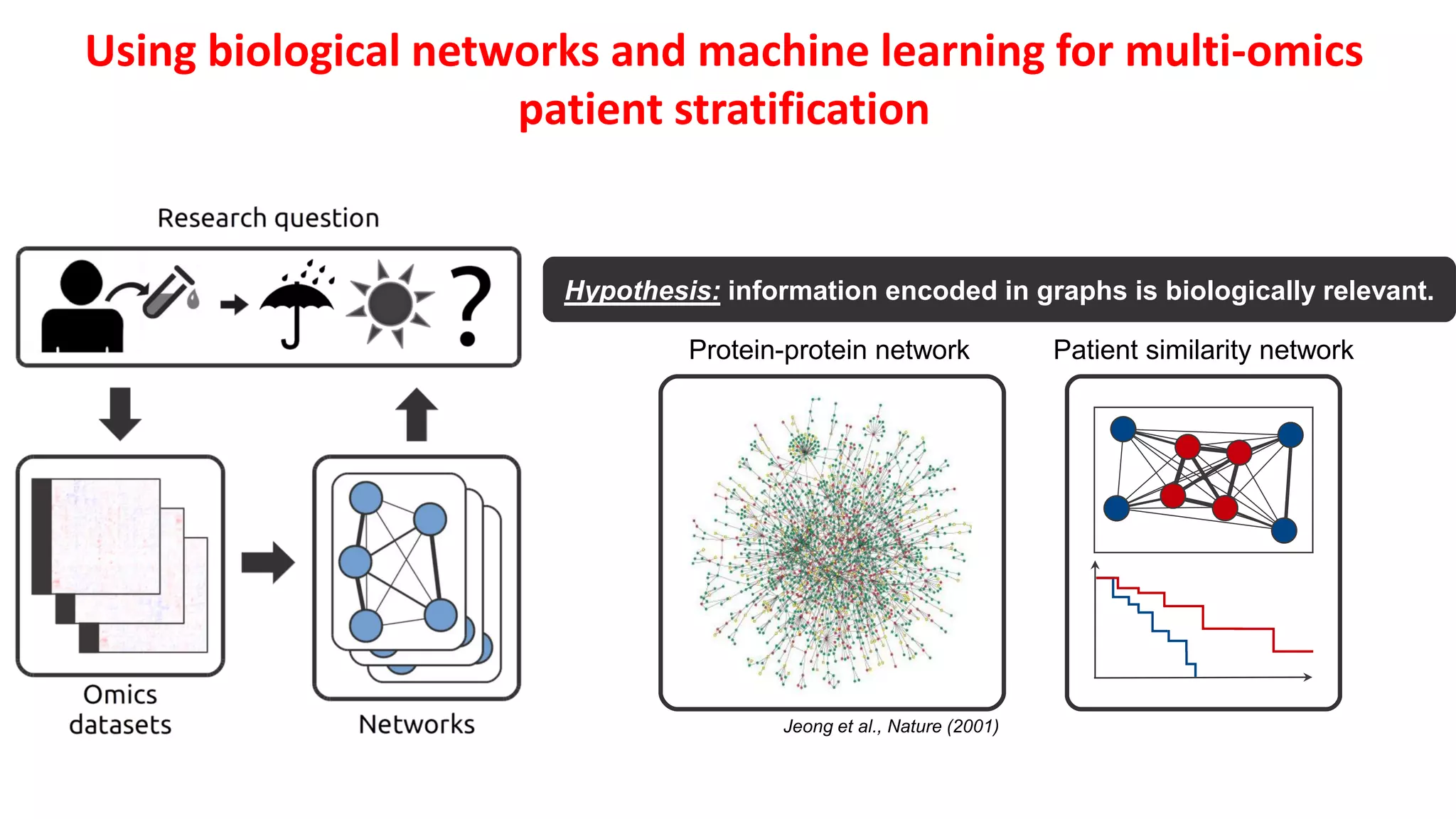
2. Combining machine learning and biological network models can help address these challenges by encoding data in biologically meaningful networks and extracting network-based features for prediction.
3. Examples applying this approach to cancer datasets showed that models based on network centrality features outperformed other methods, and deep learning using these features achieved the best prediction performance across multiple neuroblastoma datasets.















![Application example (2): Neuroblastoma multi-omics datasets
from the CAMDA challenge, a deep learning approach*
Global strategy Algorithm Parameters Balanced
accuracy
Death from disease, Fischer-M
DNN h=[8,8,8,2], o=Adam, lr=1e-3, d=0.3 87.3% *
SVM t=RBF, c=64, g=0.25 75.4%
RF n=100 75.1% *
Disease progression, Fischer
DNN h=[4,2,2,2], o=Adam, lr=1e-3, d=0.3 84.7% *
SVM t=RBF, c=16, g=0.0625 81.8%
RF n=100 78.1% *• Network features from each dataset: Centrality (12), modularity
(30 to 47) features.
• Models based on each feature category, and their combination
• Data: 498 patients (2 omic datasets, gene expression data)
• Training (50% of total data), validation and test datasets
• DNNs: multiple architectures, Rectified Linear Units (ReLU),
Softmax function (2 outputs)
Prediction performance on test
dataset (top models)
Top DNN: Input features are graph centrality measures
Fischer-M: 1 dataset only (microarrays)
Fischer: 2 datasets (microarrays + RNA-Seq)
*Article in preparation](https://image.slidesharecdn.com/franciscoazuaje-luxdsmeetup-180912223445/75/Challenges-and-opportunities-for-machine-learning-in-biomedical-research-21-2048.jpg)