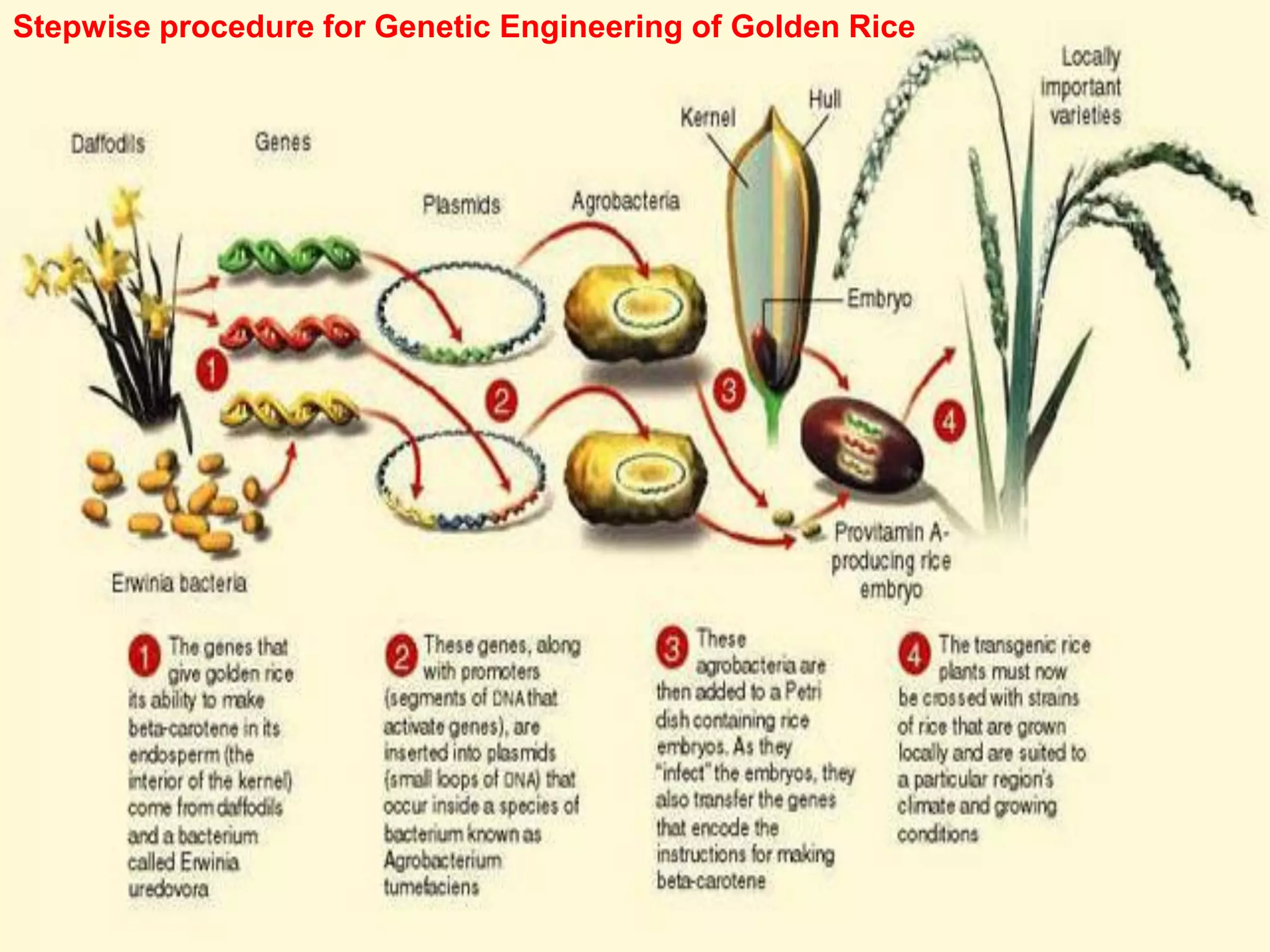
The document summarizes a workshop held on February 11, 2021 at Smt. Kasturbai Walchand College in Sangli to demonstrate experiments from the revised syllabus of the B.Sc. III practical course. It includes demonstrations and explanations of experiments on isolating plant genomic DNA and estimating the DNA using diphenylamine. It also discusses the steps involved in genetically engineering golden rice through the introduction of genes from daffodil and bacteria to allow the rice plant to produce beta-carotene.