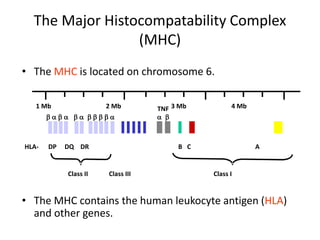
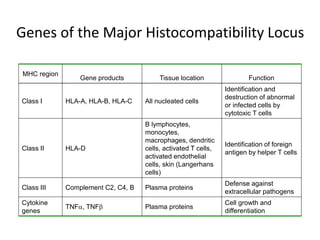
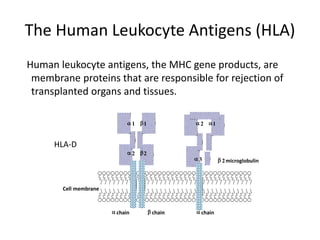
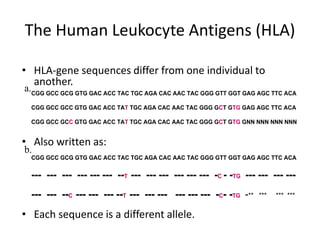
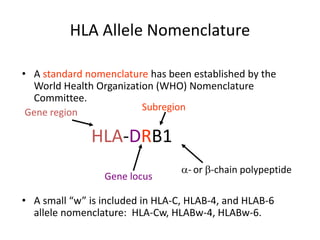
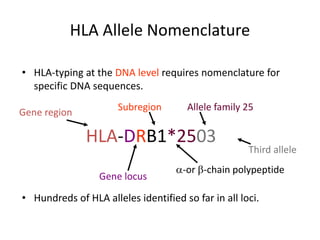
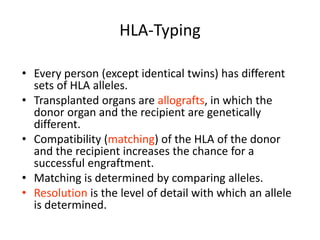
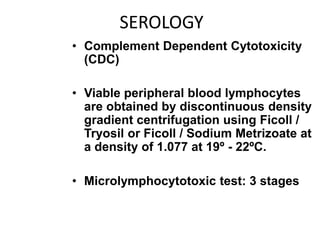
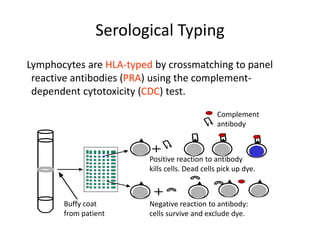
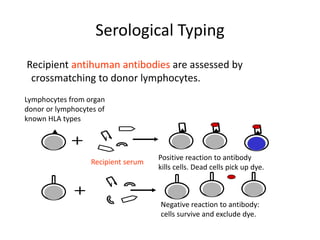
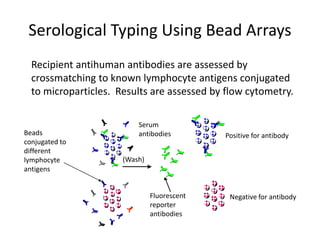
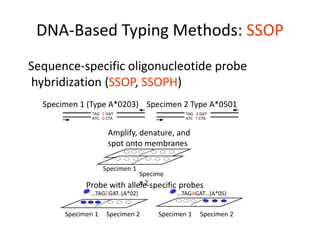
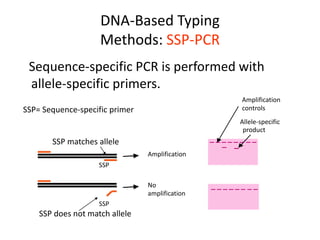
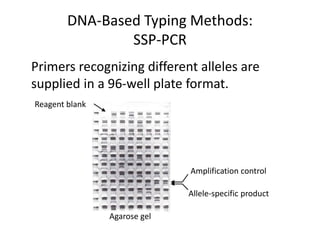
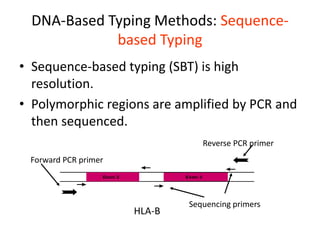
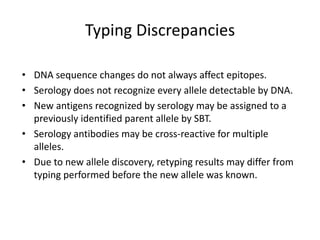
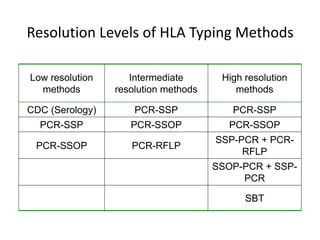
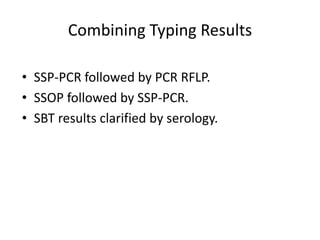
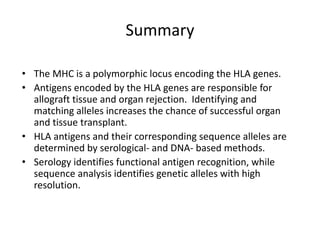
The document discusses HLA typing methods. It describes the major histocompatibility complex (MHC) which contains the human leukocyte antigen (HLA) genes. HLA typing is important for transplant matching. Methods include serology using lymphocyte cytotoxicity, as well as molecular techniques like PCR with sequence-specific primers or probes and sequence-based typing for highest resolution. Each method has advantages and limitations in resolution and reliance on viable cells. Combining methods can resolve discrepancies.