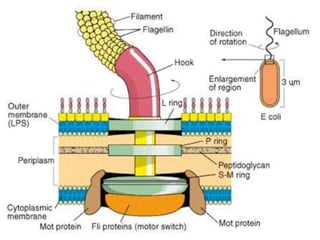
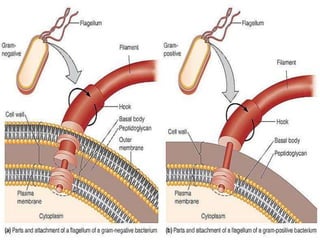
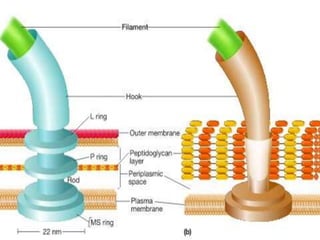
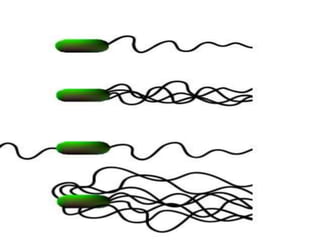
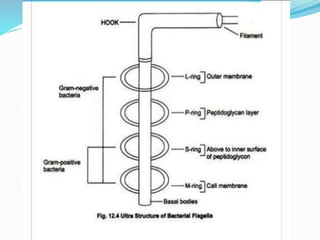
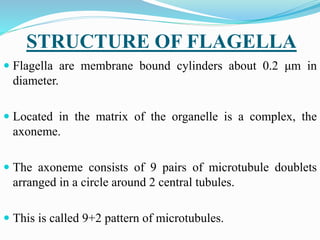
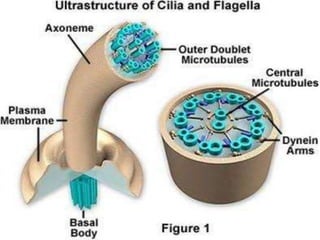
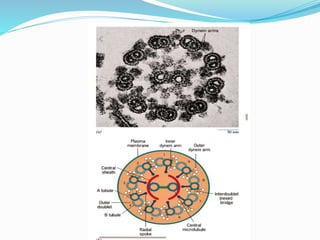
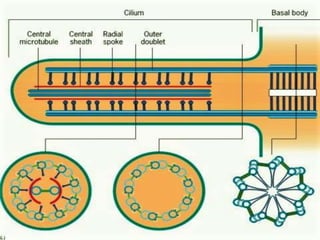
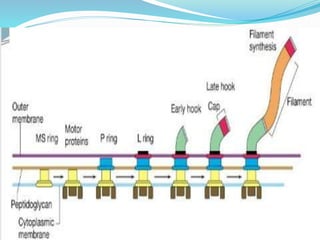
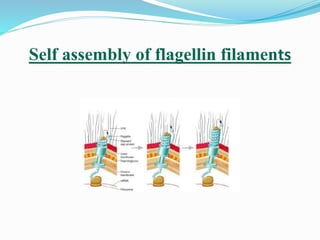
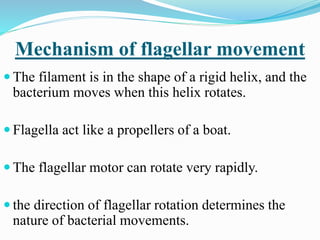
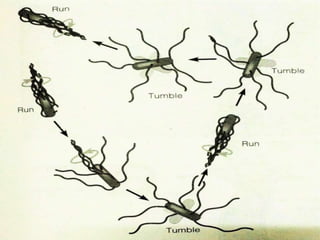
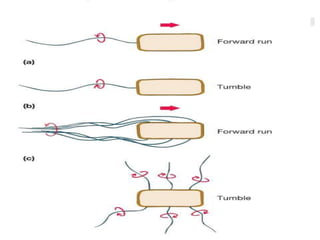
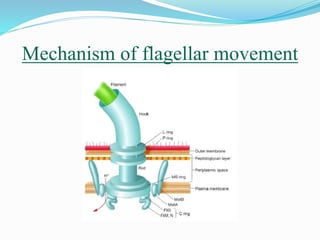
The bacterial flagellum has three main parts - the filament, basal body, and hook. The filament is the longest, rigid structure made of the protein flagellin. The basal body is embedded in the cell and contains protein rings. The hook connects the filament to the basal body. The basal body contains protein rings and a central rod that span the cell membranes. Rotation of the flagellum is driven by a motor composed of a rotor and stator. Proton motive force powers the motor and causes clockwise or counter-clockwise rotation for movement or tumbling.