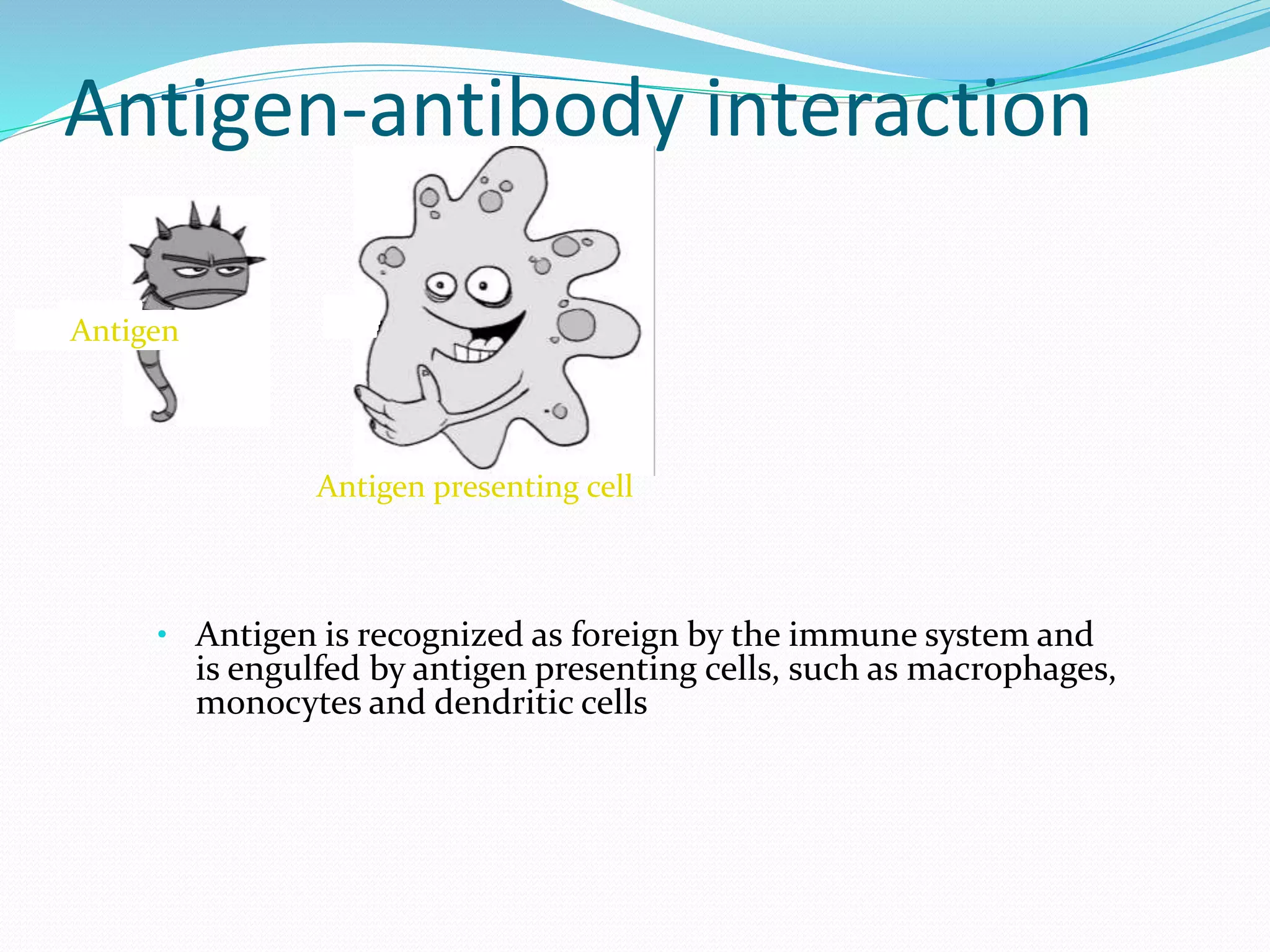
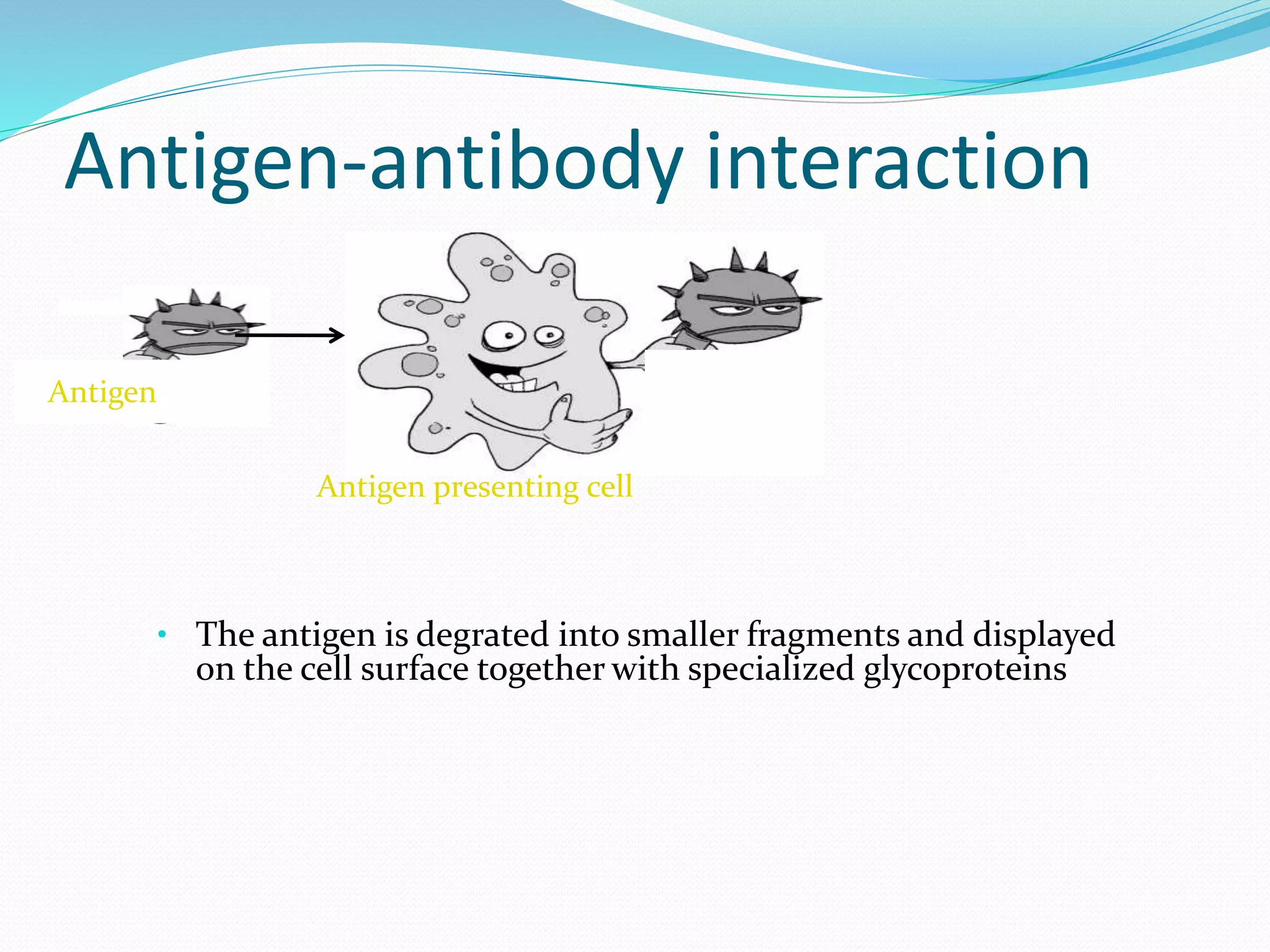
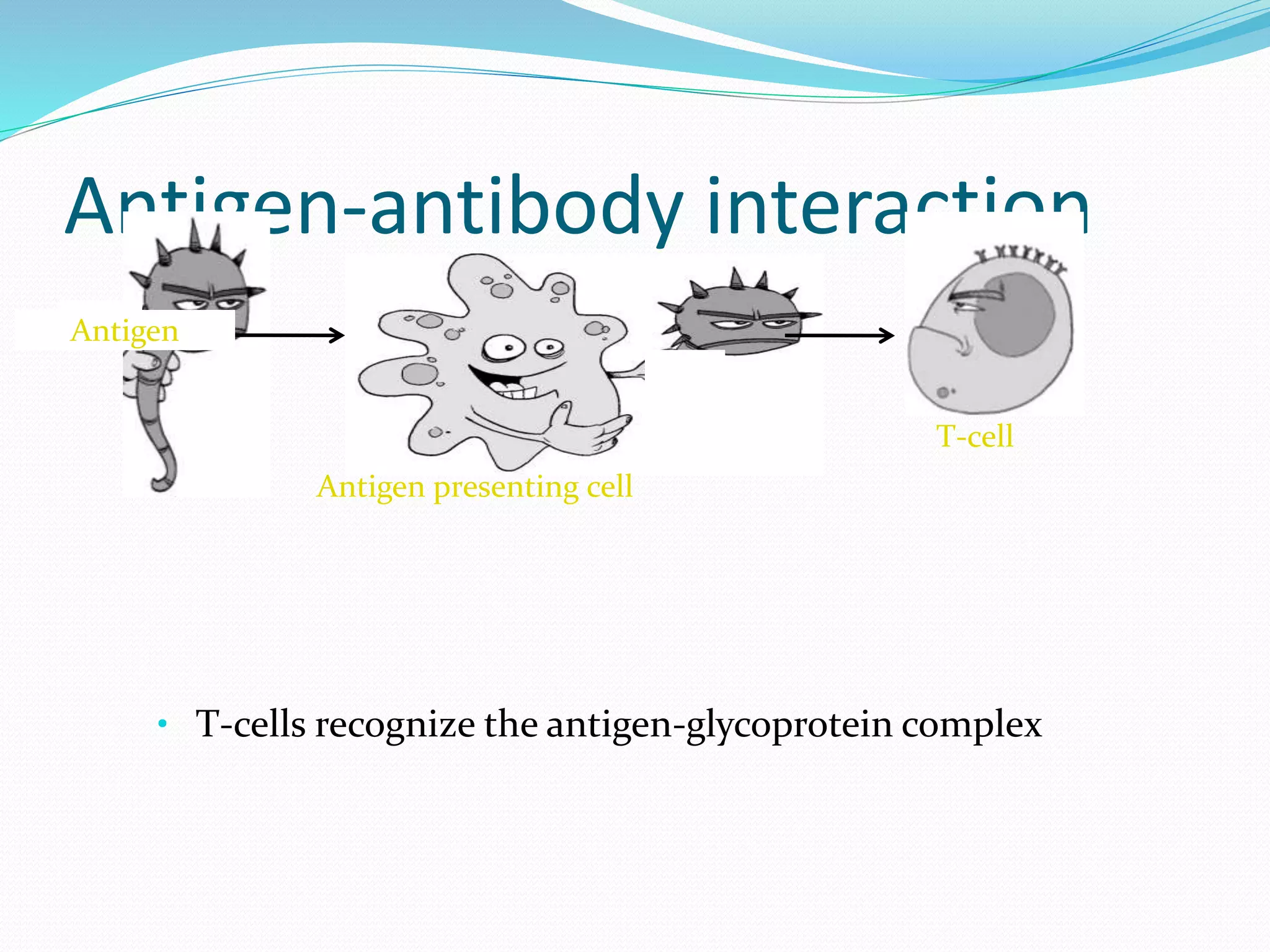
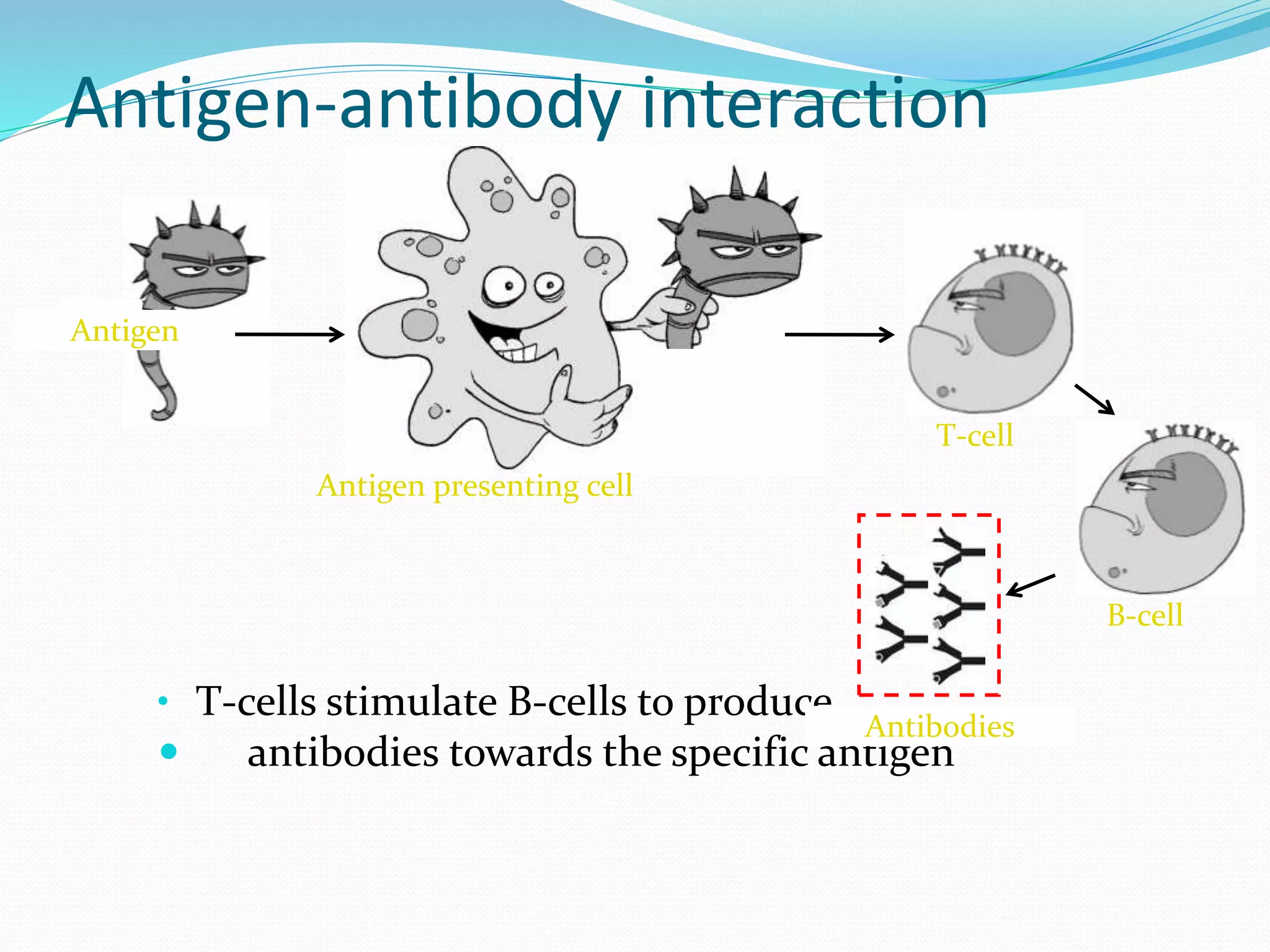
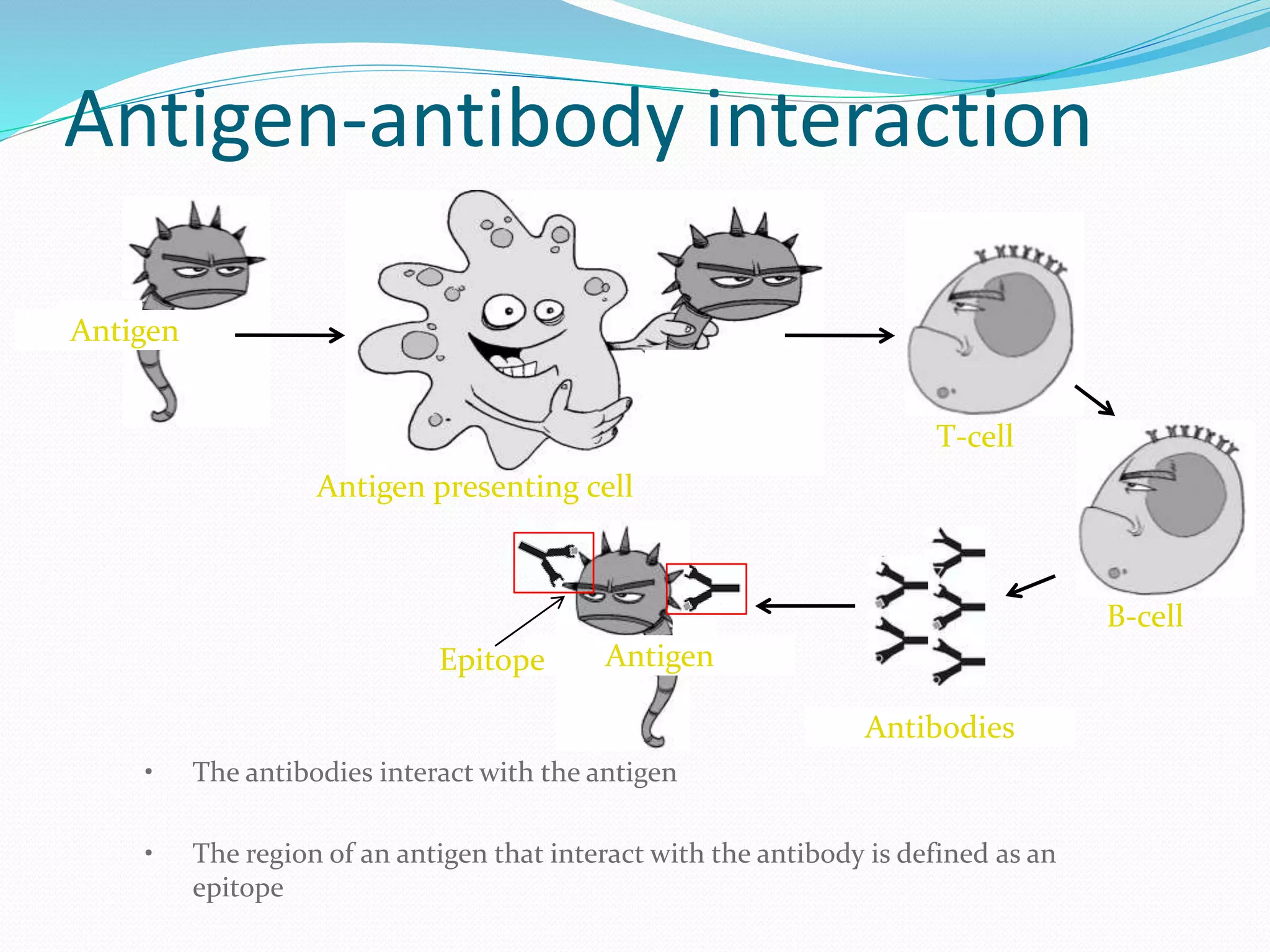
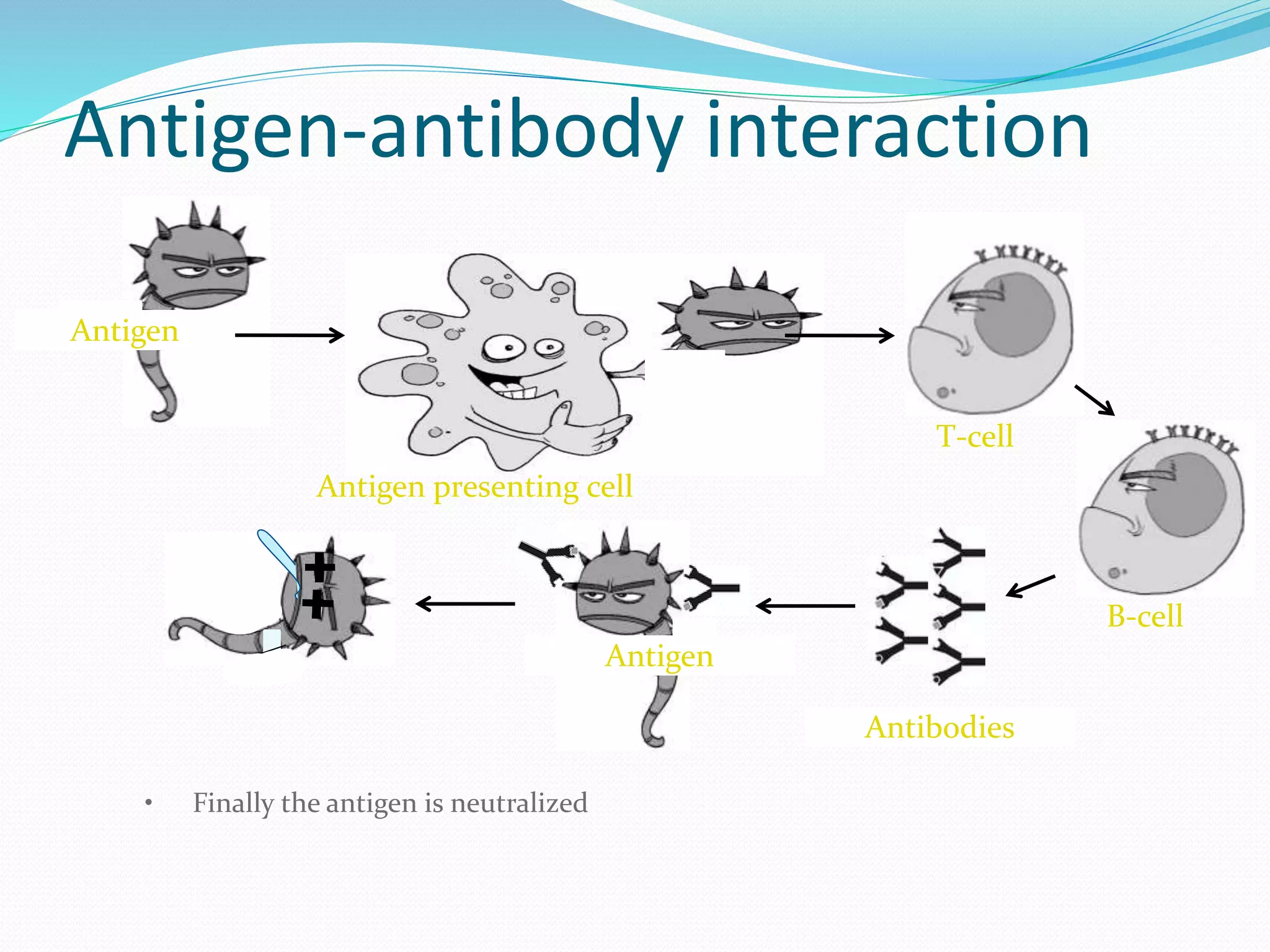
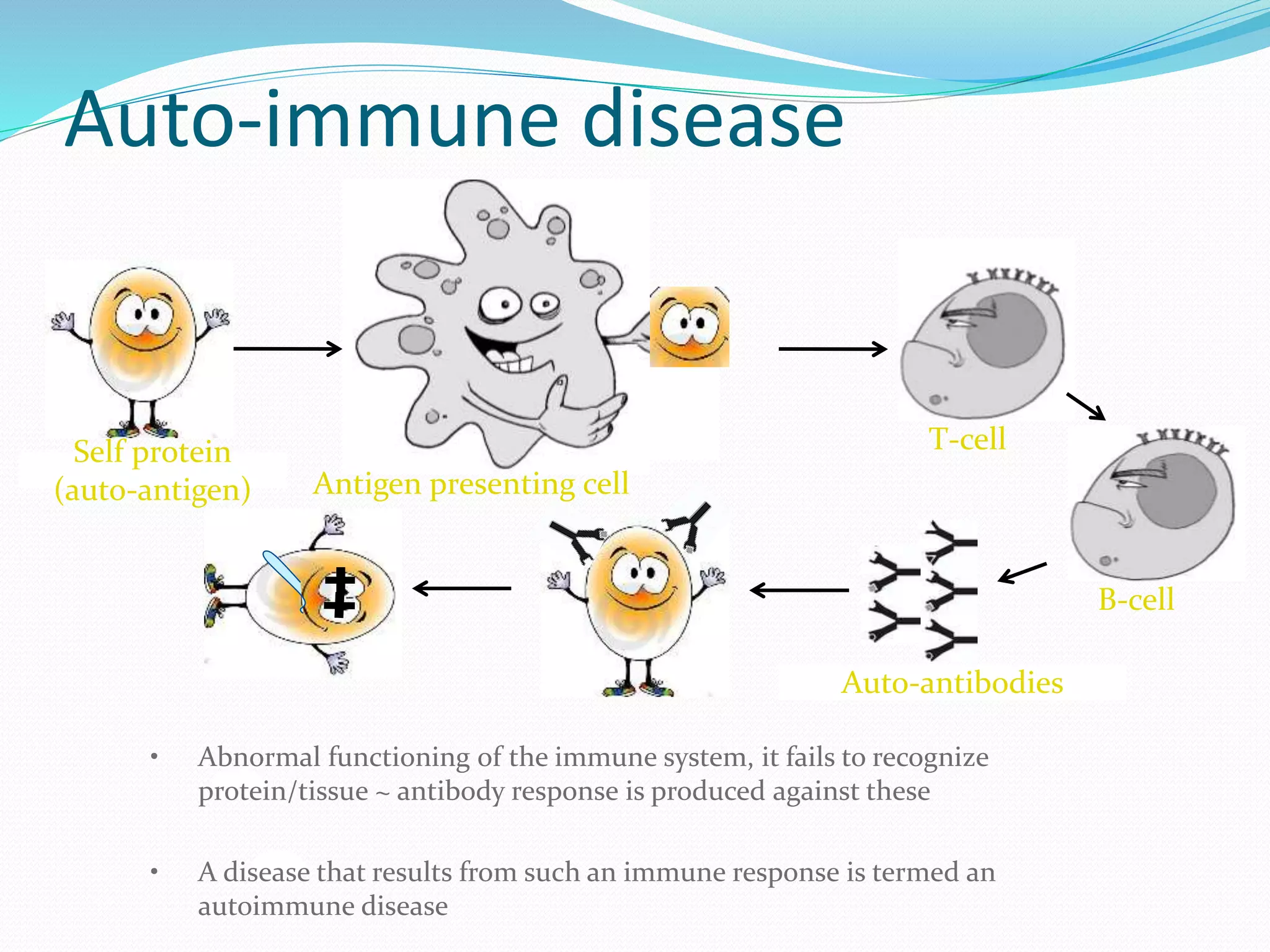
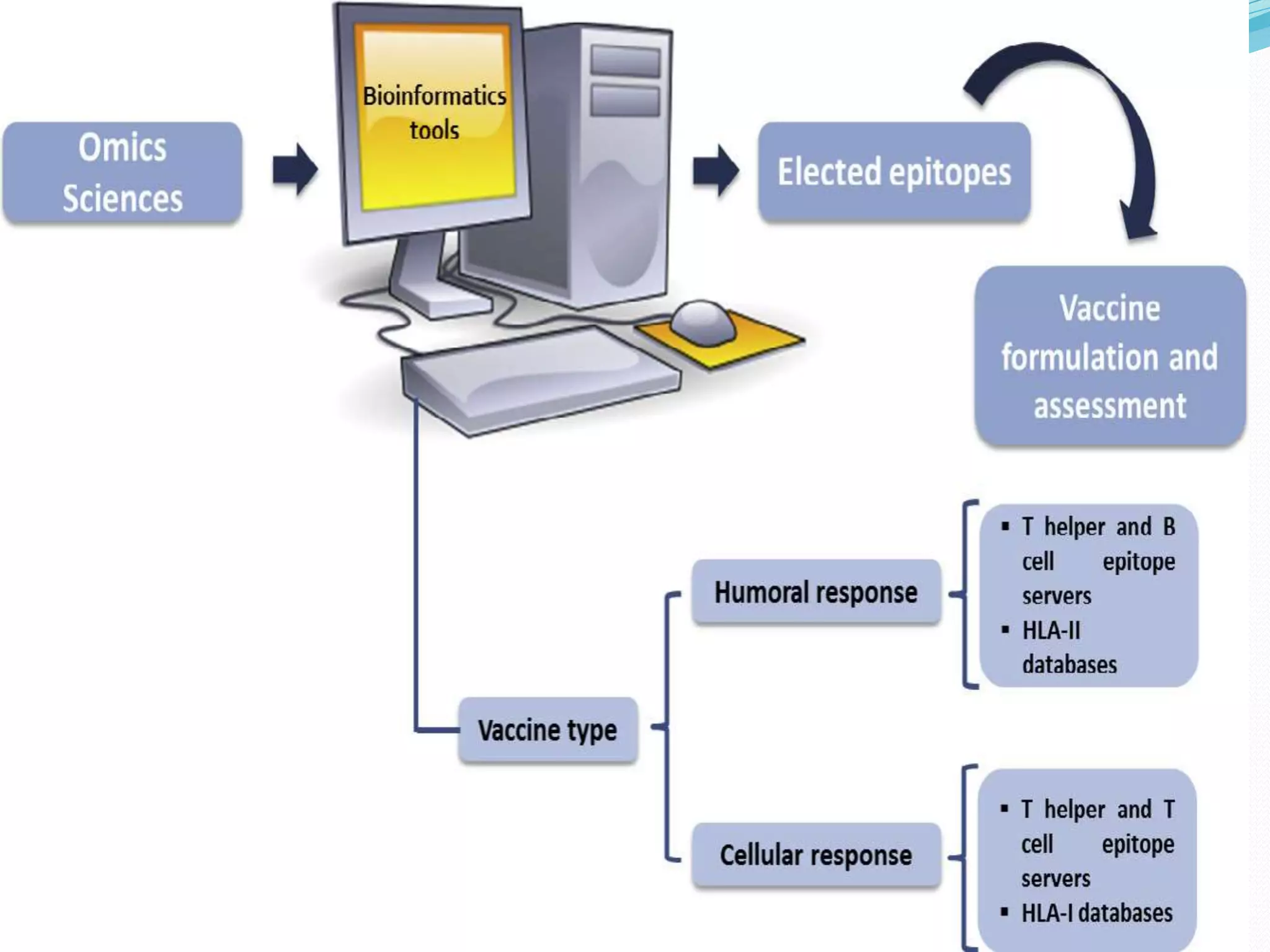
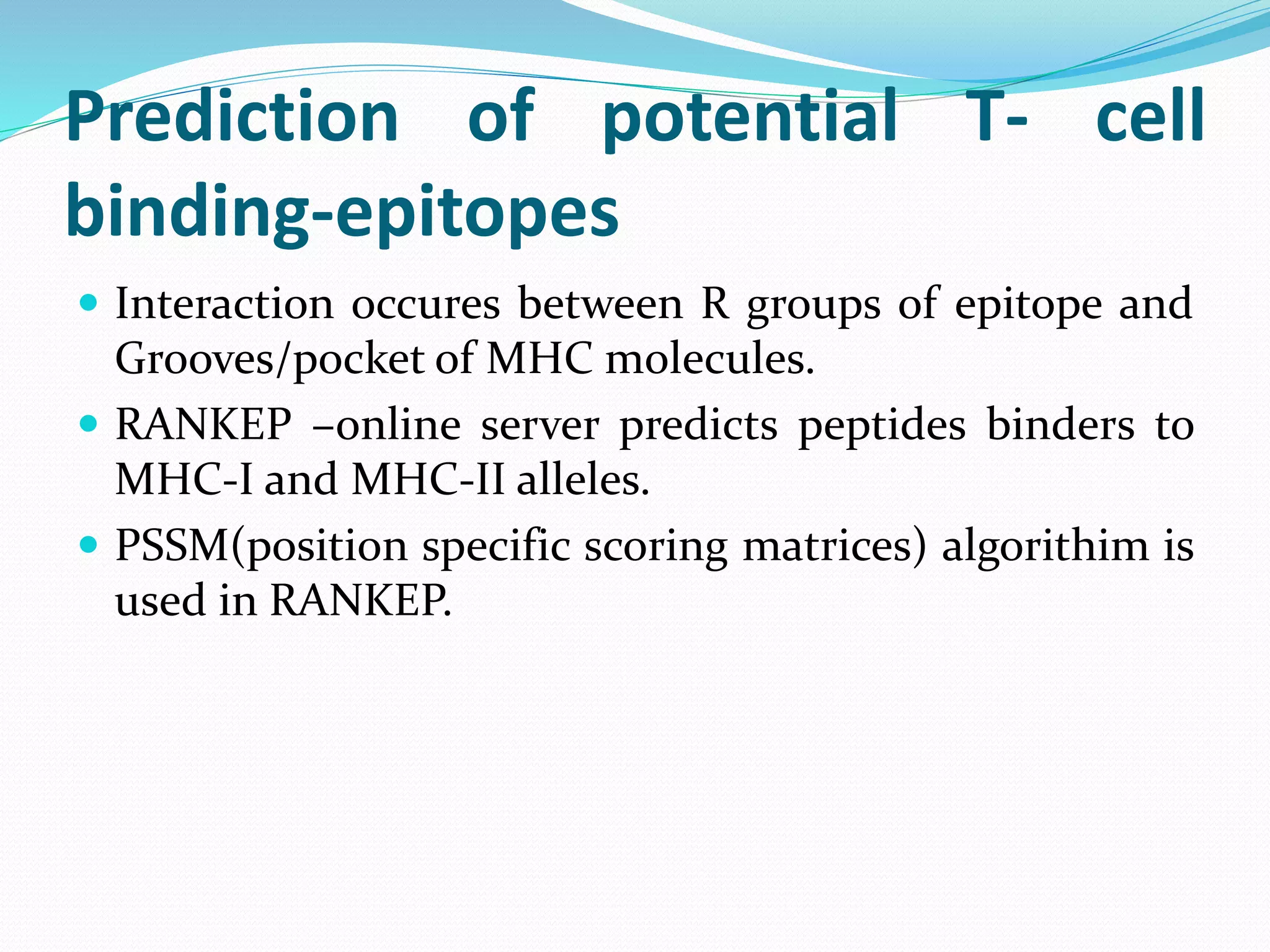
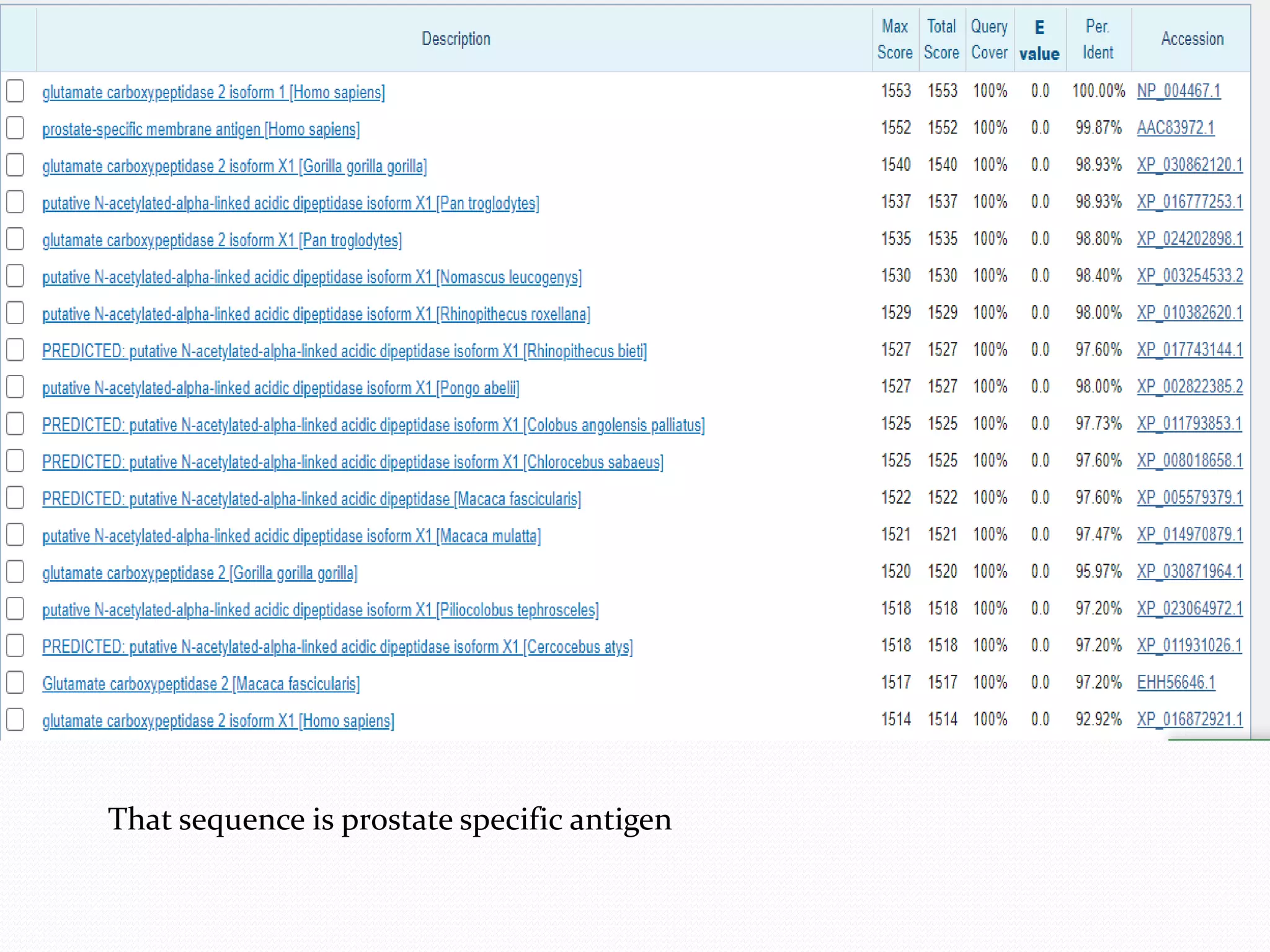
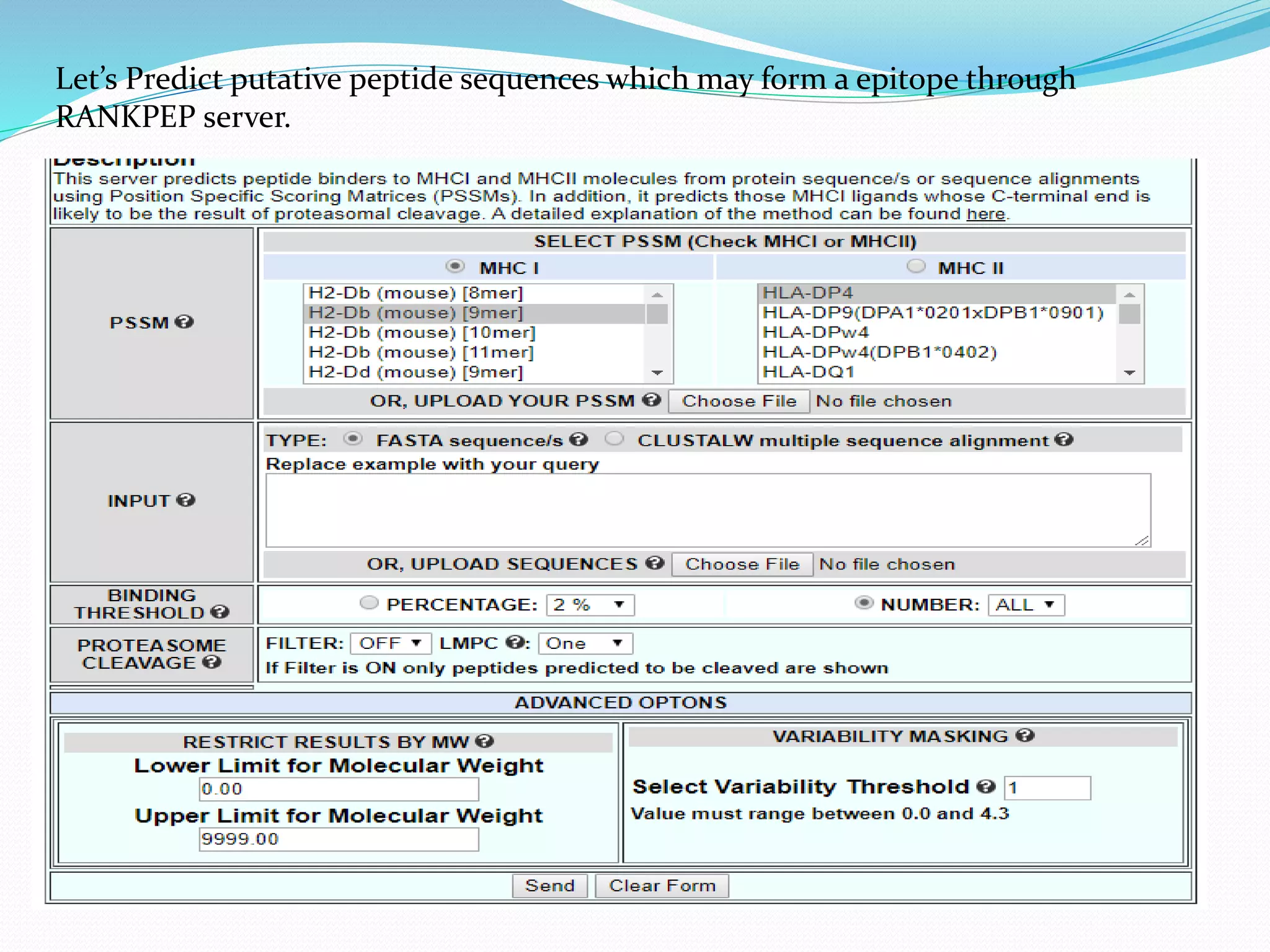
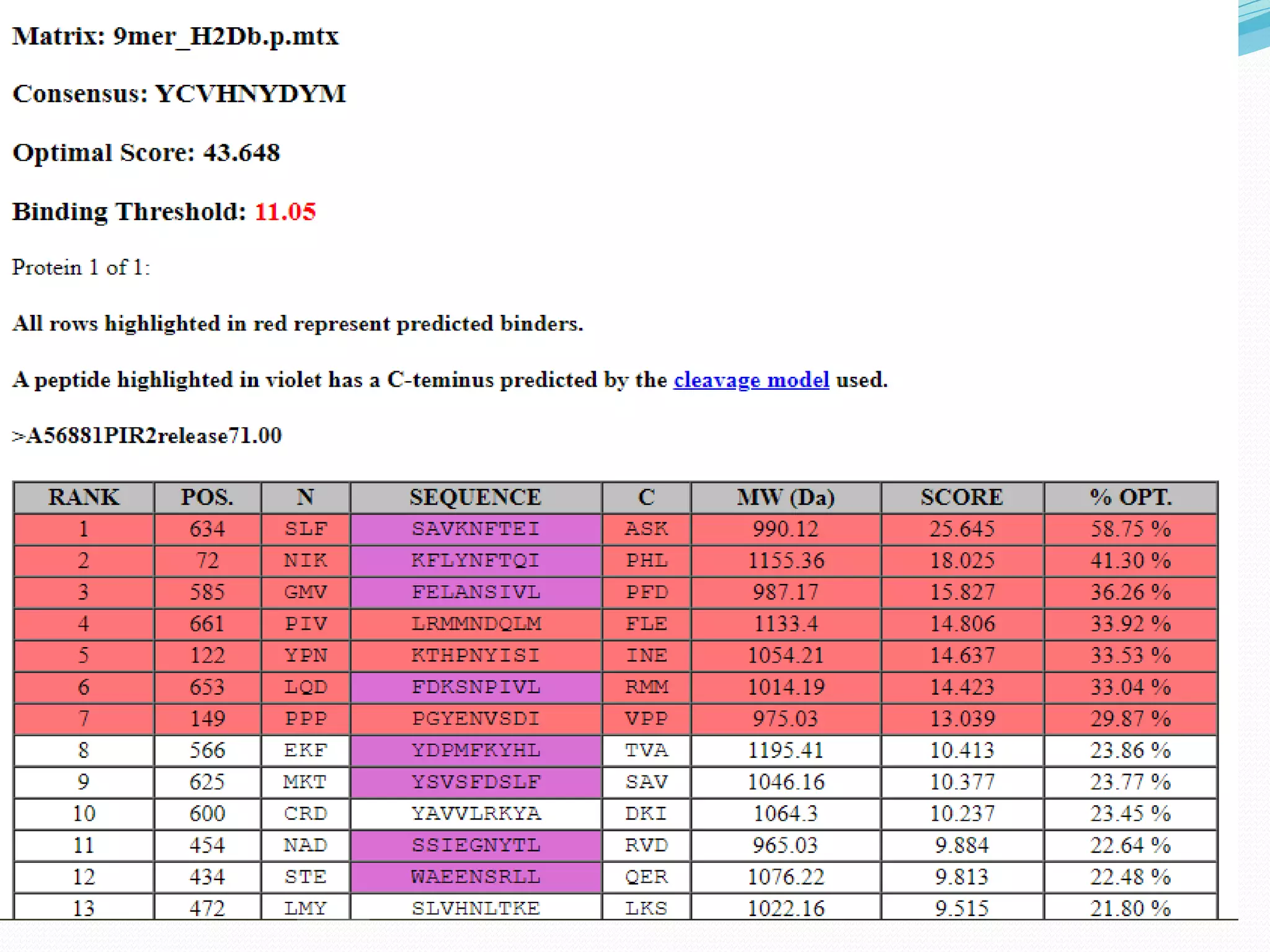
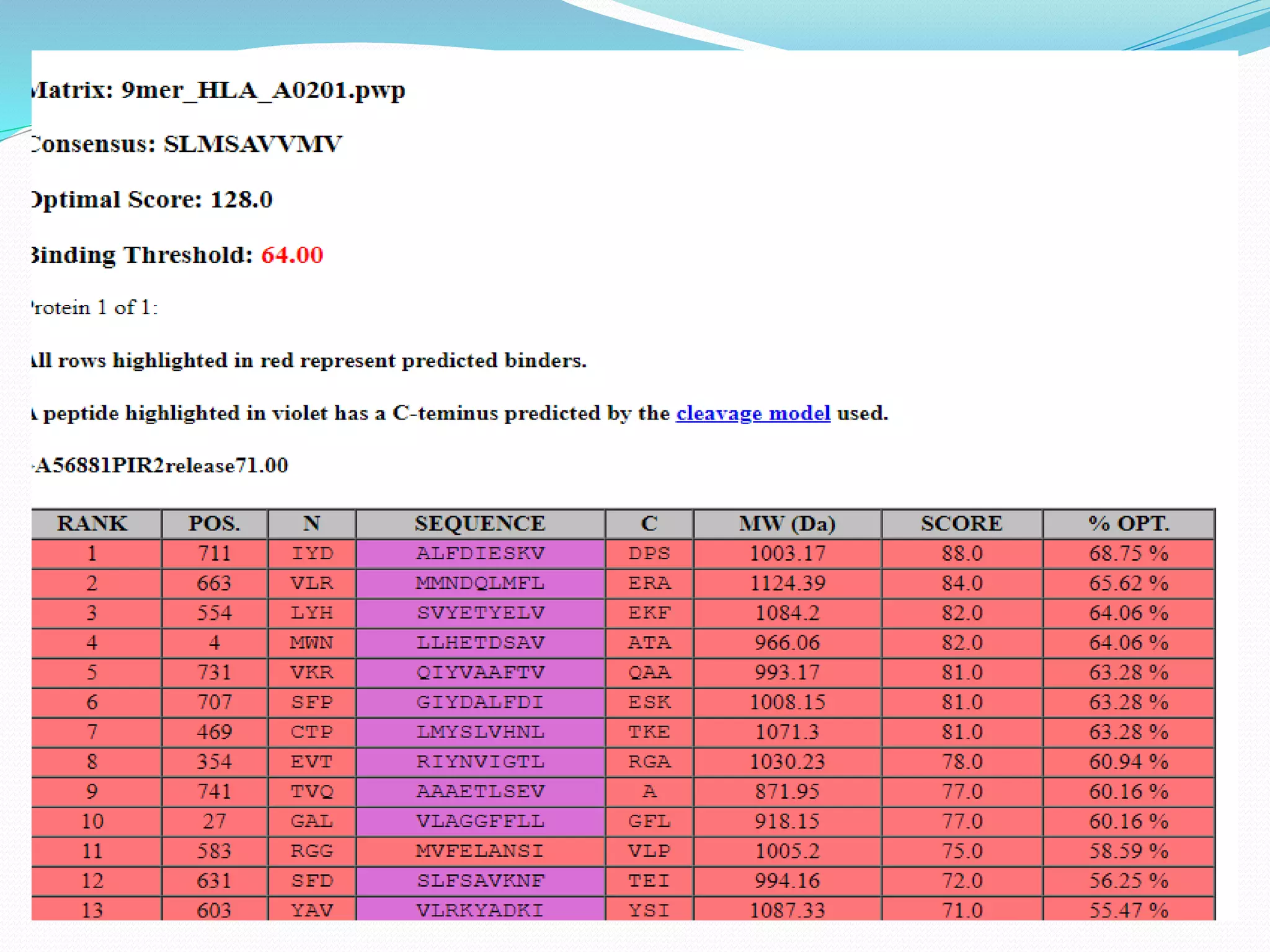
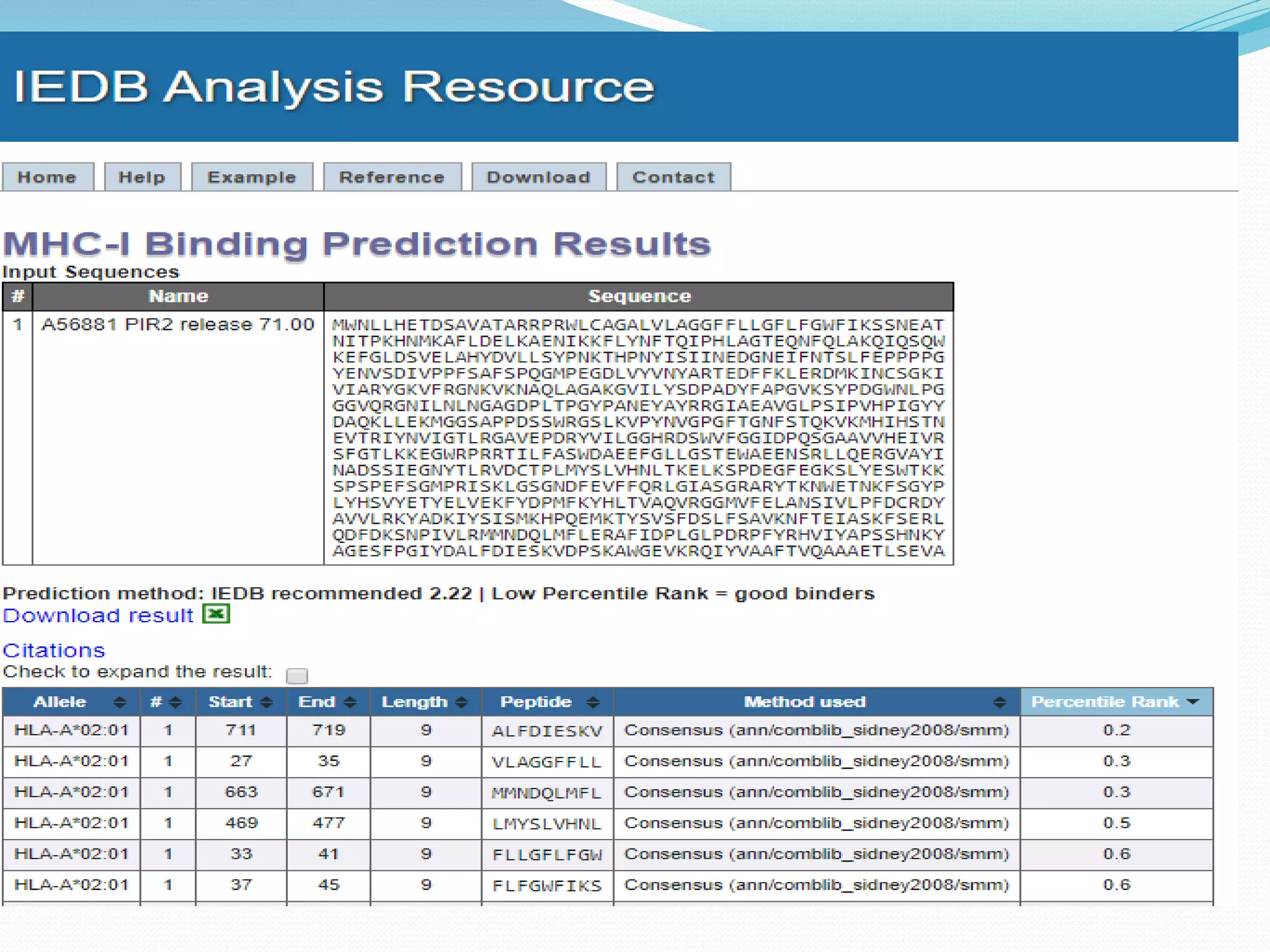
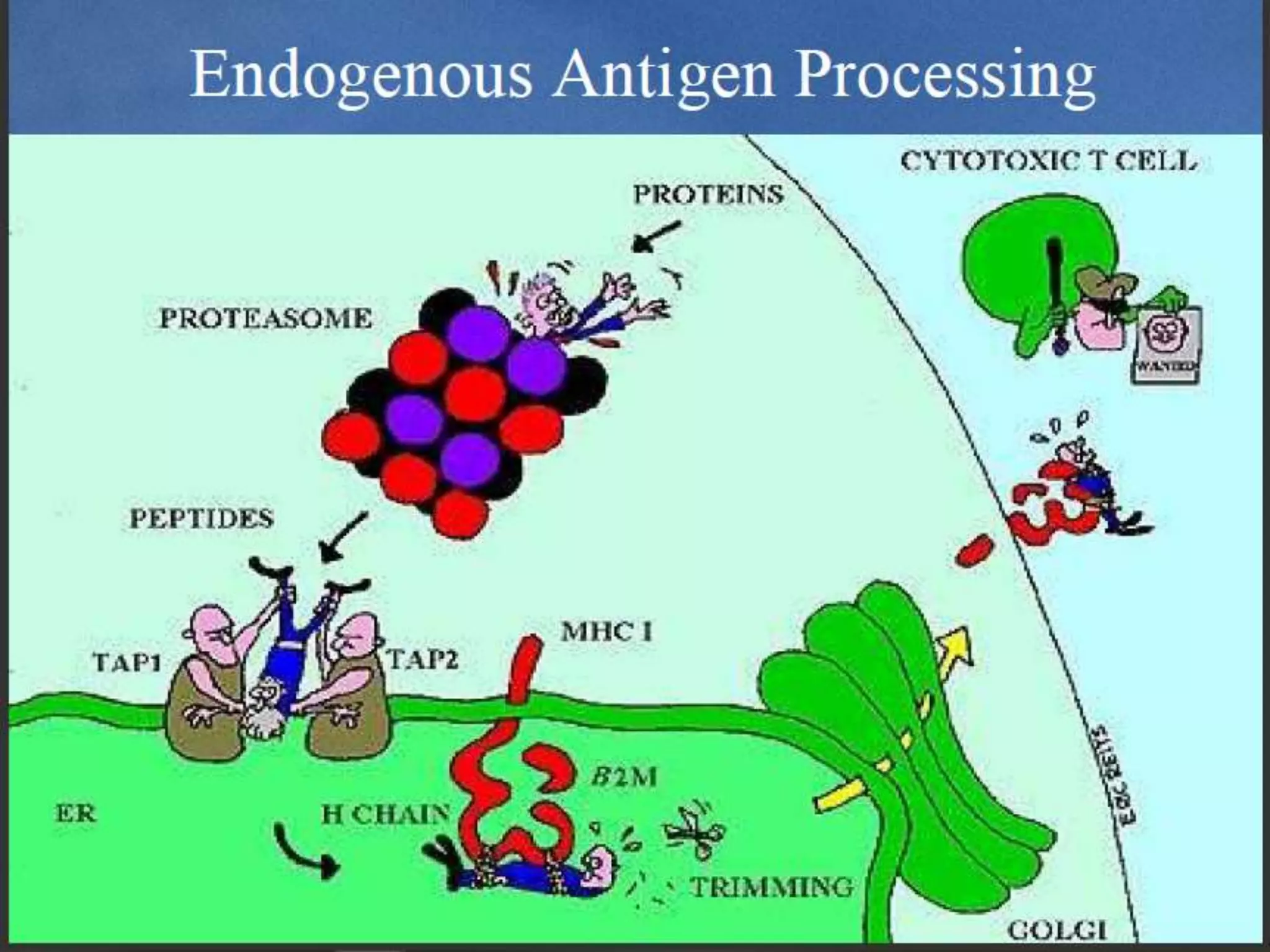
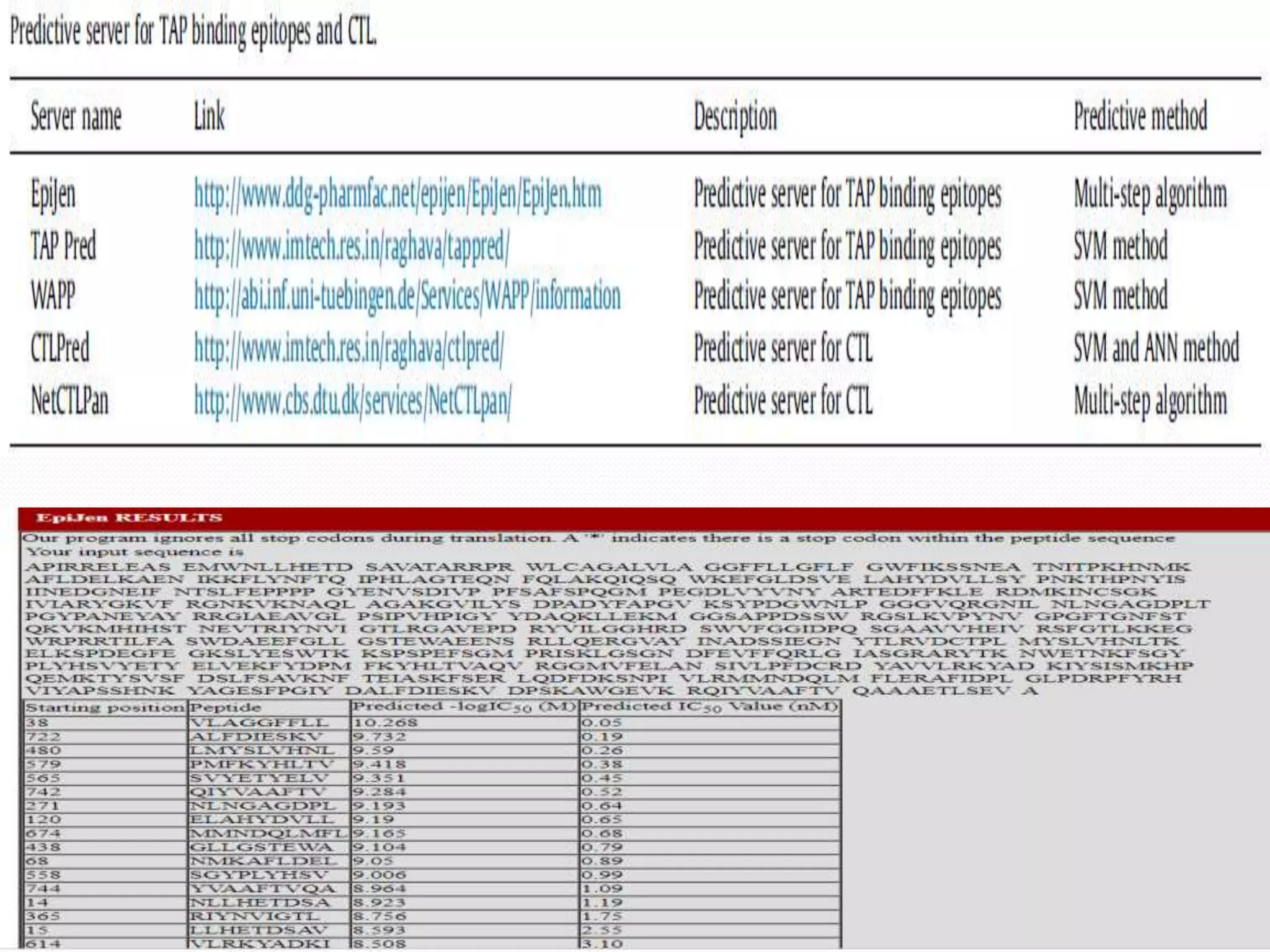
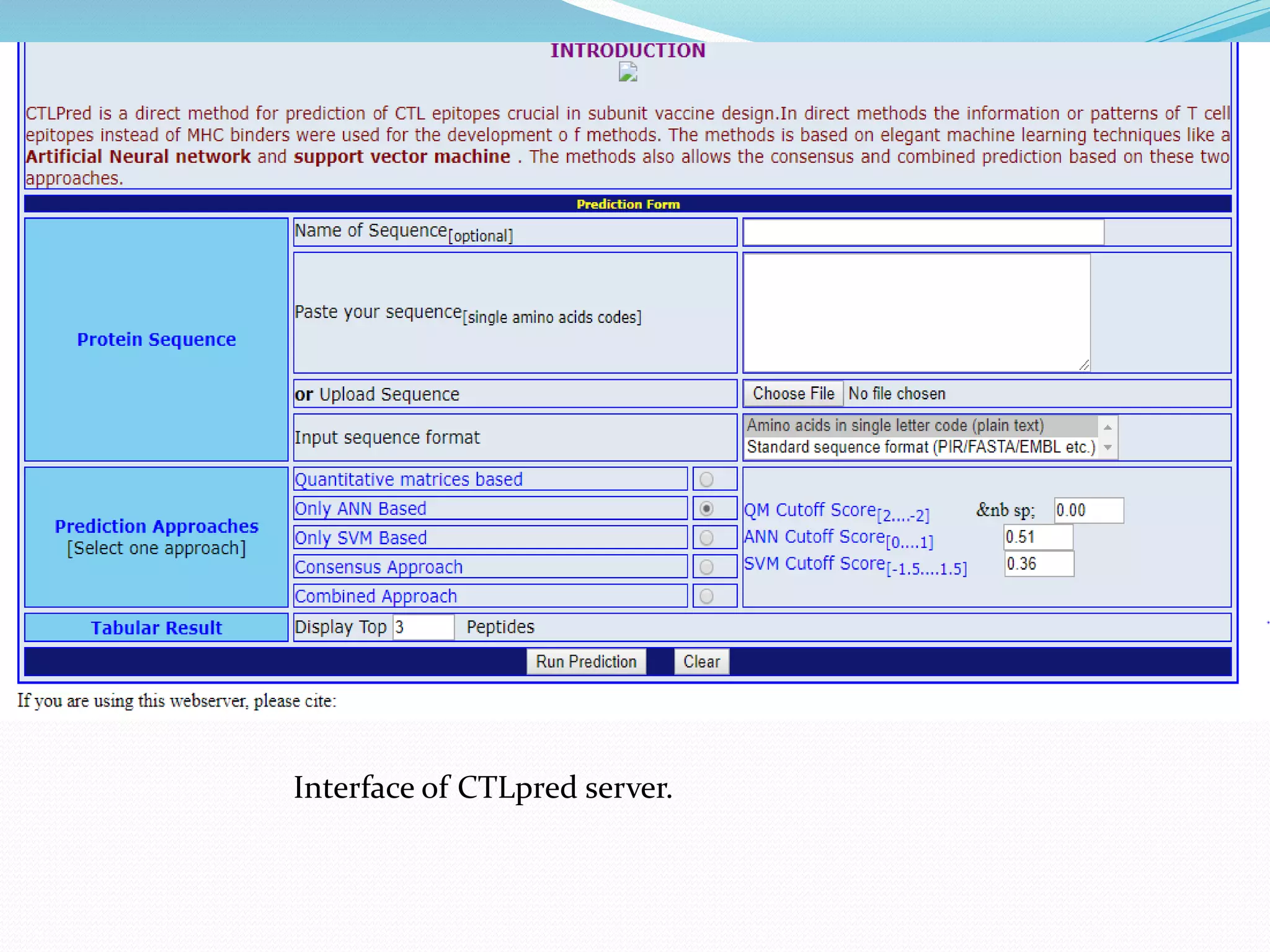
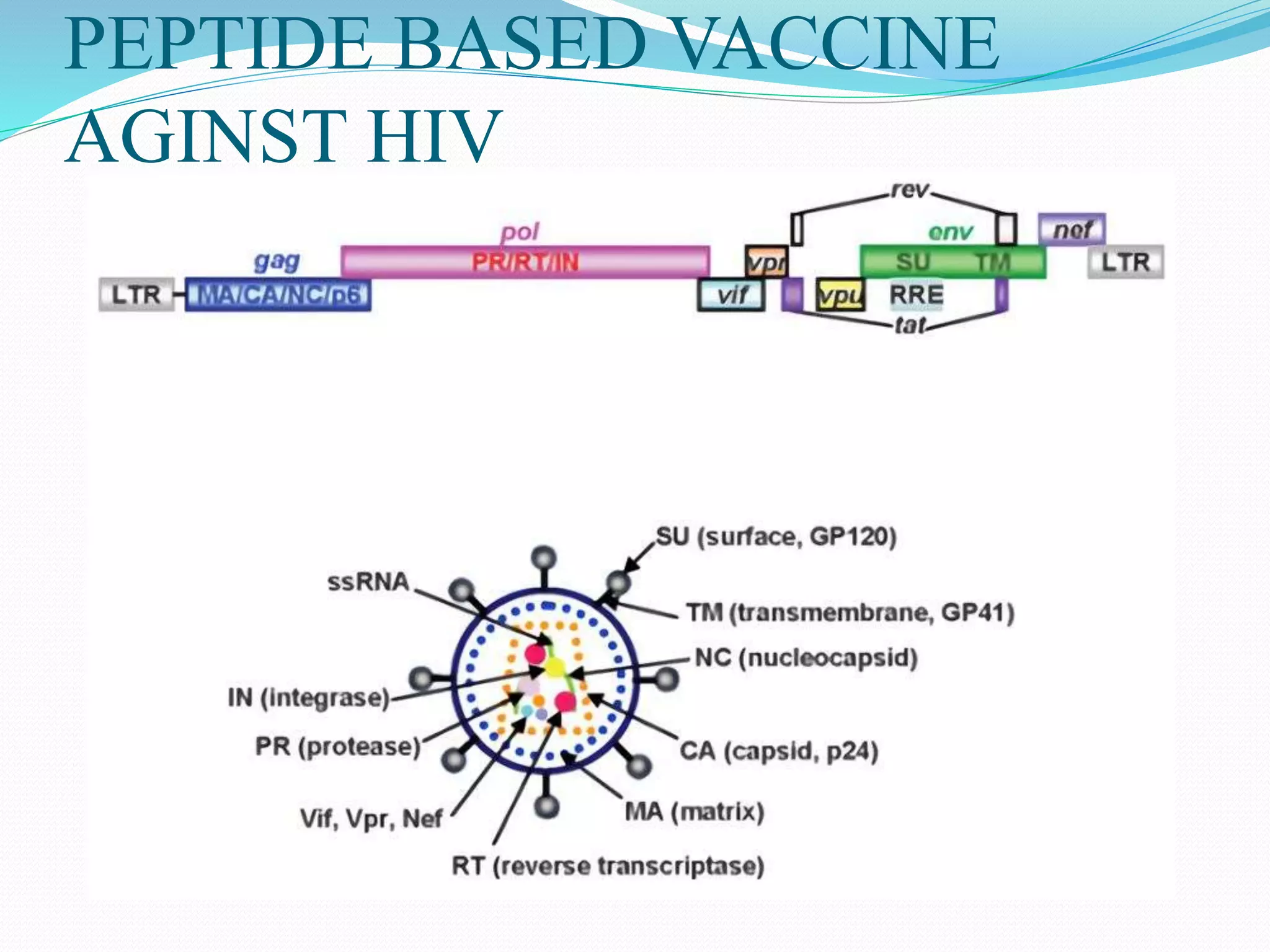
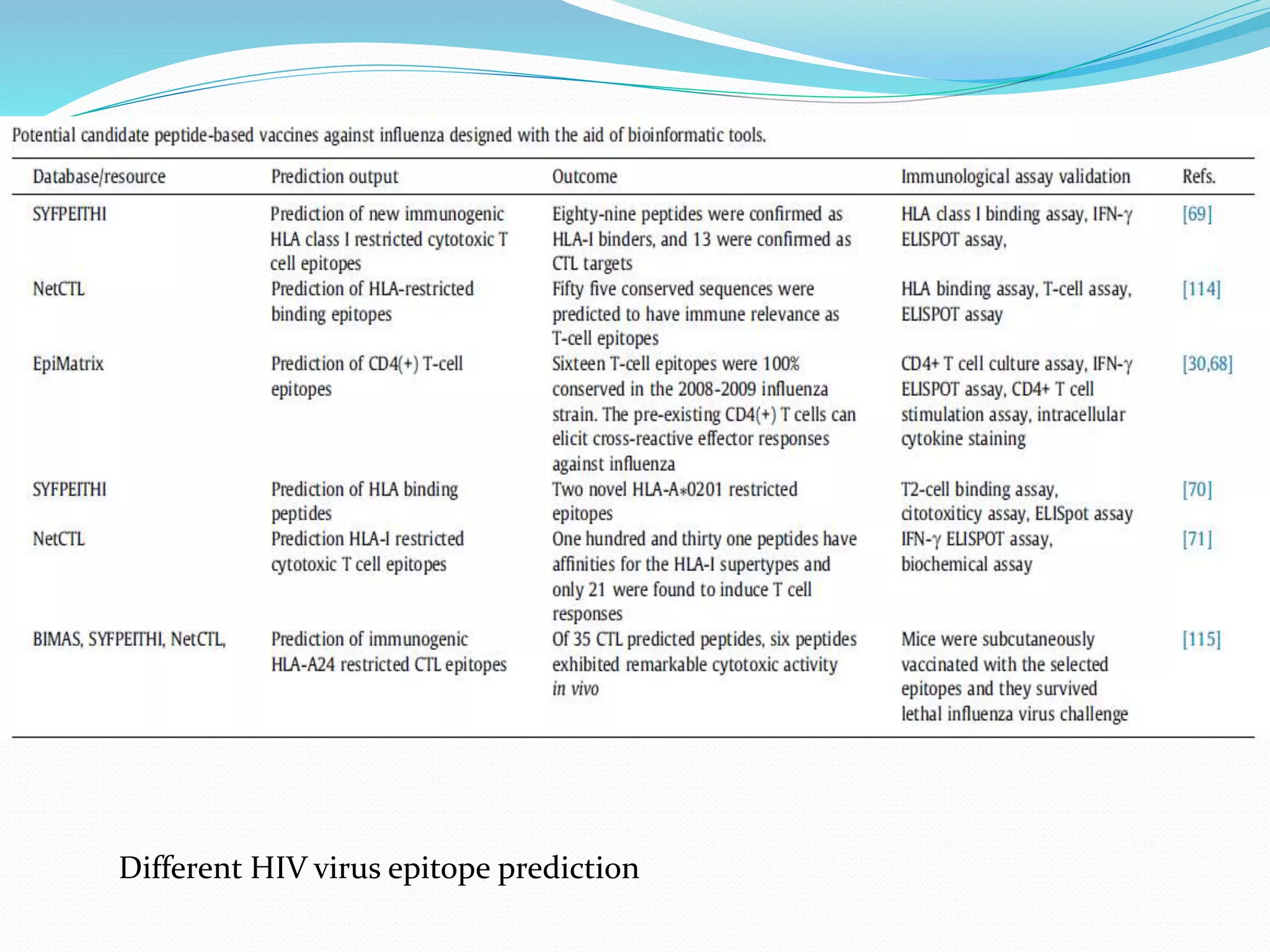
The document discusses epitope mapping, a process for identifying antibody binding sites on antigens, which is vital for vaccine development and understanding antigen-antibody interactions. It outlines various methods for epitope mapping and highlights the importance of using bioinformatics to enhance the efficiency of vaccine production against diseases like influenza and HIV. Additionally, the document describes prediction techniques for T-cell and B-cell binding epitopes, emphasizing their significance in designing effective immunotherapies.