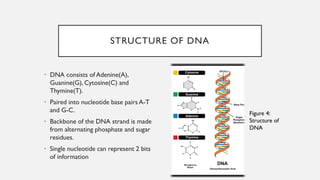
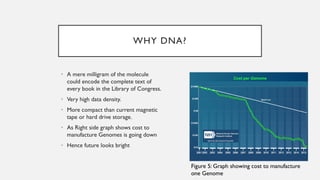
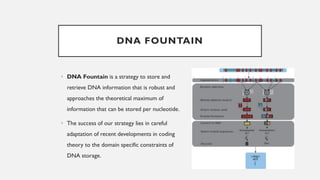
This document discusses DNA digital data storage. It explains that DNA can store vastly more data than current technologies in a smaller space and last much longer. However, writing and reading DNA data is currently much slower and more expensive than modern storage methods. The document outlines how binary data can be converted to DNA nucleotide sequences and provides examples. It also reviews recent developments that aim to improve DNA data storage methods and decrease costs.