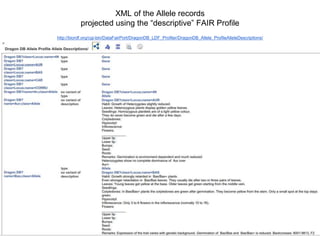
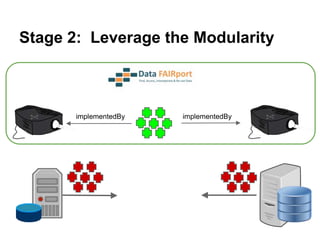
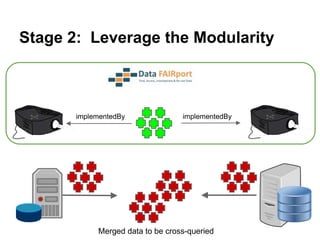
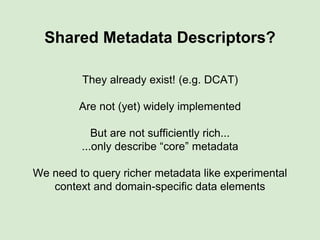
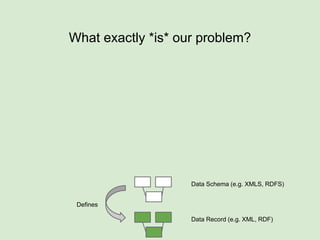
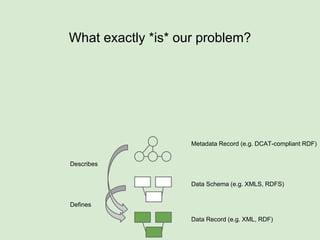
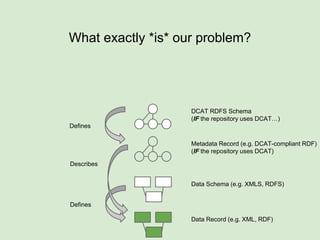
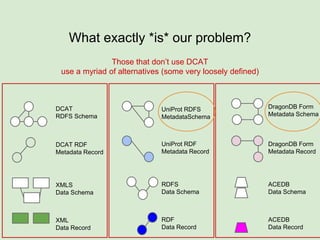
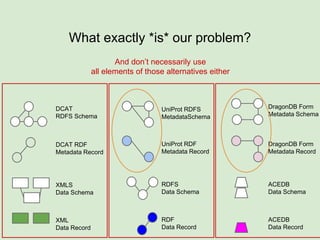
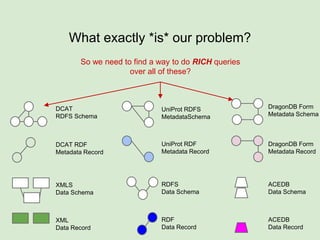
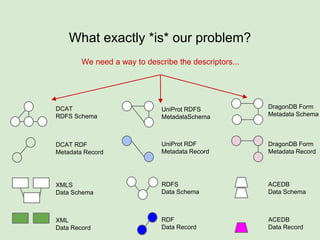
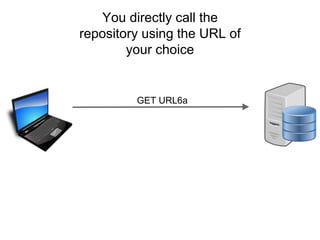
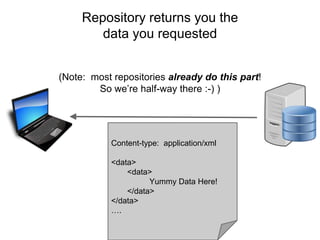
The presentation discusses the Fairport Skunkworks project, which aims to enhance the discovery and interoperability of legacy data repositories through the development of common repository access methods and meta-descriptors. It outlines the challenges faced, including a lack of harmonized metadata structures and diverse data formats, and proposes solutions such as the creation of FAIR profiles for standardized metadata description. Key components involve making data findable, accessible, interoperable, and reusable, ultimately promoting better integration across various repositories.



















![Exemplar use-cases:
● A piece of software that can generate a “sensible”
data submission form for any repository
(at the Force 2015 meeting a few months ago I gave a presentation of a working
example of this… so I won’t repeat that today…)
● A piece of software that can generate a “sensible”
query form/interface for any repository
(demonstration of this today!)
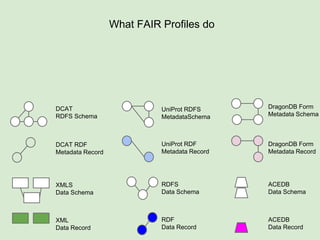
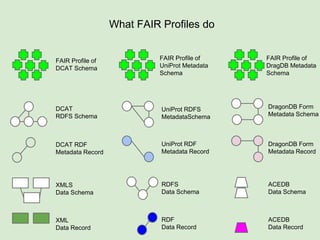
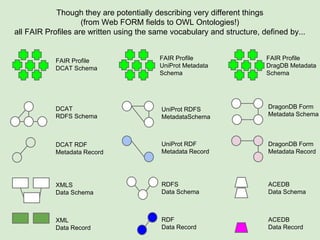
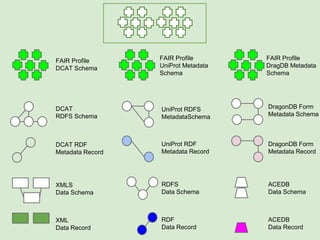
Skunkworks Task #1 - [F]indable
Invent harmonized cross-repository meta-
descriptors](https://image.slidesharecdn.com/presentationoffairprototypetoelsevier-jul102015-150713060133-lva1-app6892/85/Data-FAIRport-Prototype-Demo-Presentation-to-Elsevier-Jul-10-2015-32-320.jpg)







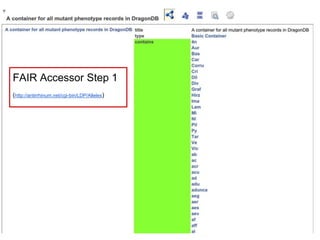
![Skunkworks Task #2 - [A]cessible
Are there already access layer definitions?](https://image.slidesharecdn.com/presentationoffairprototypetoelsevier-jul102015-150713060133-lva1-app6892/85/Data-FAIRport-Prototype-Demo-Presentation-to-Elsevier-Jul-10-2015-53-320.jpg)

![A set of behaviors for providing a unified (albeit simplistic!)
access layer for “records” contained in any Web resource
Skunkworks Task #2 - [A]cessible
Are there already access layer definitions?](https://image.slidesharecdn.com/presentationoffairprototypetoelsevier-jul102015-150713060133-lva1-app6892/85/Data-FAIRport-Prototype-Demo-Presentation-to-Elsevier-Jul-10-2015-55-320.jpg)









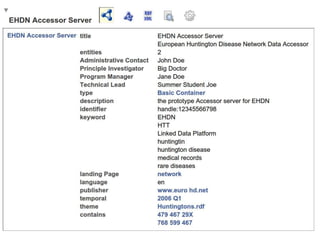
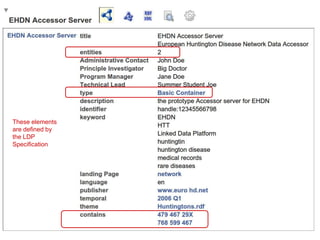
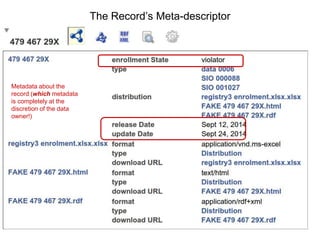
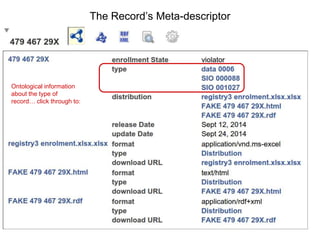
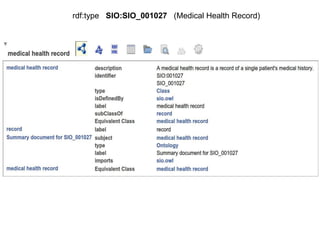
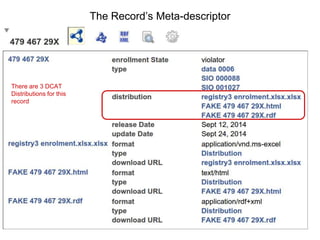




![package EHDN_Accessor; # this should be the same as your filename!
use strict;
use warnings;
use JSON;
use base 'FAIR::Accessor';
my $config = { title => 'European Huntington Disease Network Data Accessor',
serviceTextualDescription => 'Server for some ERDN Data',
textualAccessibilityInfo => "The information from this server requries no authentication", # this could also be a $URI describing the accessibiltiy
mechanizedAccessibilityInfo => "", # this must be a URI to an RDF document
textualLicenseInfo => "CC-BY", # this could also be a URI to the license info
mechanizedLicenseInfo => "", # this must be a URI to an RDF document
baseURI => "", # I don't know what this is used for yet, but I have a feeling I will need it!
ETAG_Base => "EHDN_Accessor_For_RegInfo",
localNamespaces => {ehdn => 'http://ehdn.org/some/items/', ehdnpred => 'http://ehdn.org/some/predicates/'},
localMetadataElements => [qw(erdnpred:fromHospital erdnpred:lastevaluatedDate) ], };
my $service = EHDN_Accessor->new(%$config);
$service->handle_requests;
sub get_all_meta_URIs {
my ($starting_at_record, $path_info) = @_;
$path_info ||="";
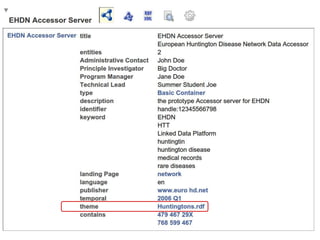
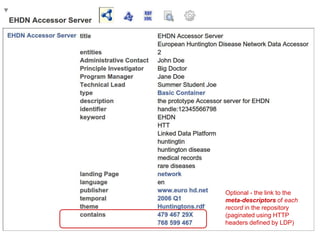
my %result = ( 'dc:title' => "EHDN Accessor Server",
'dcat:description' => "the prototype Accessor server for EHDN",
'dcat:identifier' => "handle:12345566798",
'dcat:keyword' => ["medical records", "rare diseases", "EHDN", "Linked Data Platform", 'HTT', 'huntington'],
'dcat:landingPage' => 'http://www.euro-hd.net/html/network',
'dcat:language' => 'en',
'dcat:publisher' => 'http://www.euro-hd.net',
'dcat:temporal' => 'http://reference.data.gov.uk/id/quarter/2006-Q1',
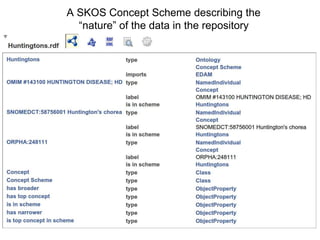
'dcat:theme' => 'http://biordf.org/DataFairPort/ConceptSchemes/Huntingtons.rdf',
'daml:has-Technical-Lead' => "Summer Student Joe",
'daml:has-Administrative-Contact' => "John Doe",
'daml:has-Program-Manager' => "Jane Doe",
'daml:has-Principle-Investigator' => "Big Doctor", );
my $BASE_URL = "http://" . $ENV{'SERVER_NAME'} . $ENV{'REQUEST_URI'} . $path_info;
my @known_records = ($BASE_URL . "/479-467-29X", $BASE_URL . "/768-599-467", );
$result{'void:entities'} = scalar(@known_records); # THE TOTAL *NUMBER* OF RECORDS THAT CAN BE SERVED
$result{'ldp:contains'} = @known_records; # the listref of record ids
return encode_json(%result);
}
sub get_distribution_URIs {
my ($self, $ID, $PATH_INFO) = @_;
my (%response, %formats, %metadata);
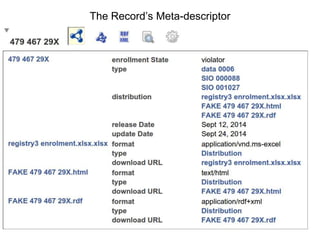
$formats{'text/html'} = 'http://myserver.org/ThisScript/record/479-467-29X.html';
$formats{'application/rdf+xml'} = 'http://myserver.org/ThisScript/record/479-467-29X.rdf';
$metadata{'rdf:type'} = ['edam:data_0006', 'sio:SIO_000088'];
extractDataFromSpreadsheet(%metadata, $ID);
$response{distributions} = %formats;
$response{metadata} = %metadata if (keys %metadata); # only set it if you can provided something
my $response = encode_json(%response);
return $response;
}
sub extractDataFromSpreadsheet{
my ($metadata, $ID) = @_;
use Spreadsheet::XLSX::Reader::LibXML;
my $db_file = "registry3-enrolment.xlsx.xlsx";
my $excel = Spreadsheet::XLSX::Reader::LibXML->new();
my $workbook = $excel->parse($db_file);
my ($sheet) = $workbook->worksheets;
my ($first, $last) = $sheet->row_range;
foreach my $row ($first .. $last) {
next unless ($sheet->get_cell($row, 0)->value eq $ID);
my $cell = $sheet->get_cell($row, 5);
$metadata->{'dcat:updateDate'} = $cell->value;
$cell = $sheet->get_cell($row, 1);
$metadata->{'dcat:releaseDate'} = $cell->value;
$cell = $sheet->get_cell($row, 3);
This is the only code that a provider
must implement… and much (almost
half!) of it is just tag/value definitions
If they don’t want to implement the full
set of drill-down behaviors then the
code is even smaller!
(This is the actual code - 68 lines -
running the demo you just saw. Most
of the heavy-lifting is handled by the
libraries I published yesterday)](https://image.slidesharecdn.com/presentationoffairprototypetoelsevier-jul102015-150713060133-lva1-app6892/85/Data-FAIRport-Prototype-Demo-Presentation-to-Elsevier-Jul-10-2015-84-320.jpg)
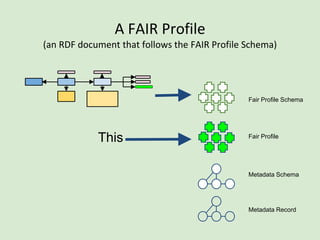
![Skunkworks Task #3 - [I]nteroperable
This is “the holy grail”!!](https://image.slidesharecdn.com/presentationoffairprototypetoelsevier-jul102015-150713060133-lva1-app6892/85/Data-FAIRport-Prototype-Demo-Presentation-to-Elsevier-Jul-10-2015-85-320.jpg)
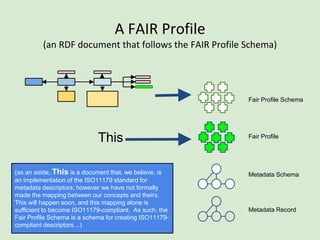
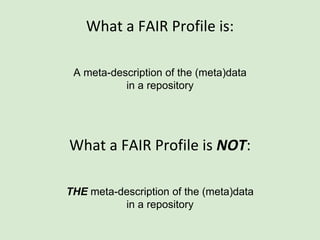
![Skunkworks Task #3 - [I]nteroperable
This is “the holy grail”!!
This is where the FAIR Profile reveals its utility
“what it IS” vs. “what it IS NOT”](https://image.slidesharecdn.com/presentationoffairprototypetoelsevier-jul102015-150713060133-lva1-app6892/85/Data-FAIRport-Prototype-Demo-Presentation-to-Elsevier-Jul-10-2015-86-320.jpg)

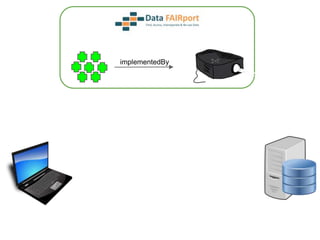
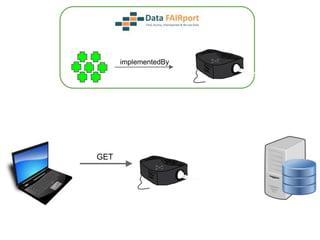
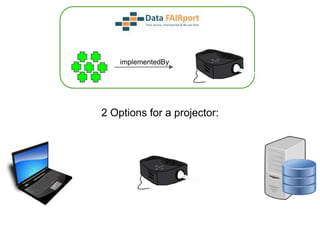
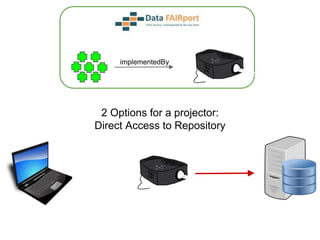
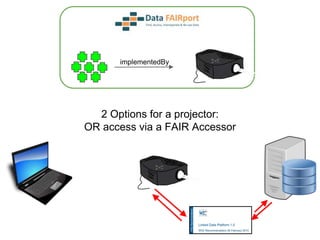
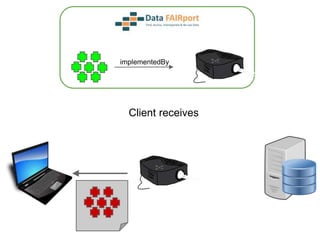
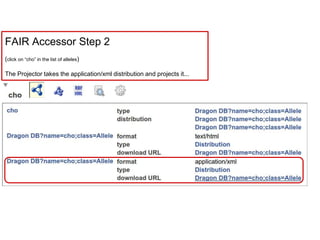
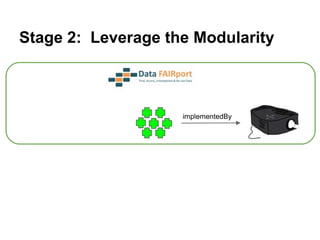
![Skunkworks Task #3 - [I]nteroperable
“FAIR Projectors”
A FAIR Projector is a (potentially) small, modular,
reusable Web based service that “projects” data
from a repository into the format
described by a FAIR Profile](https://image.slidesharecdn.com/presentationoffairprototypetoelsevier-jul102015-150713060133-lva1-app6892/85/Data-FAIRport-Prototype-Demo-Presentation-to-Elsevier-Jul-10-2015-88-320.jpg)
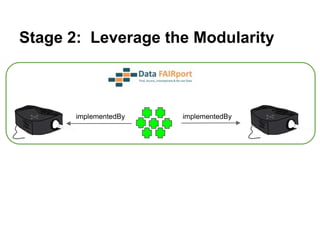
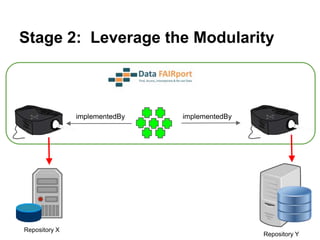
![Skunkworks Task #3 - [I]nteroperable
“FAIR Projectors”
A FAIR Projector is a (potentially) small, modular,
reusable Web based service that “projects” data
from a repository into the format
described by a FAIR Profile
http://linkeddatafragments.org/](https://image.slidesharecdn.com/presentationoffairprototypetoelsevier-jul102015-150713060133-lva1-app6892/85/Data-FAIRport-Prototype-Demo-Presentation-to-Elsevier-Jul-10-2015-89-320.jpg)