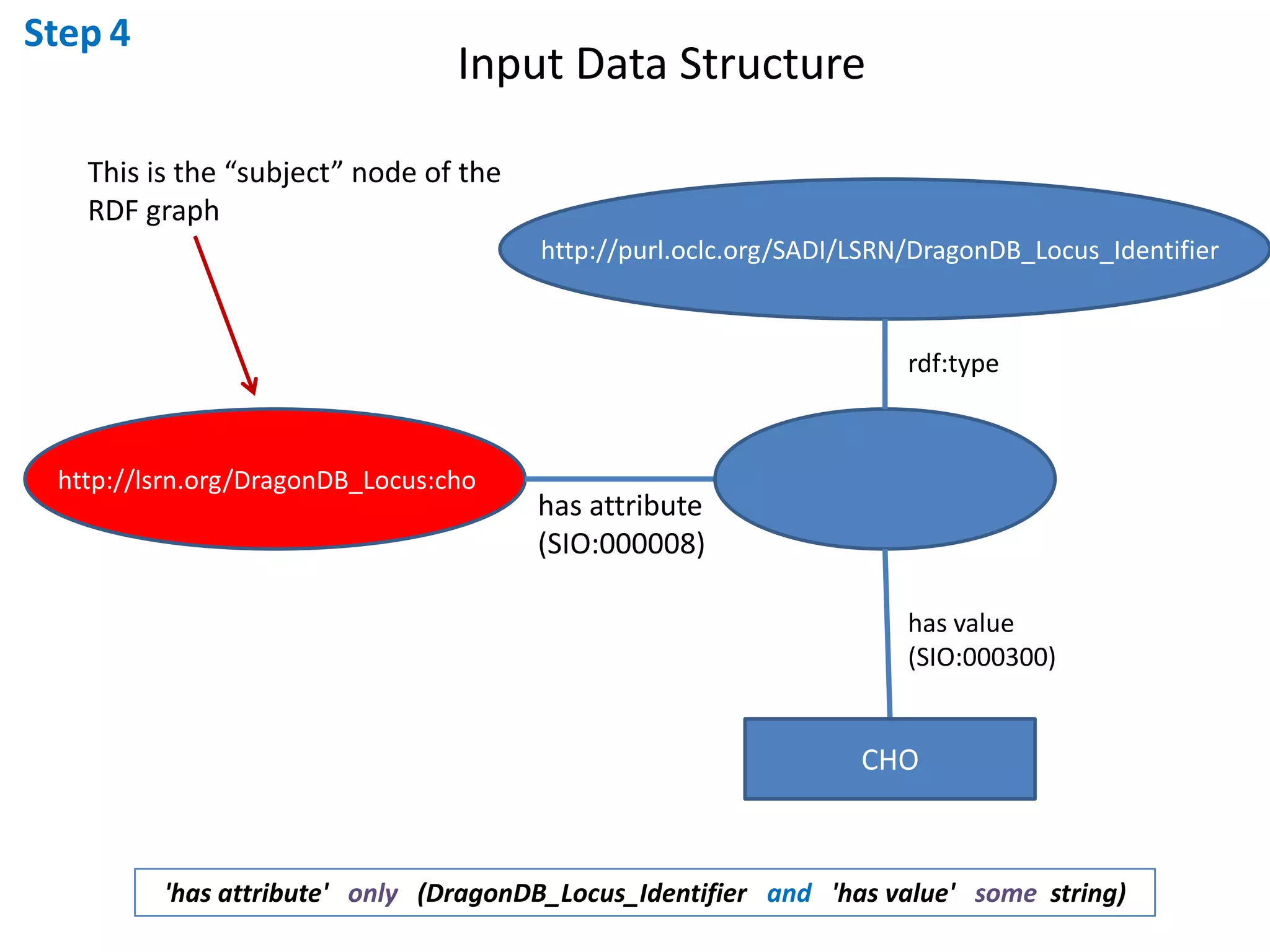
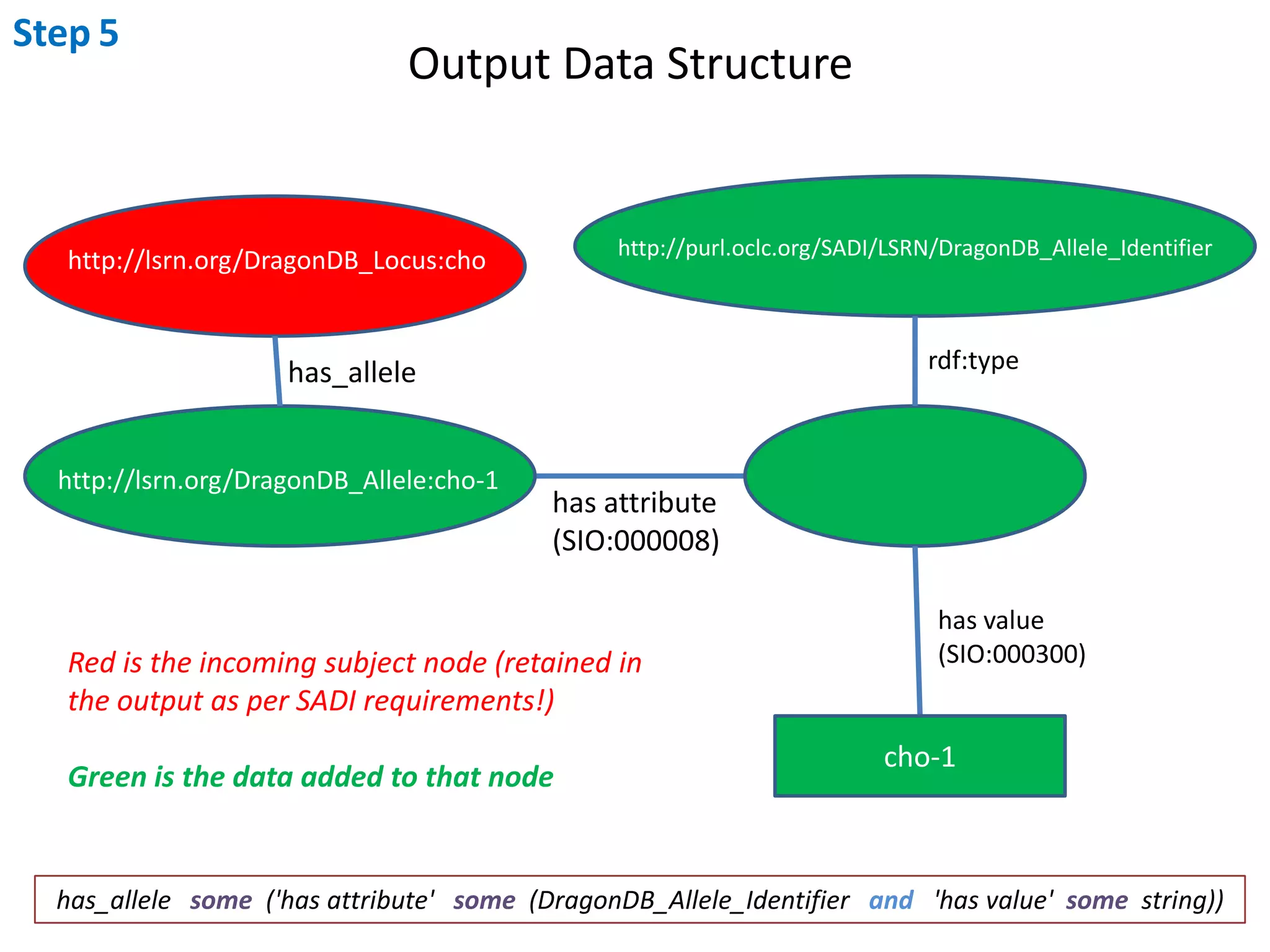
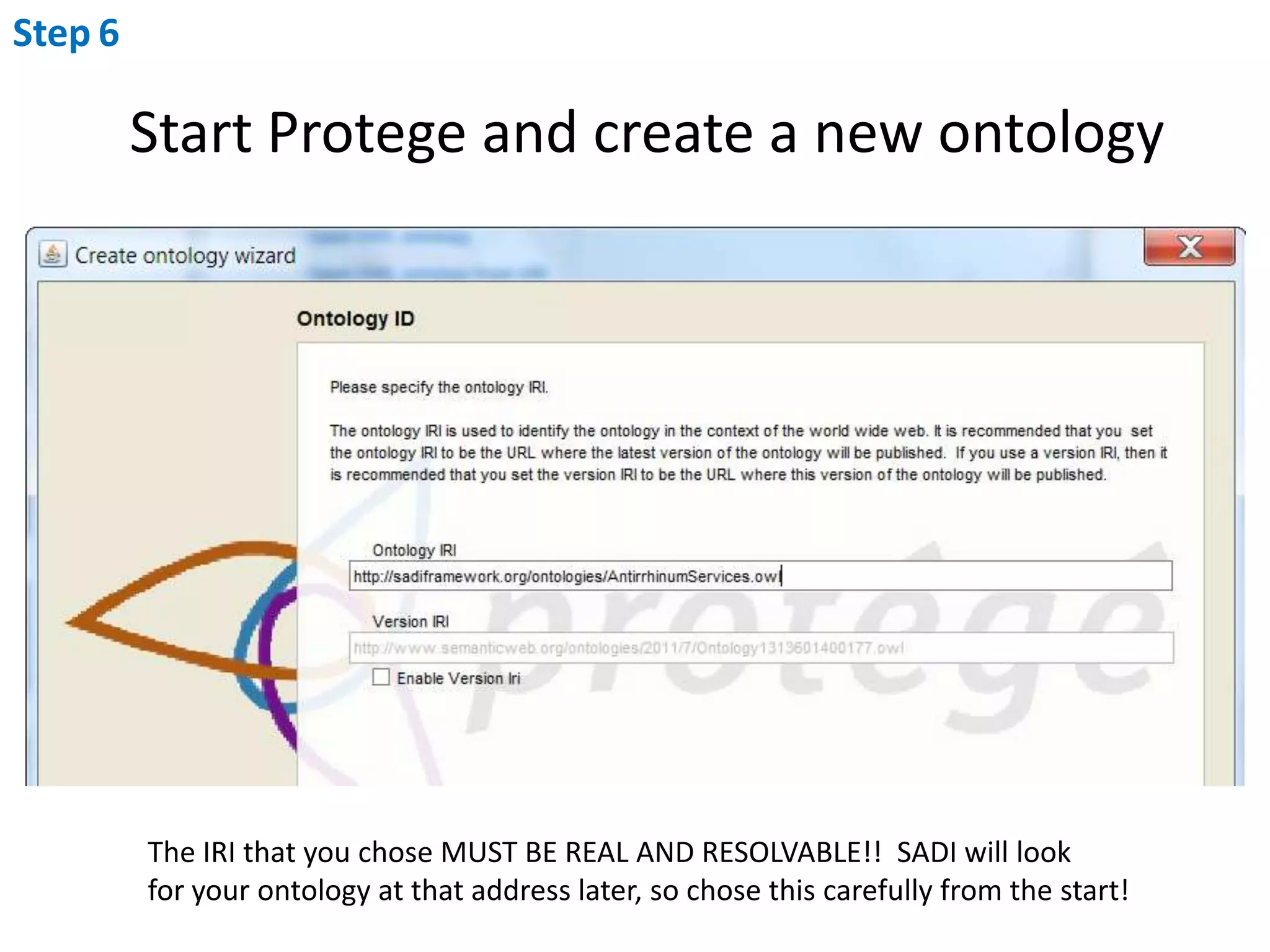
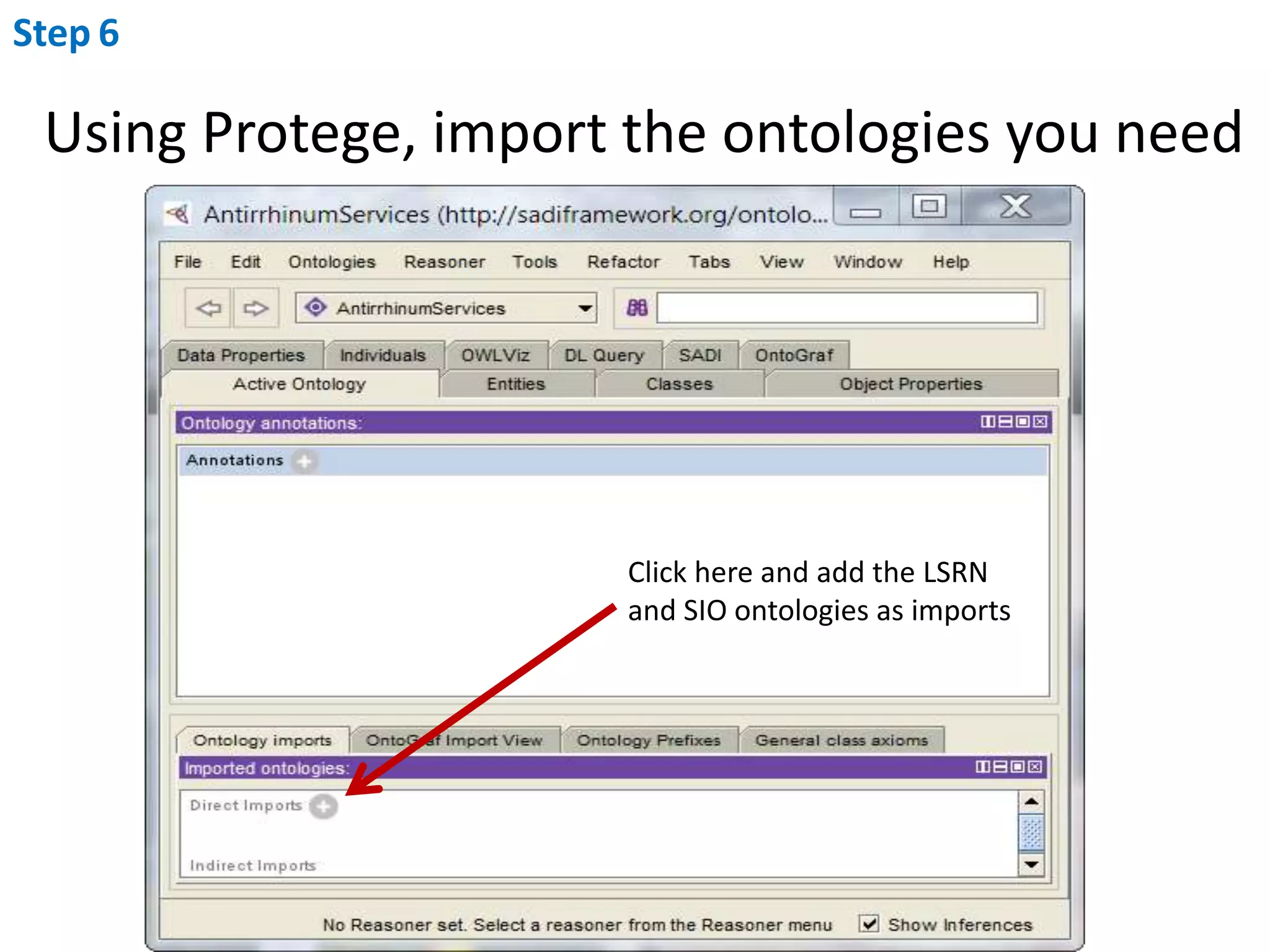
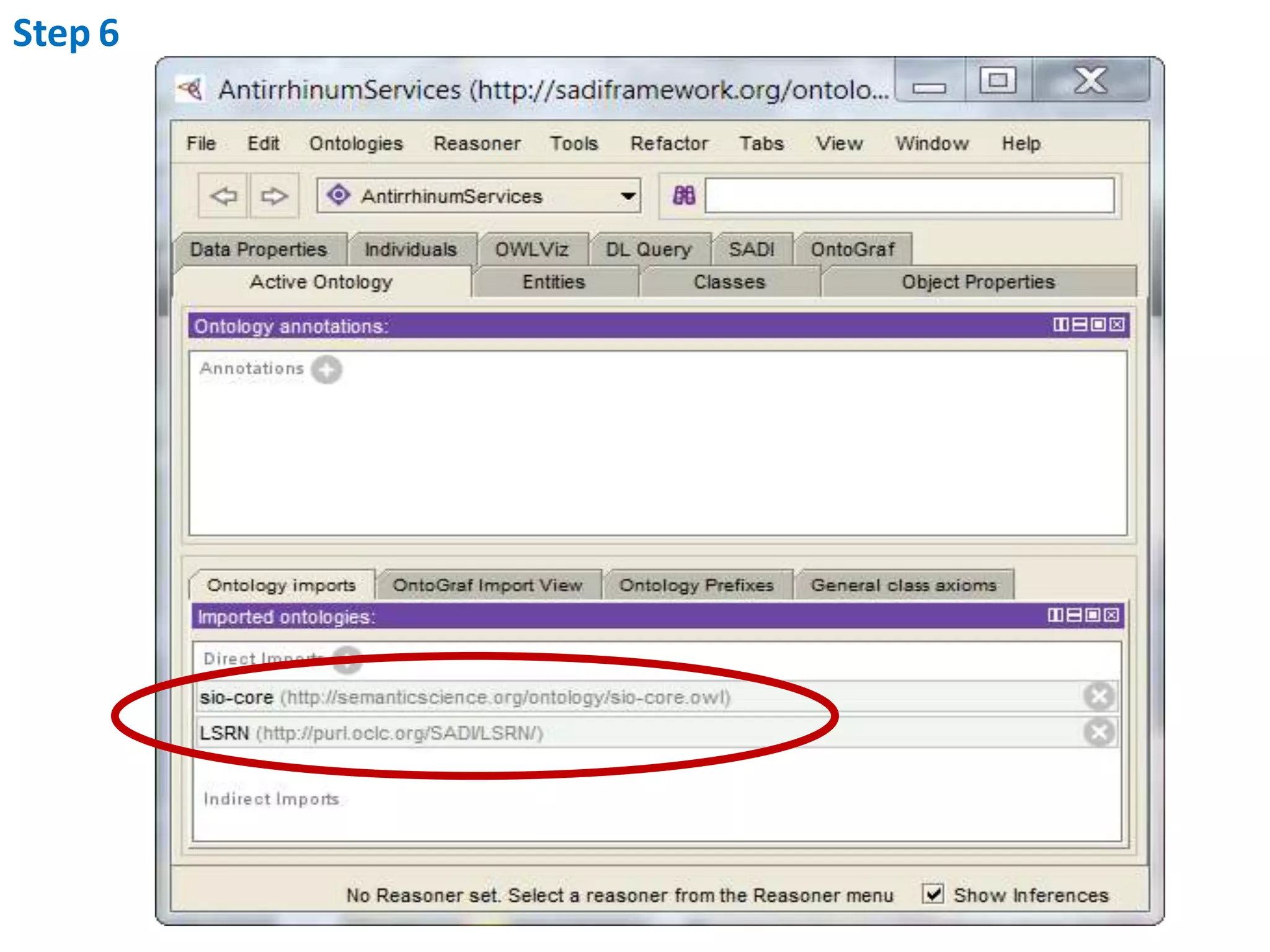
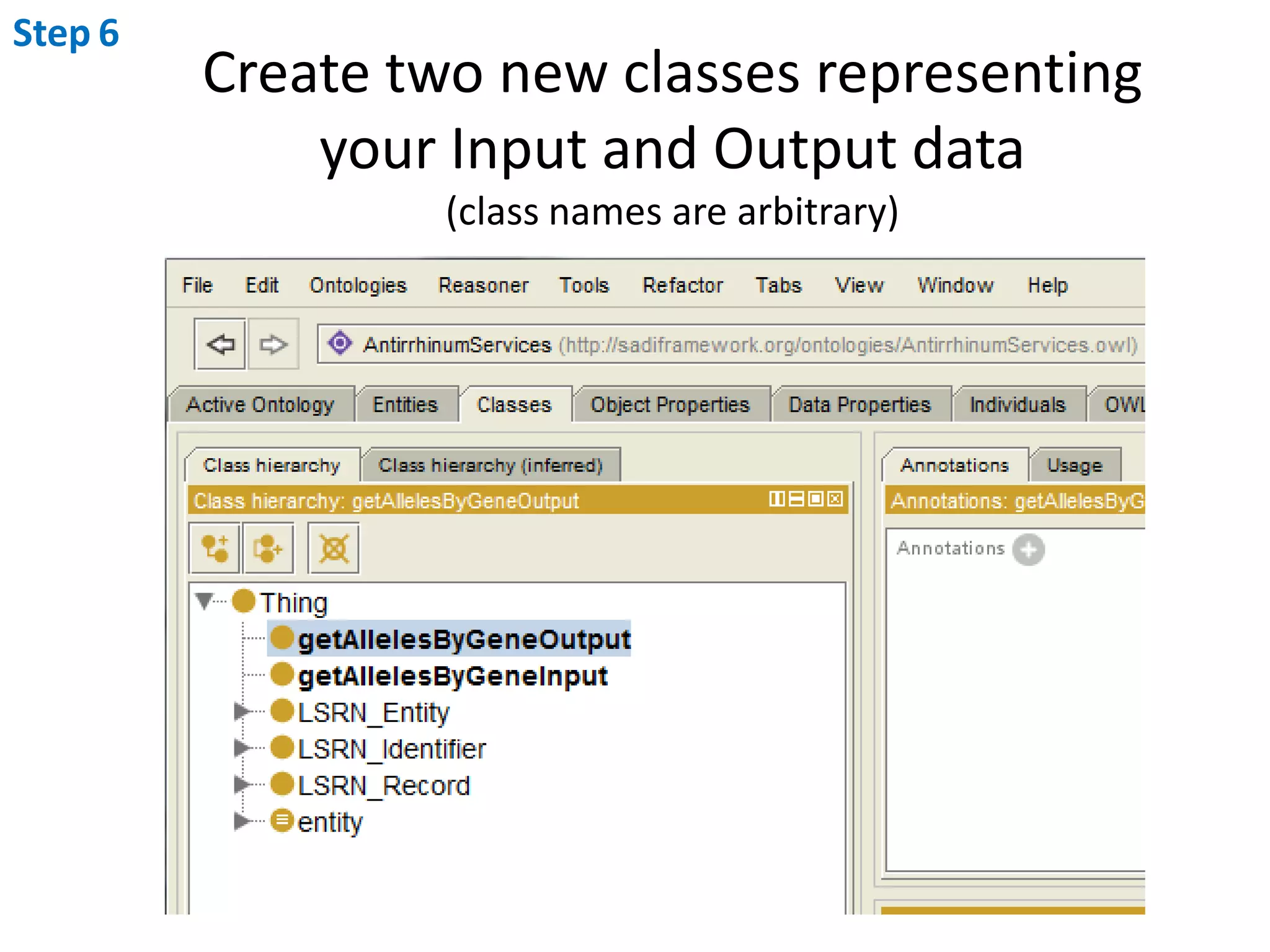
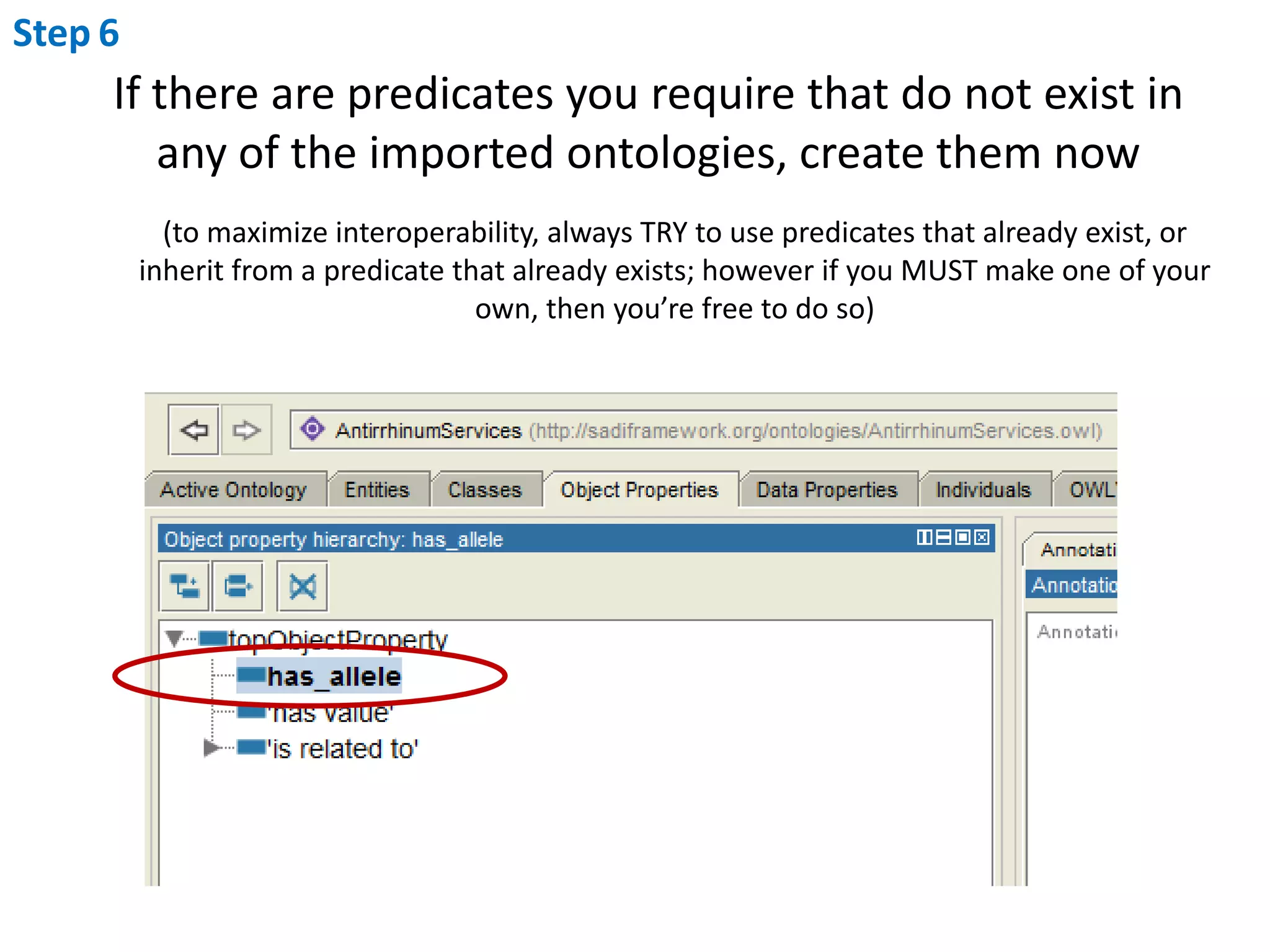
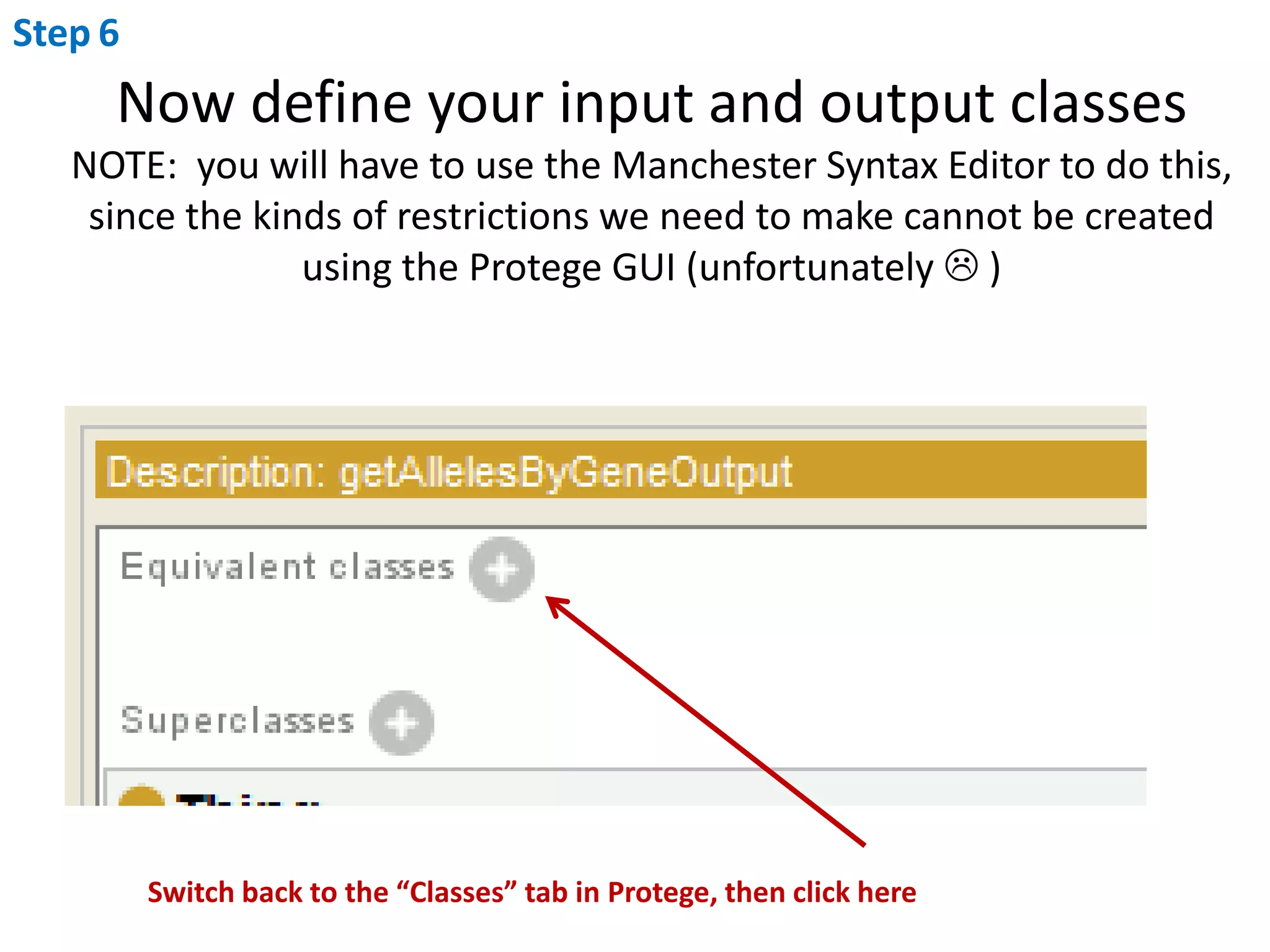
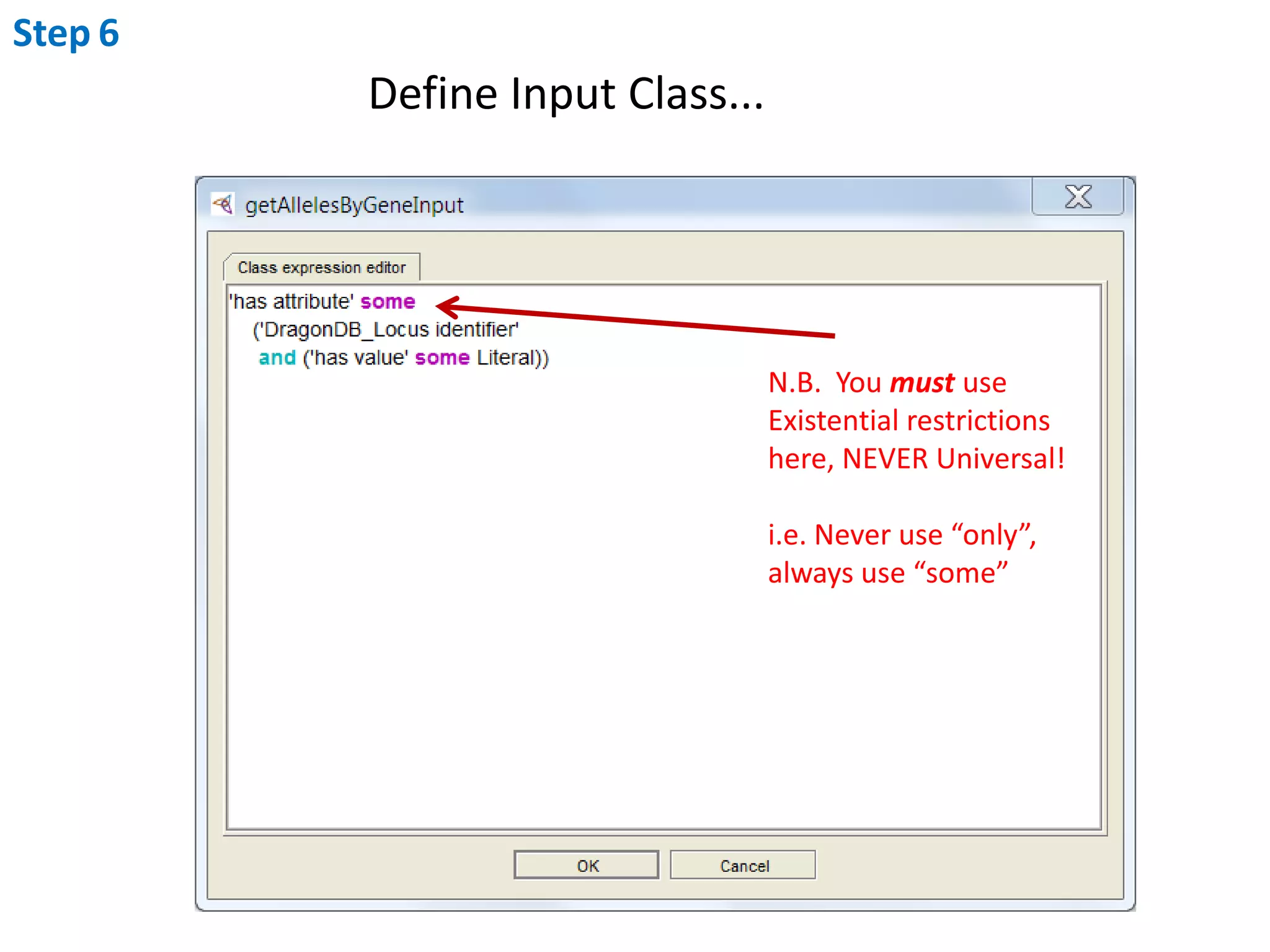
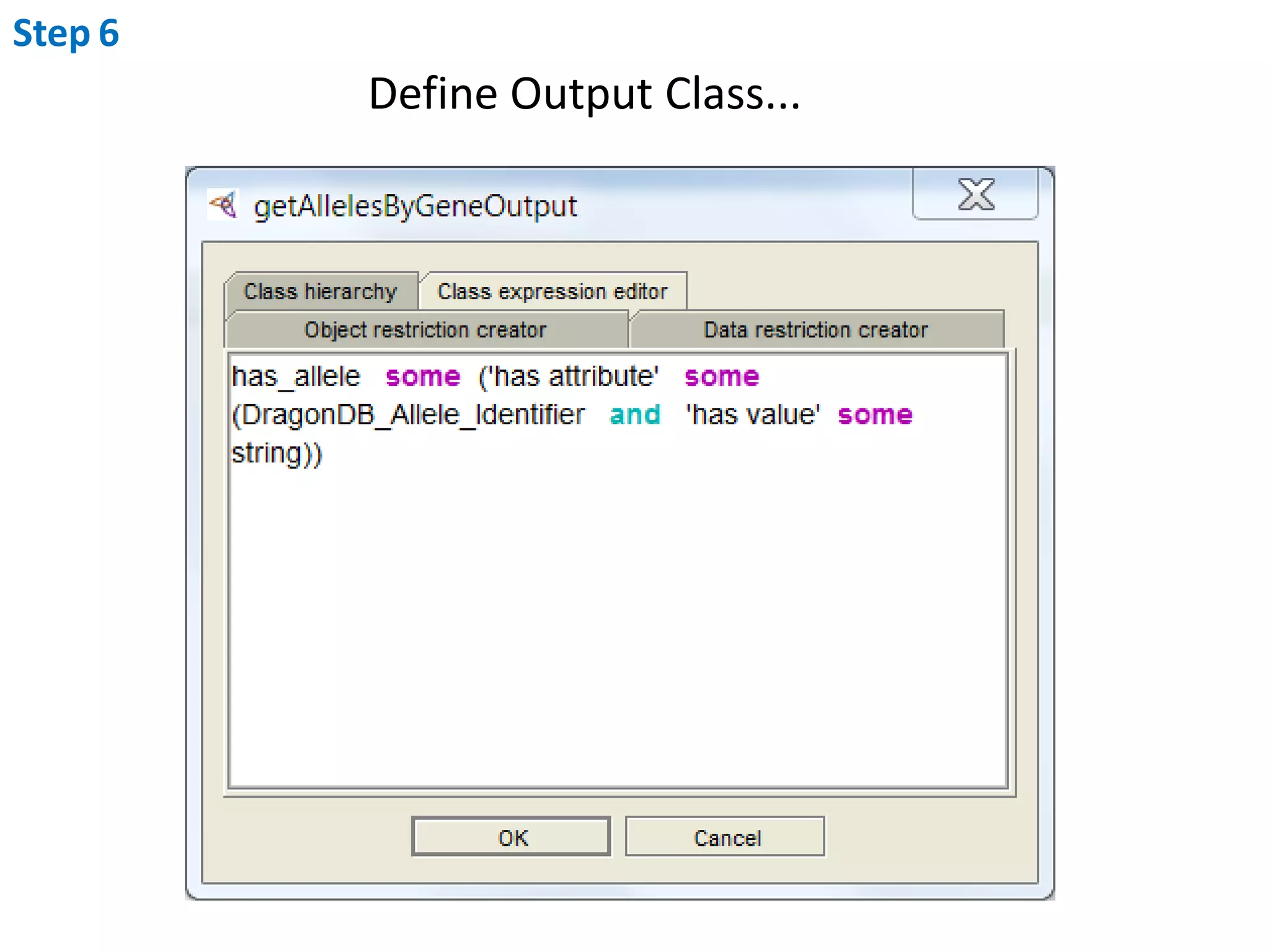
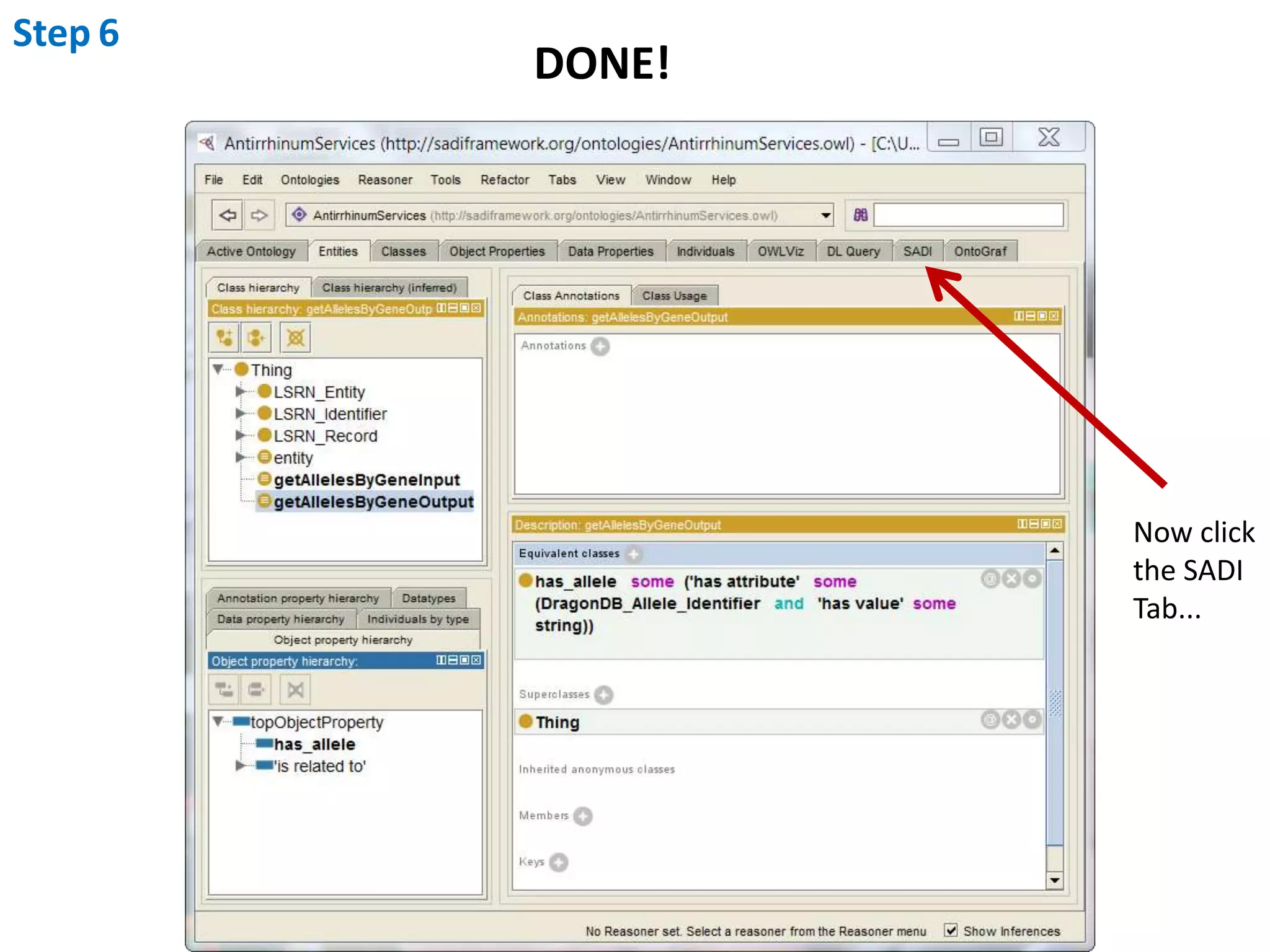
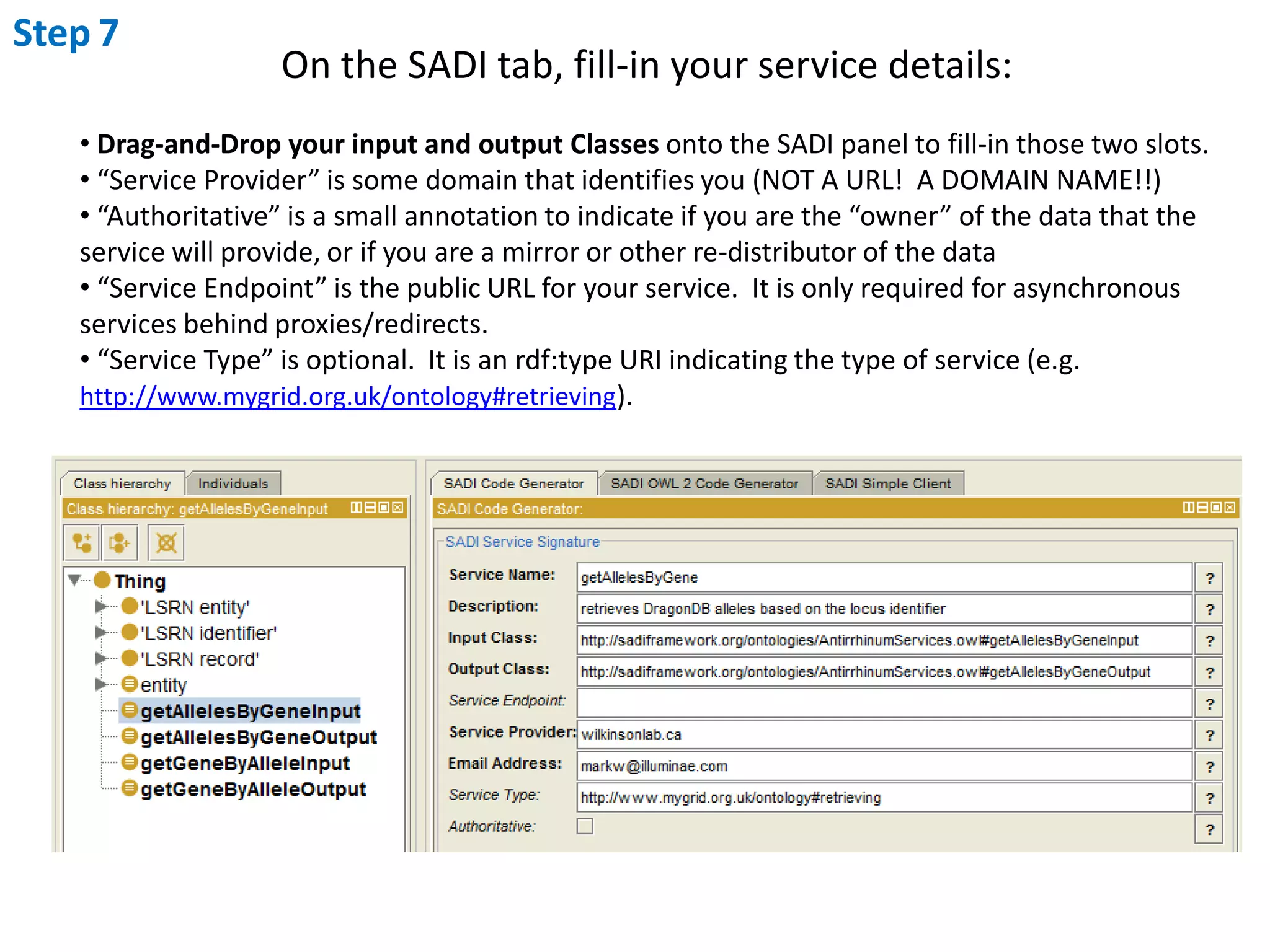
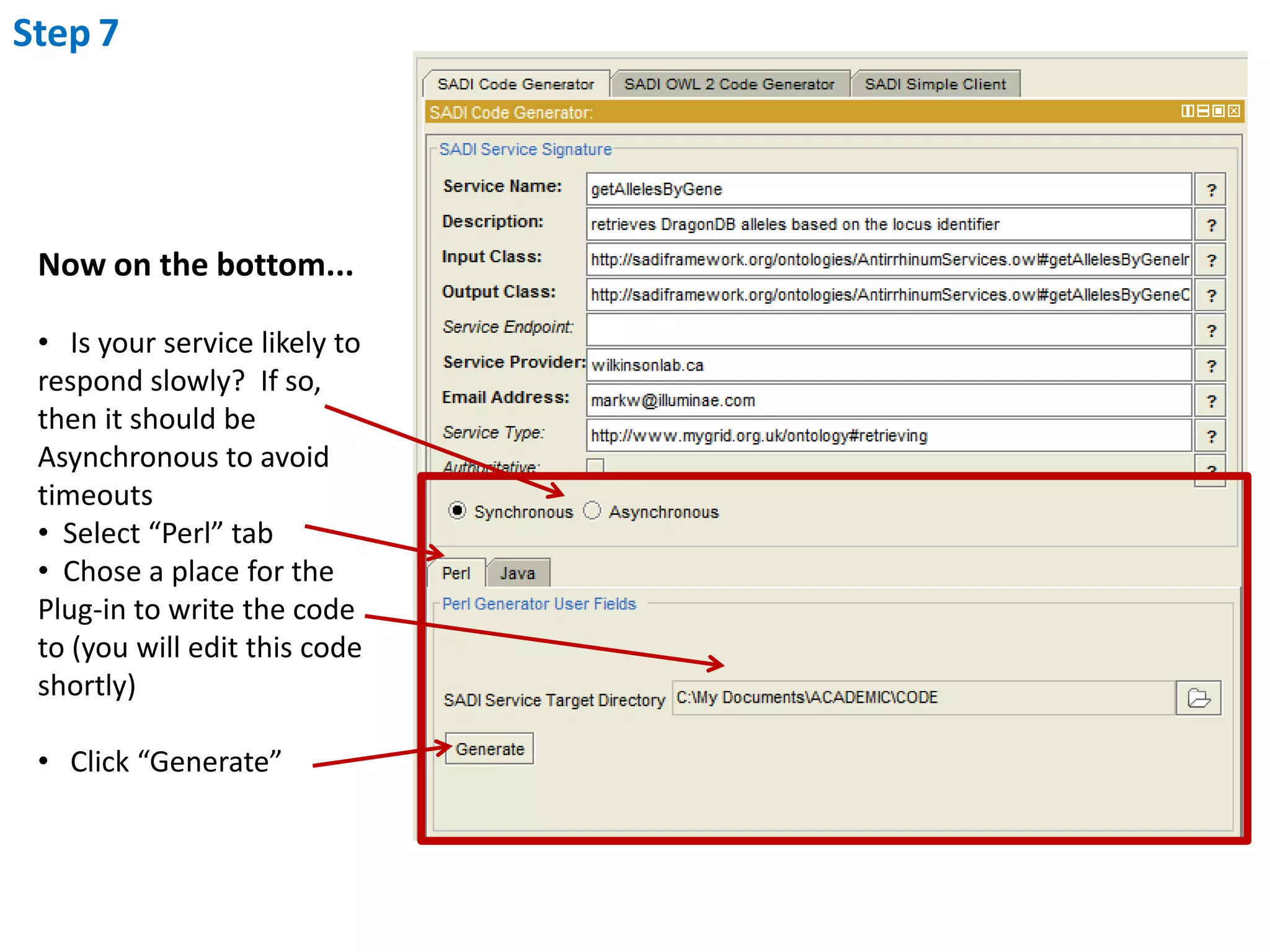
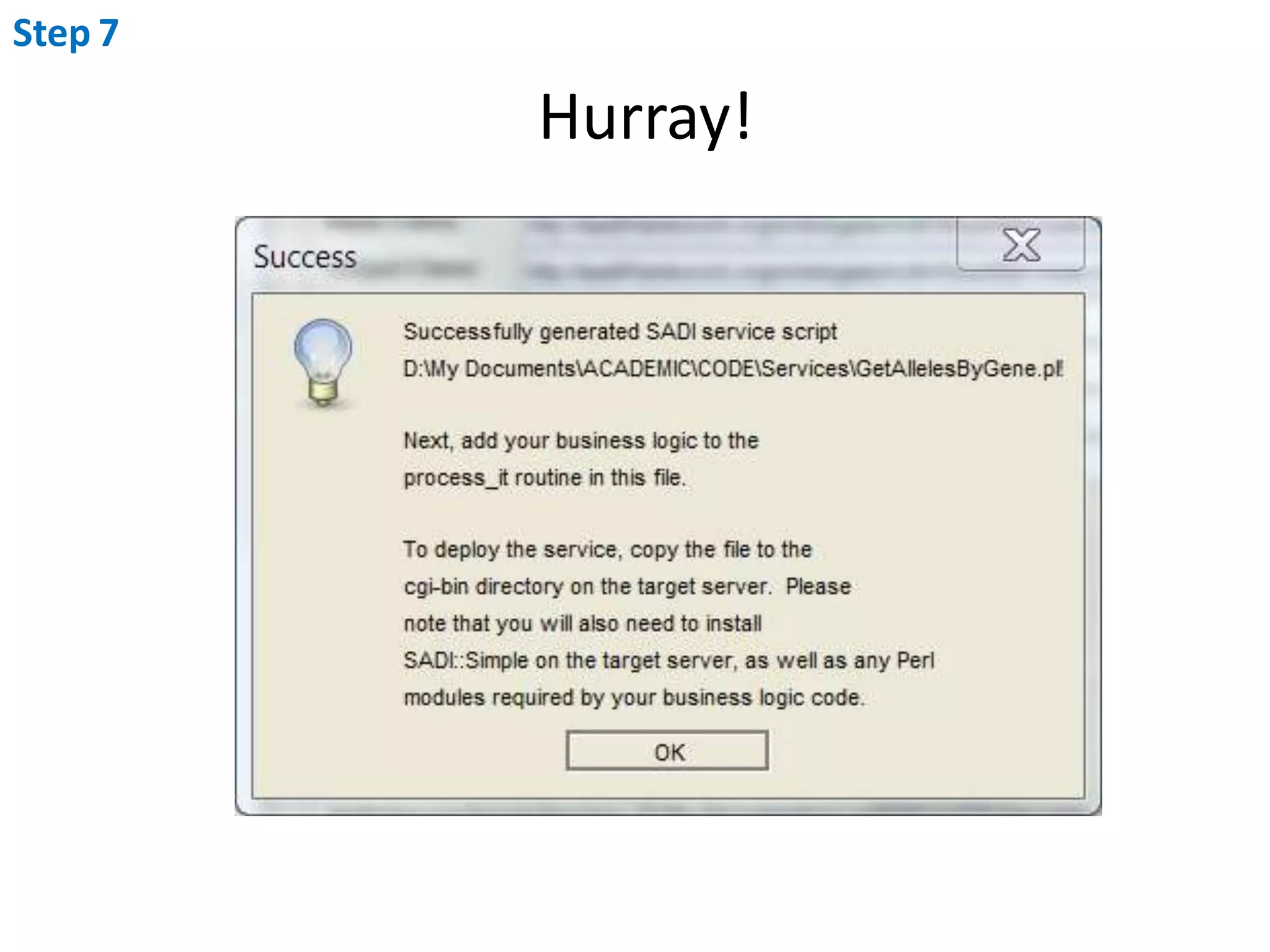
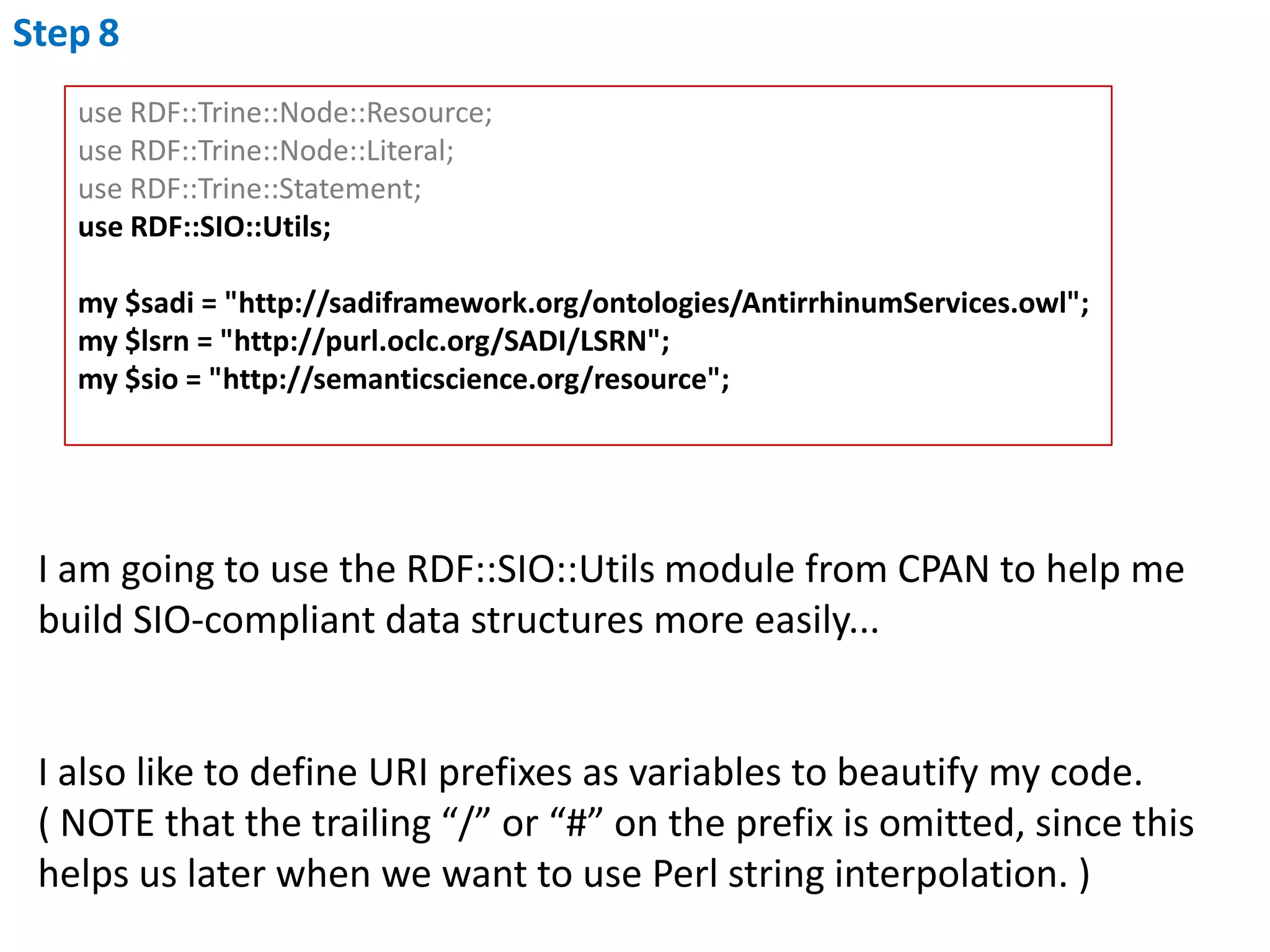
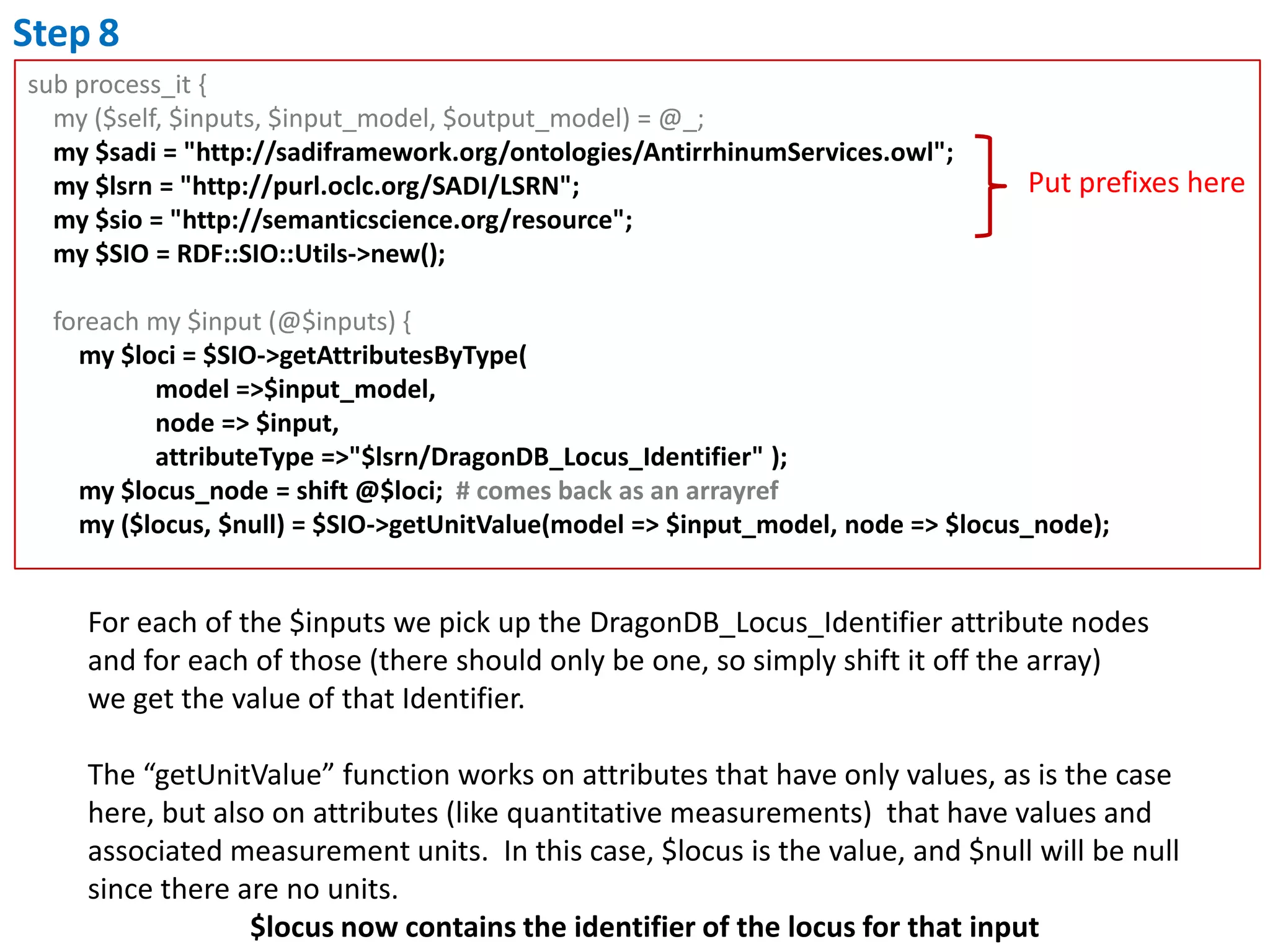
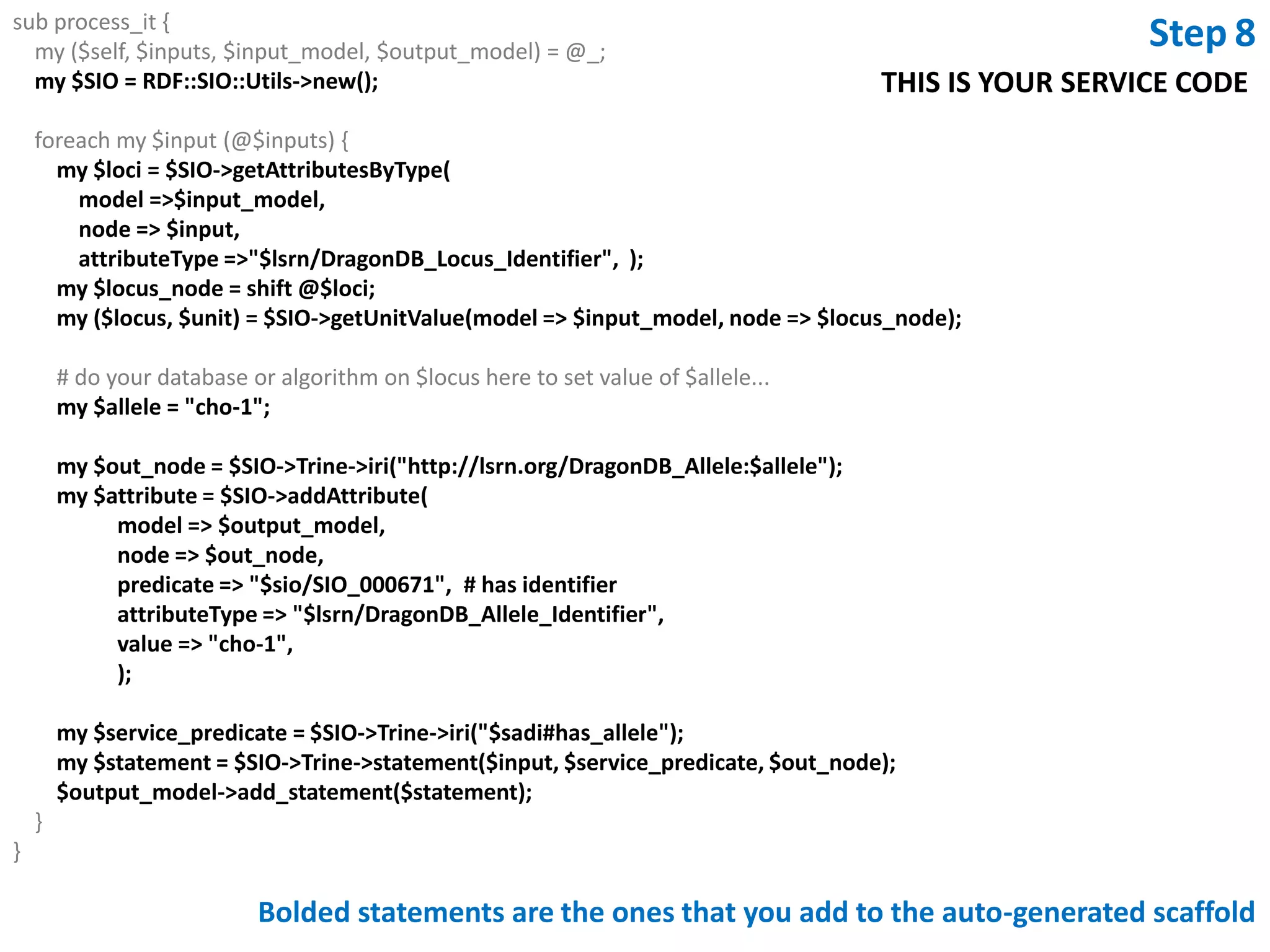
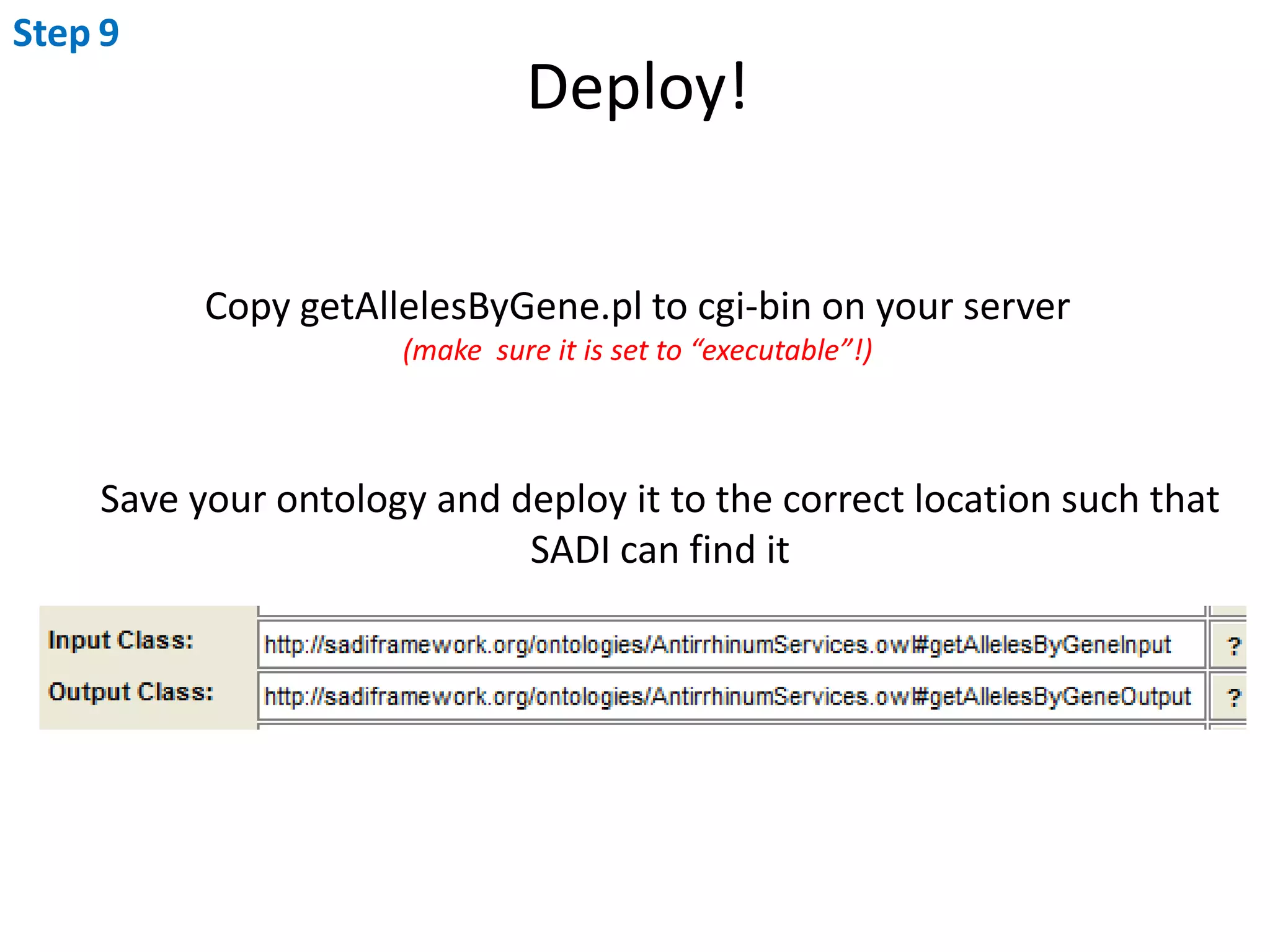
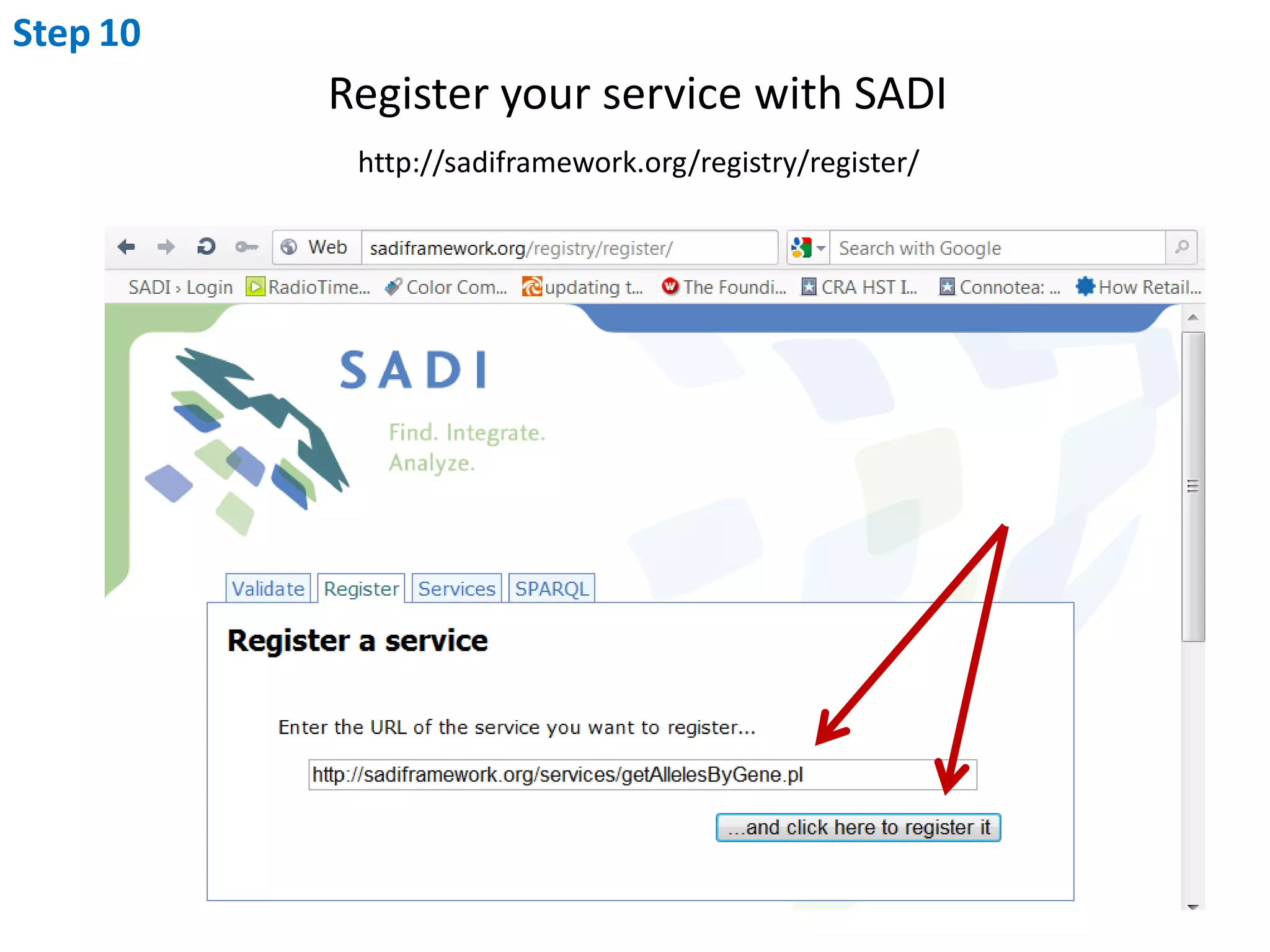
This document outlines the process of creating a SADI service in Perl using the Protege SADI plug-in, detailing steps from identifying data consumption and production to deploying the service. It includes specific instructions on modeling input and output data structures using ontologies, coding the service logic, and testing the service before registration. The example service described retrieves allele information from a genomic database based on specified gene loci.