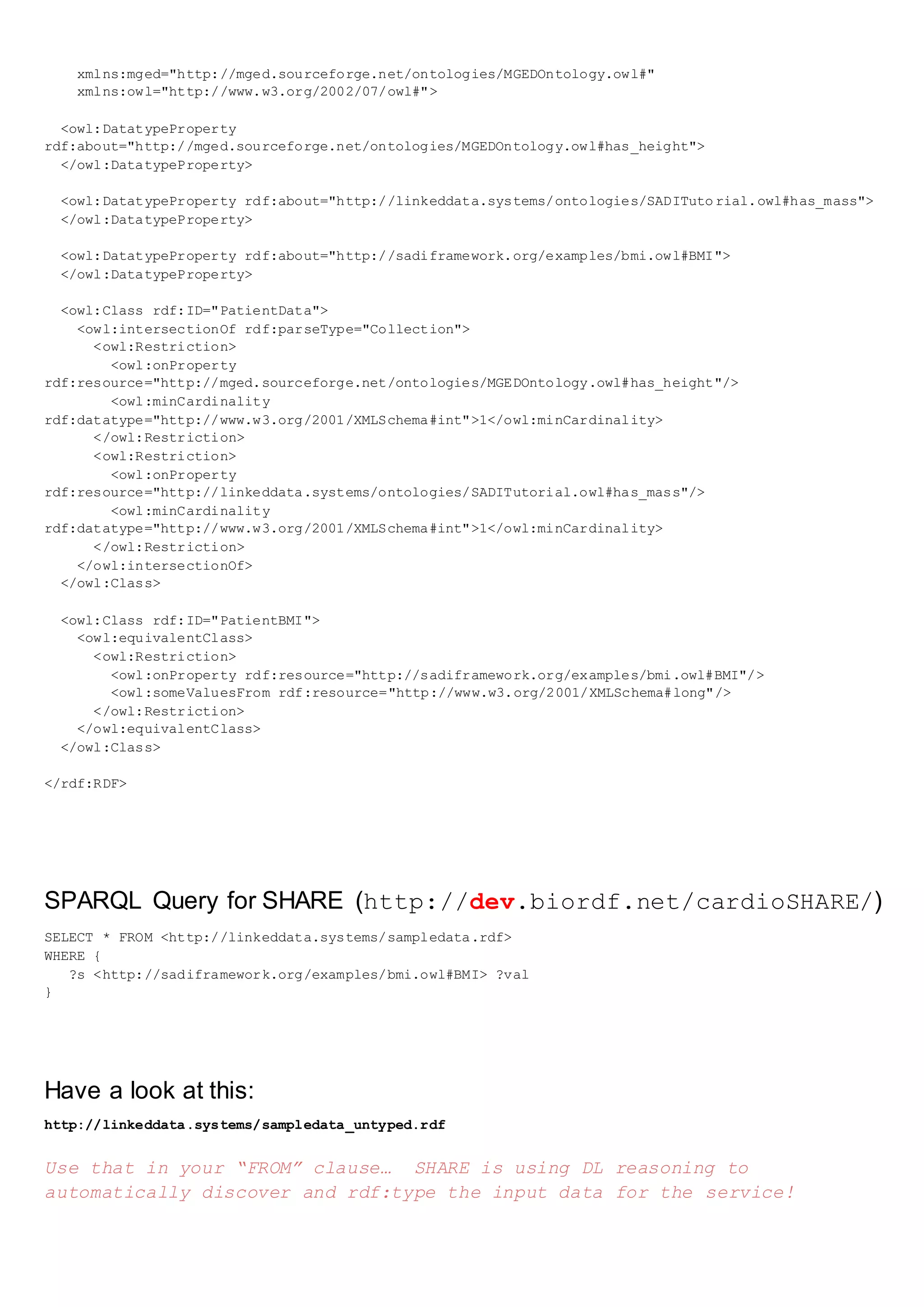
The document describes sample patient data provided in N3 and XML formats. It also describes an ontology defining classes like PatientData and properties like height and mass. Additionally, it provides a SPARQL query and discusses how a SADI service can calculate BMI from untyped input data by using description logic reasoning to discover the data types.