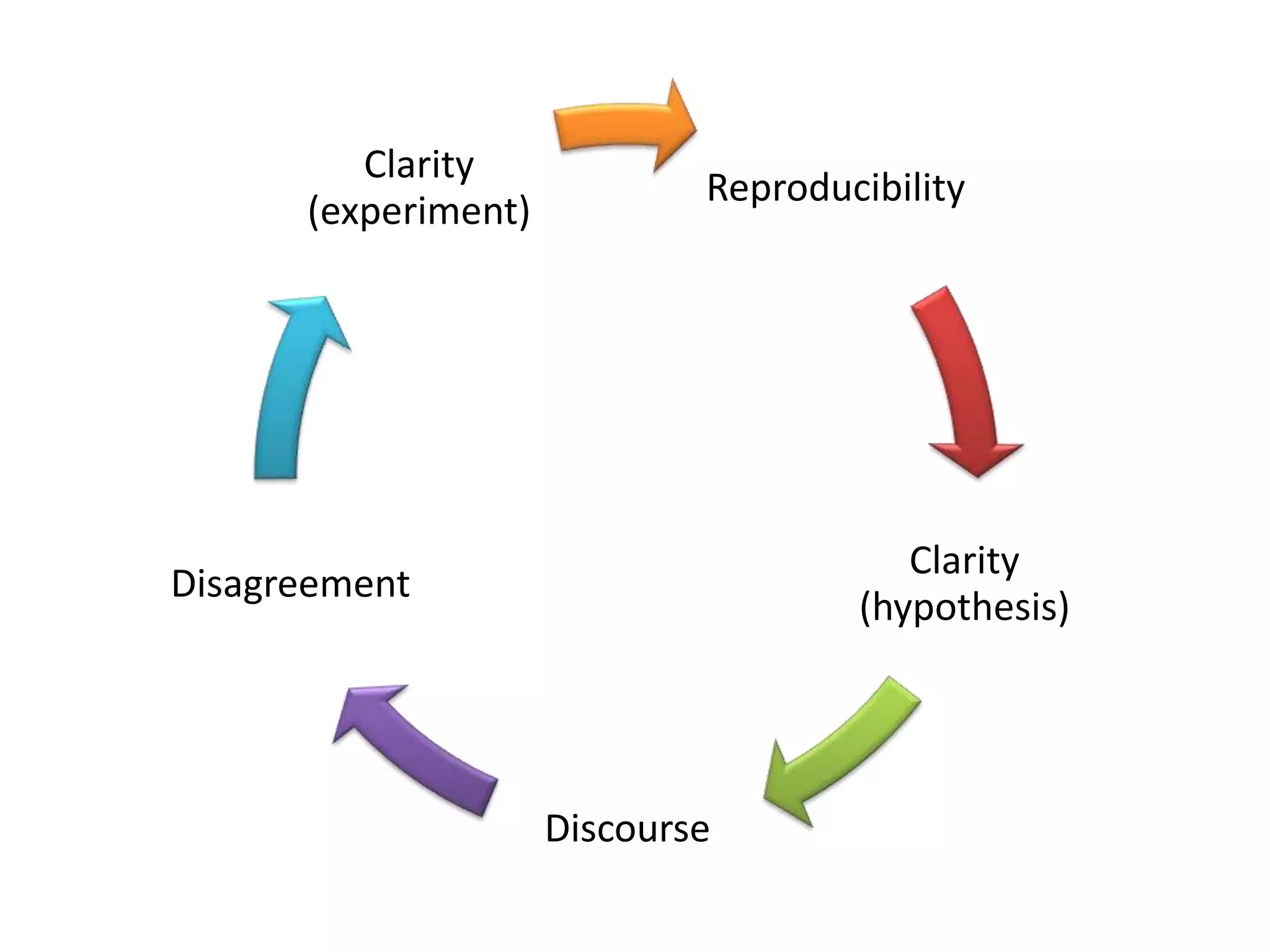
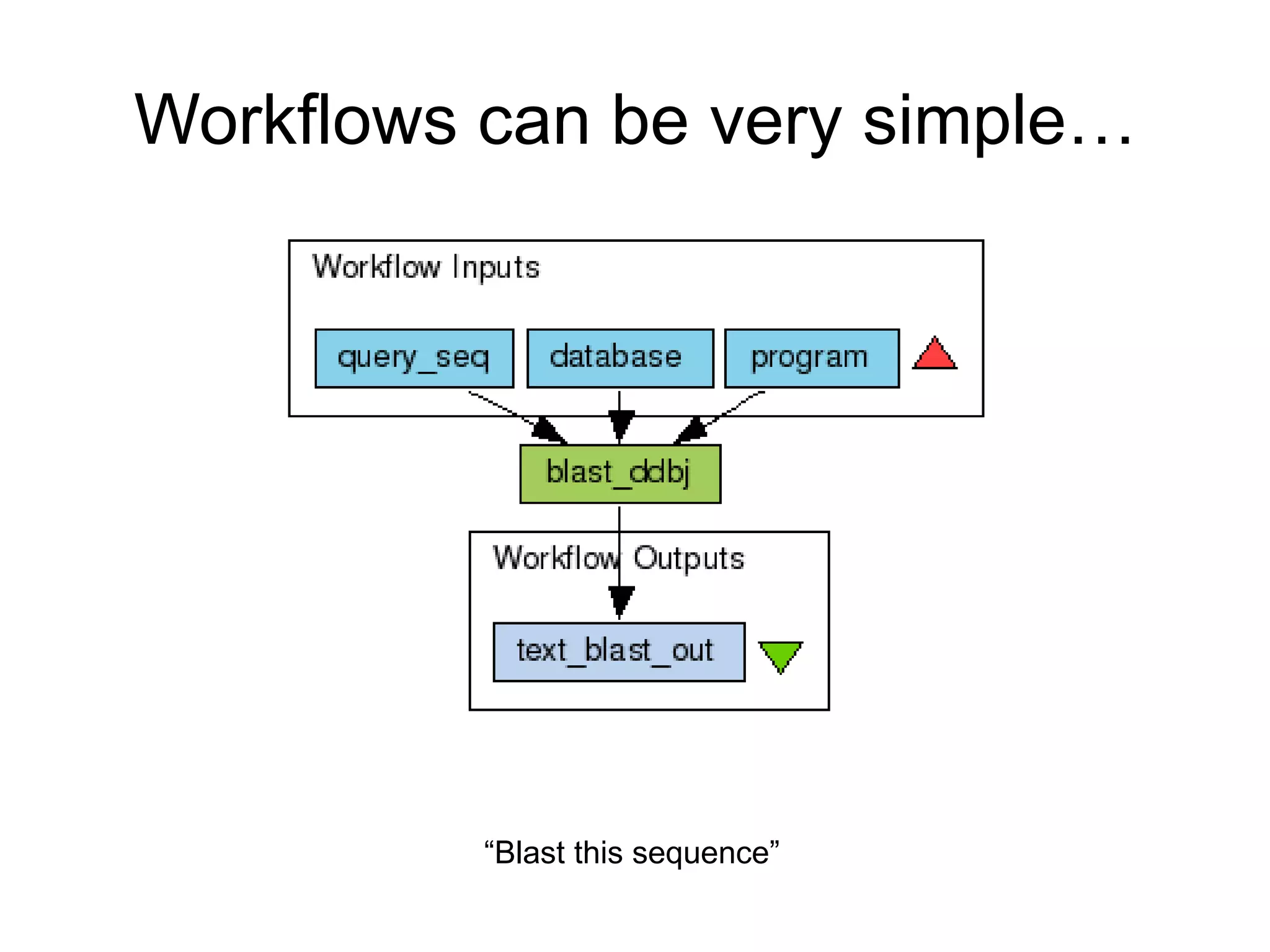
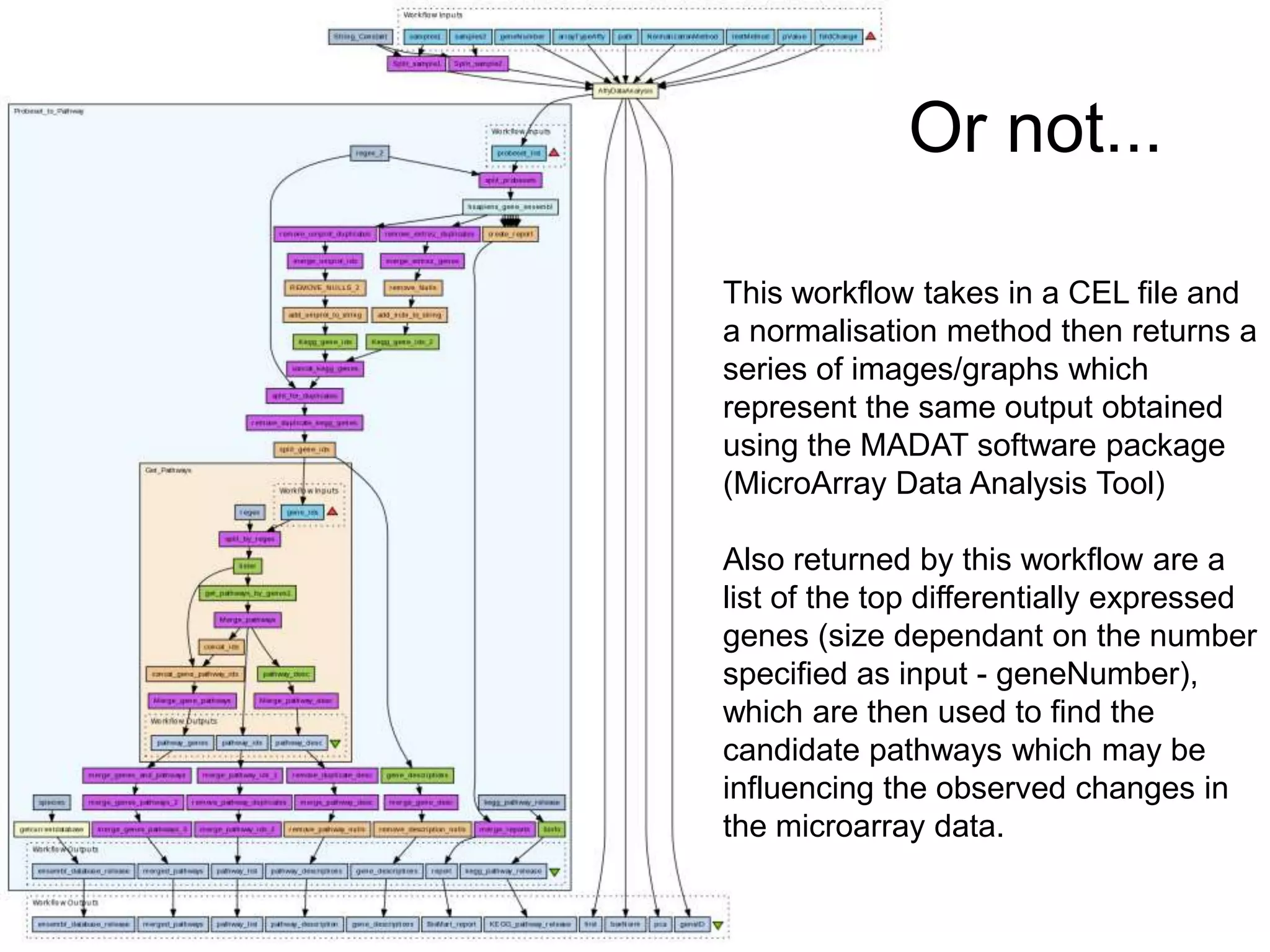
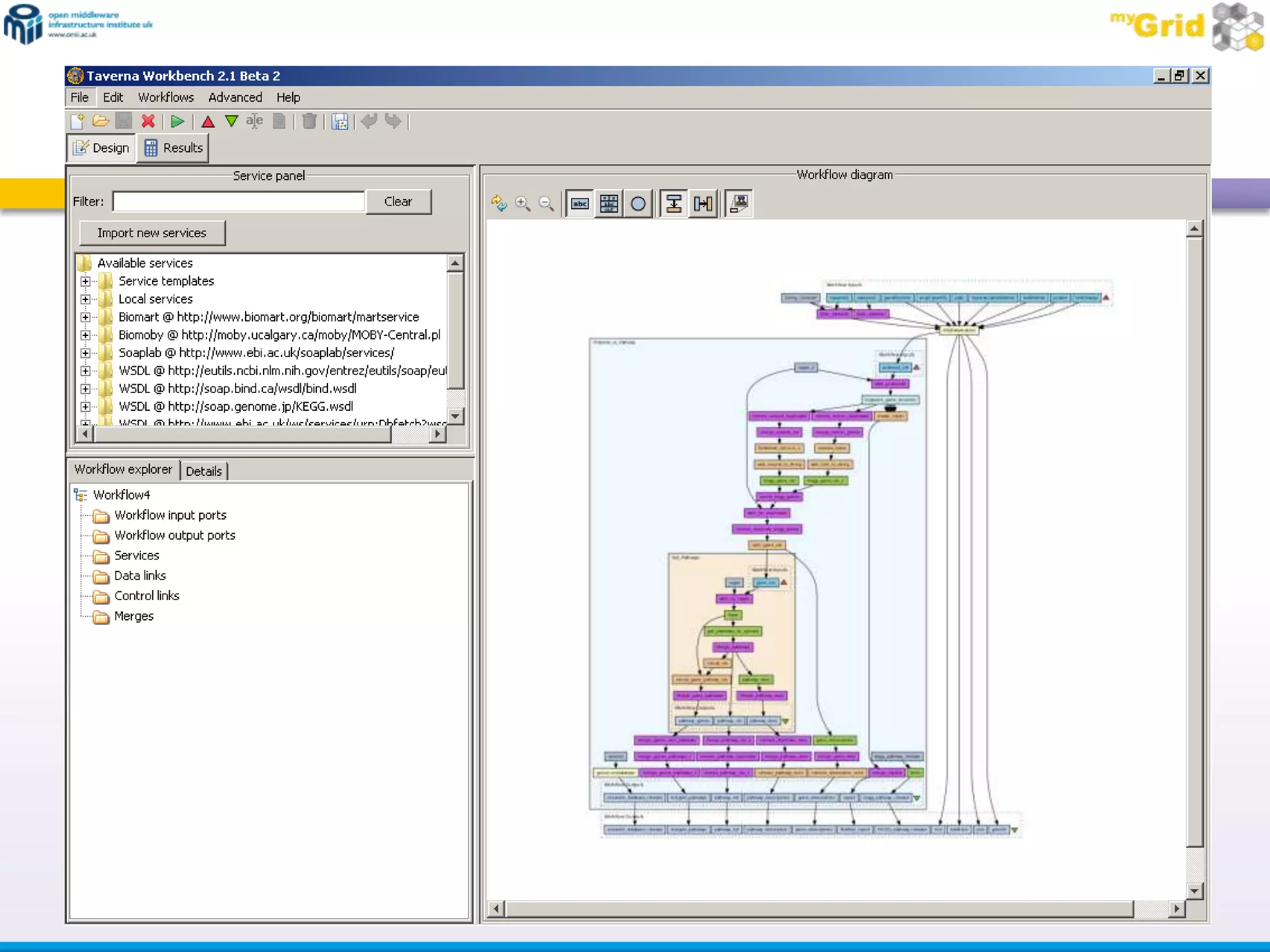
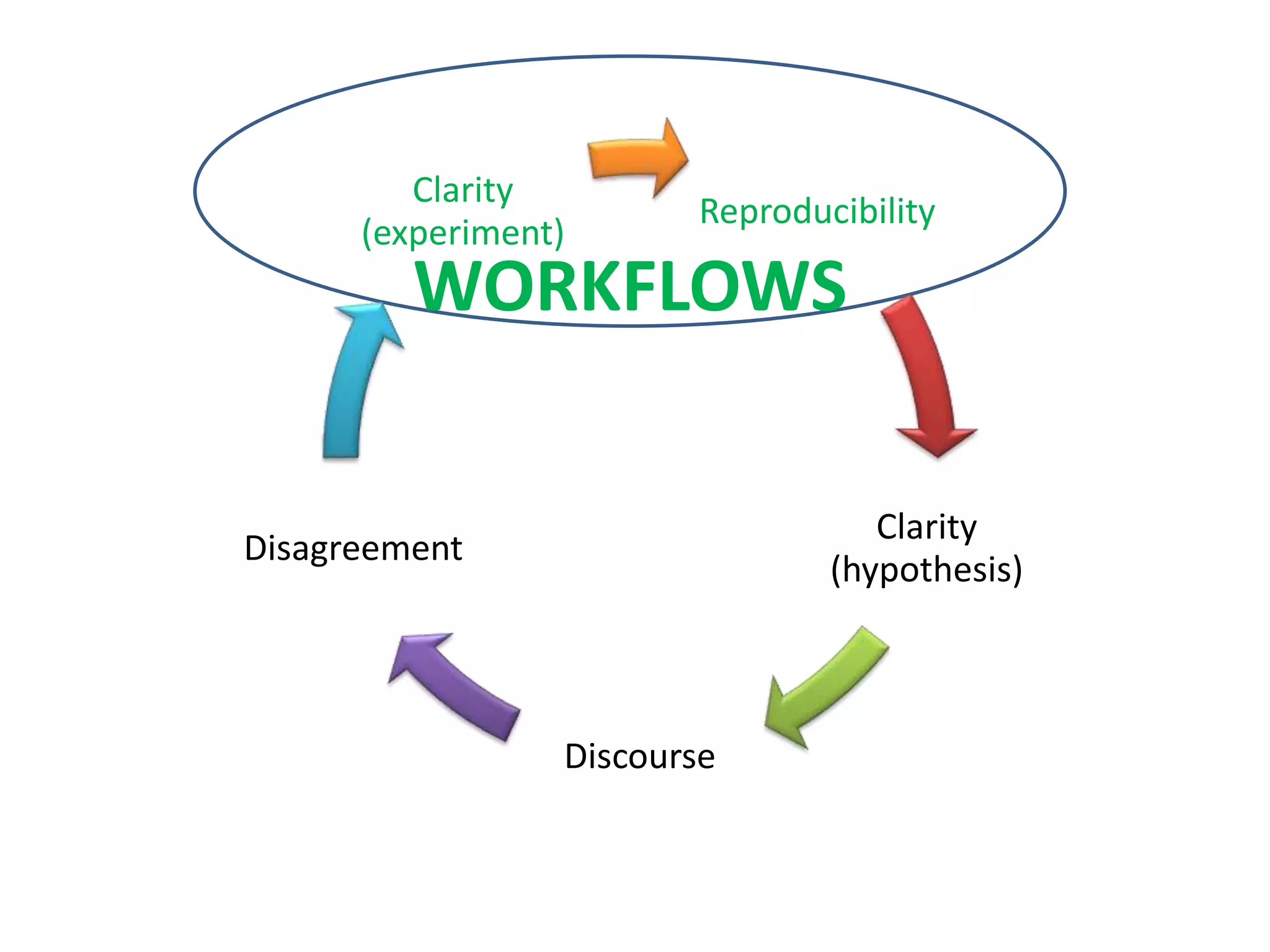
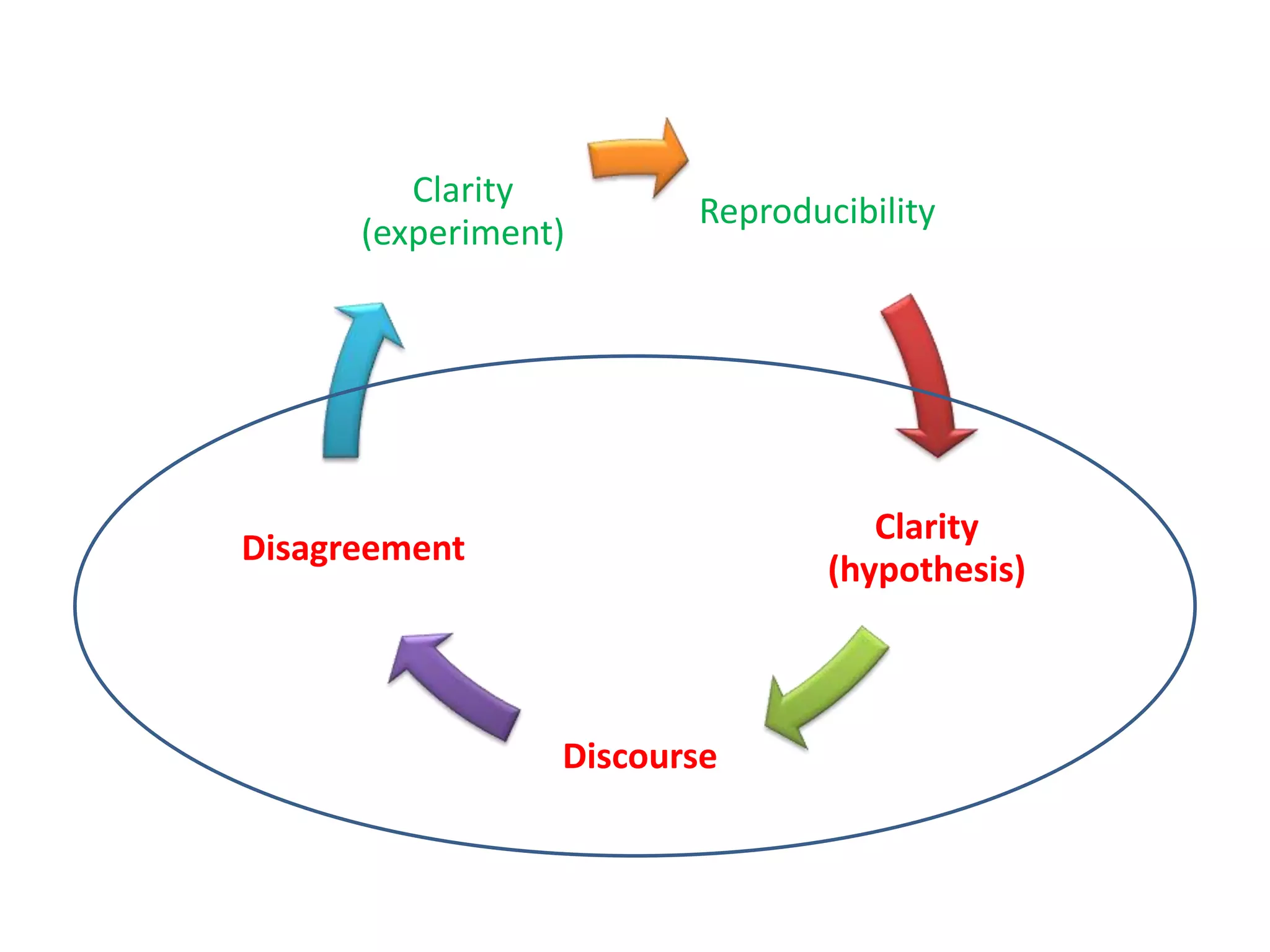
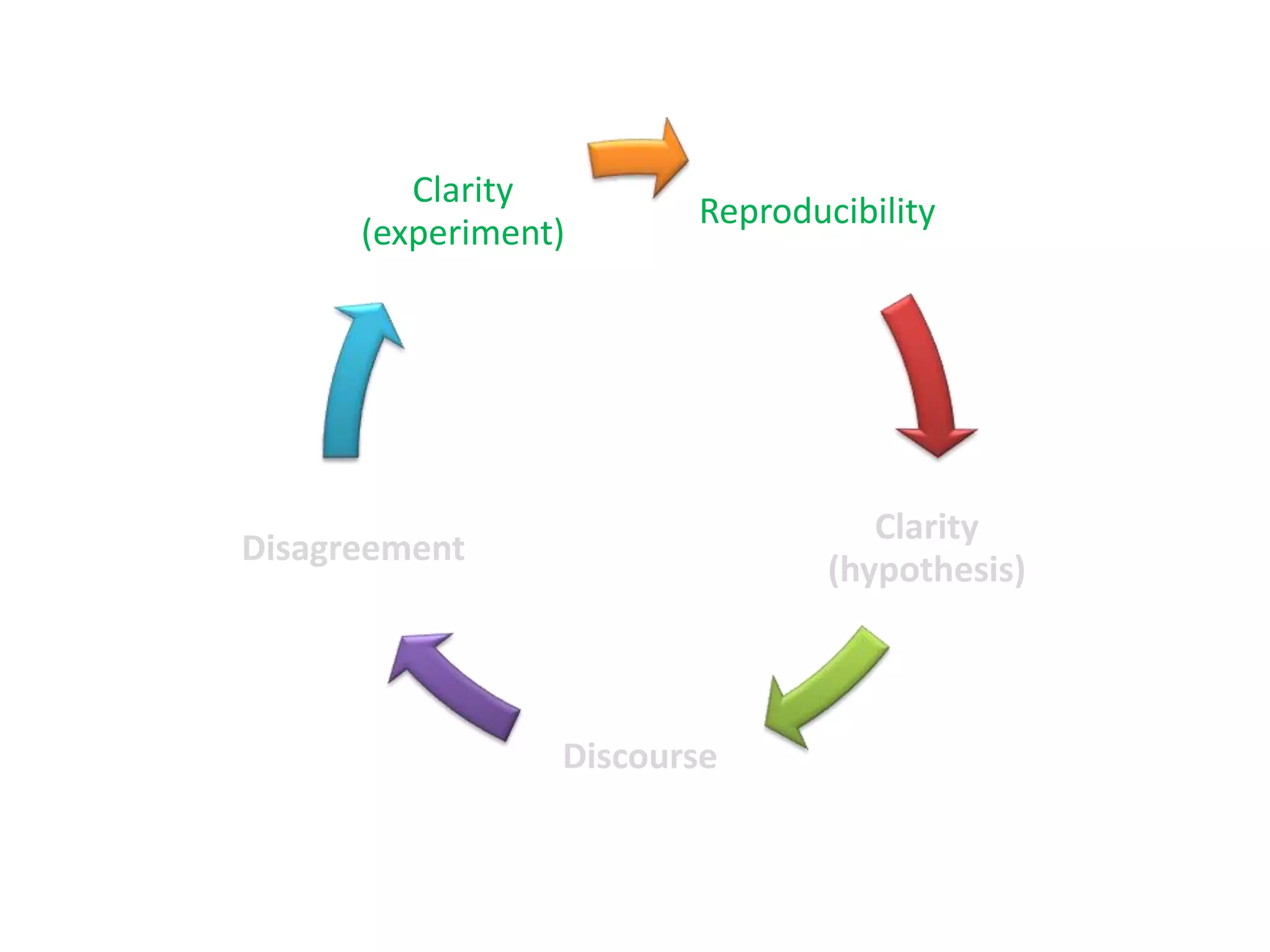
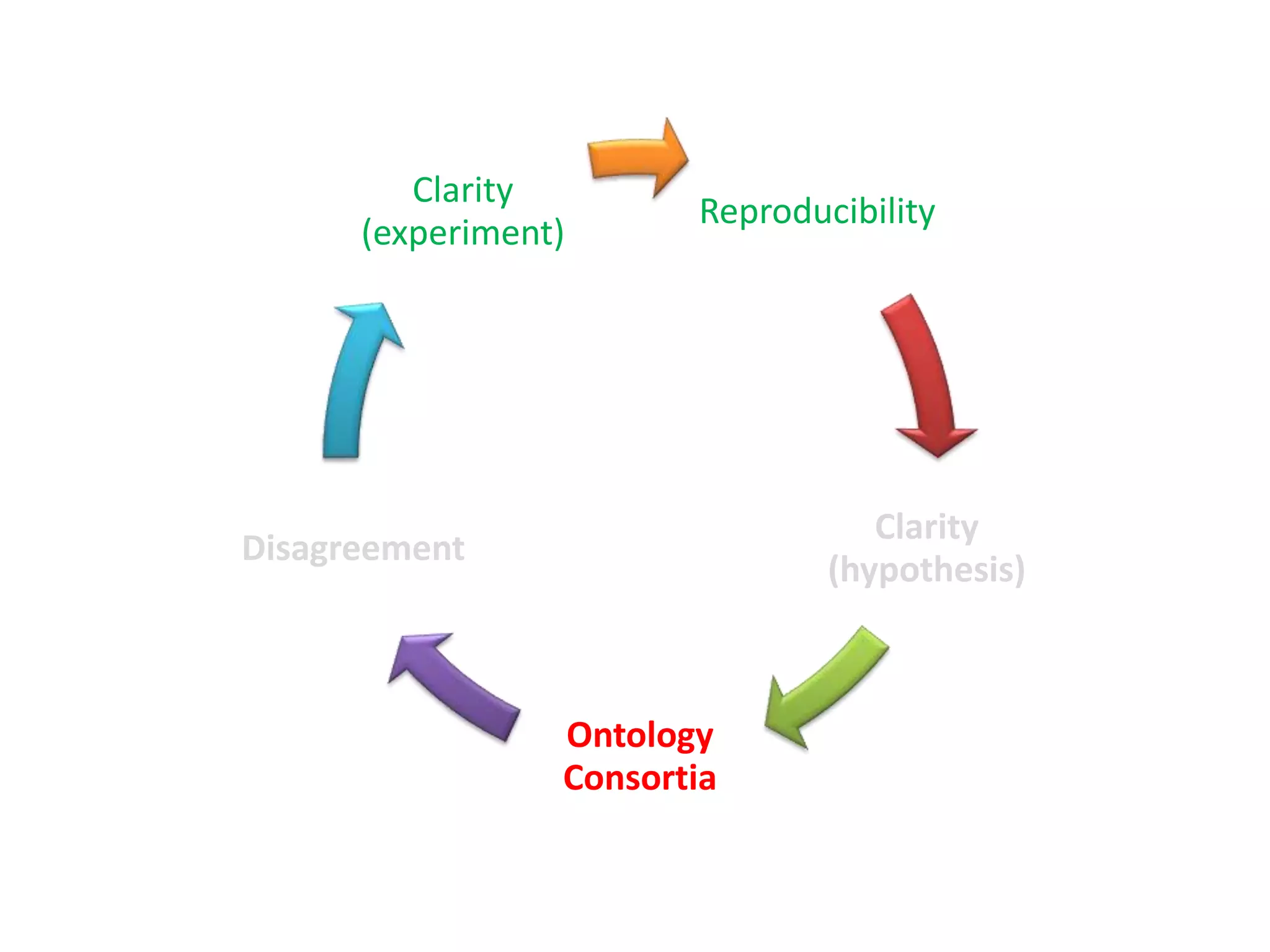
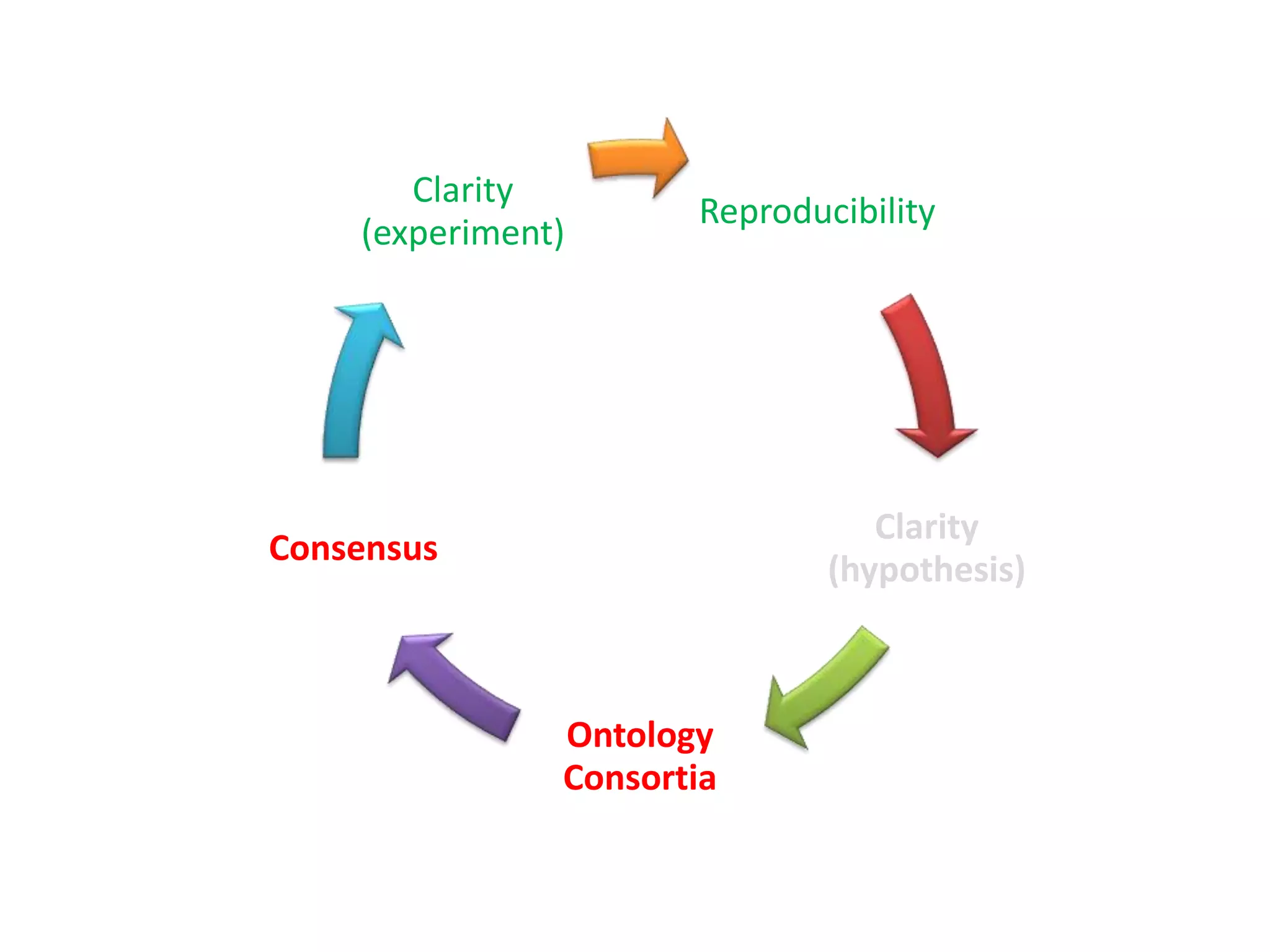
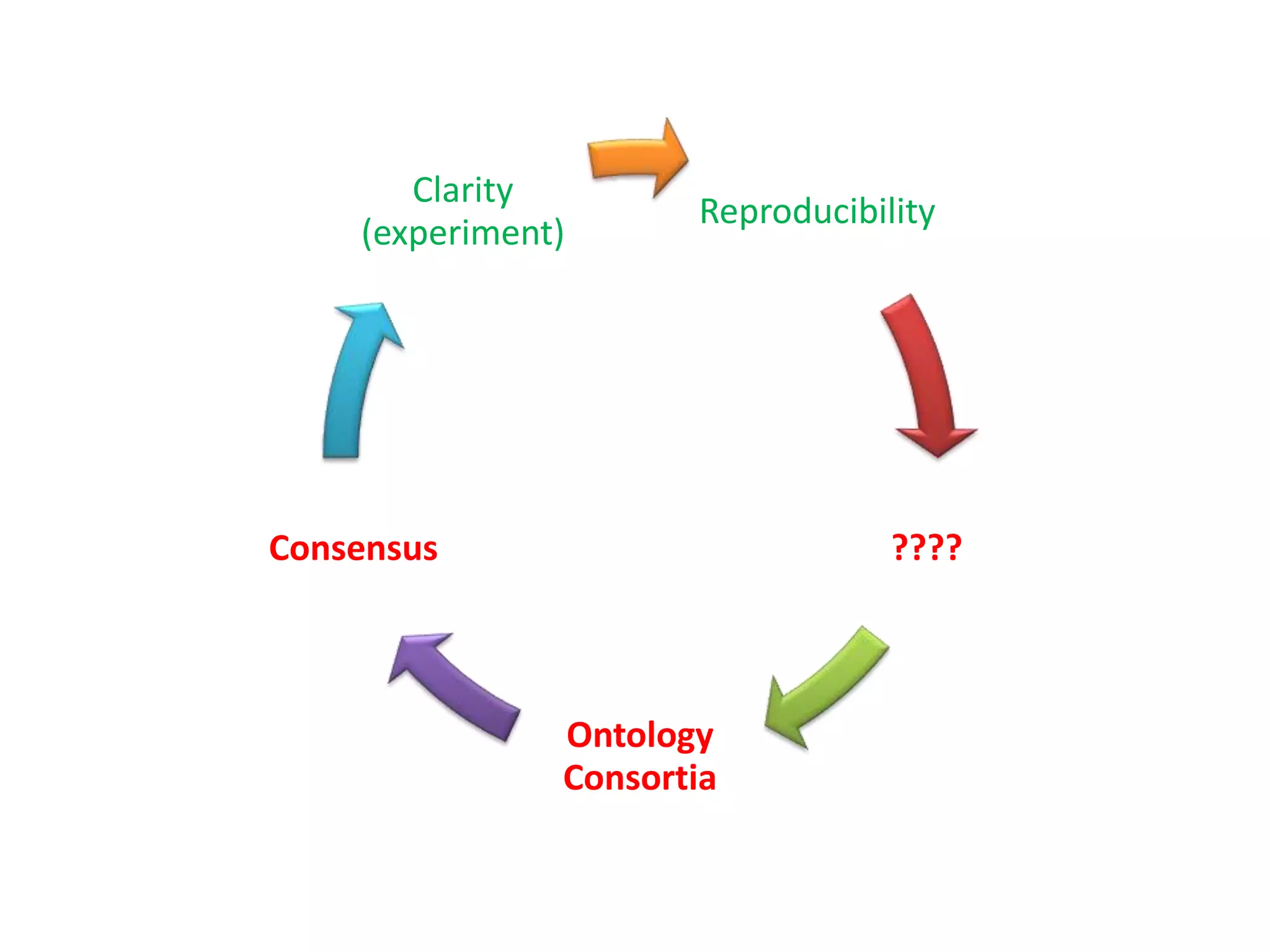
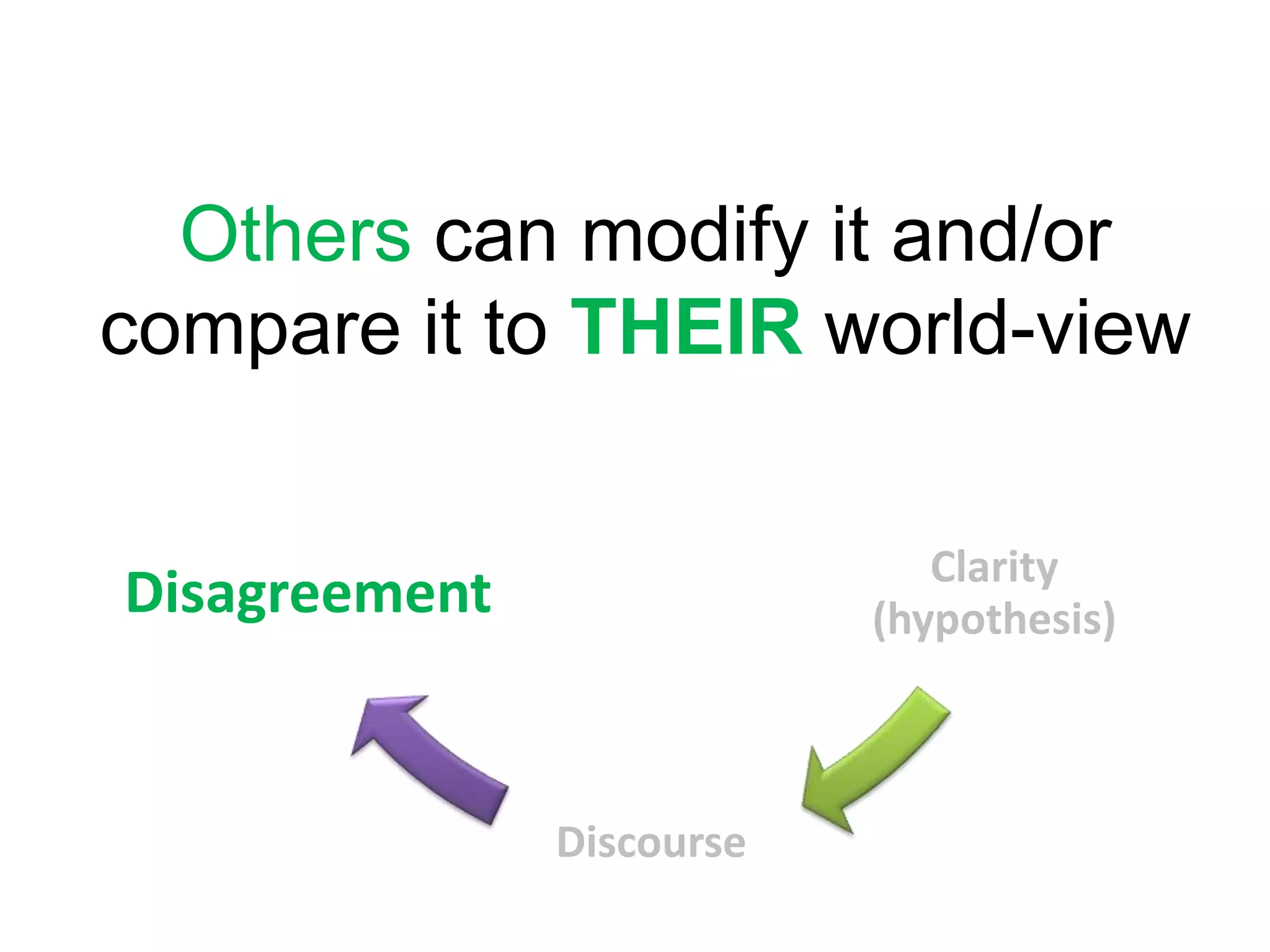
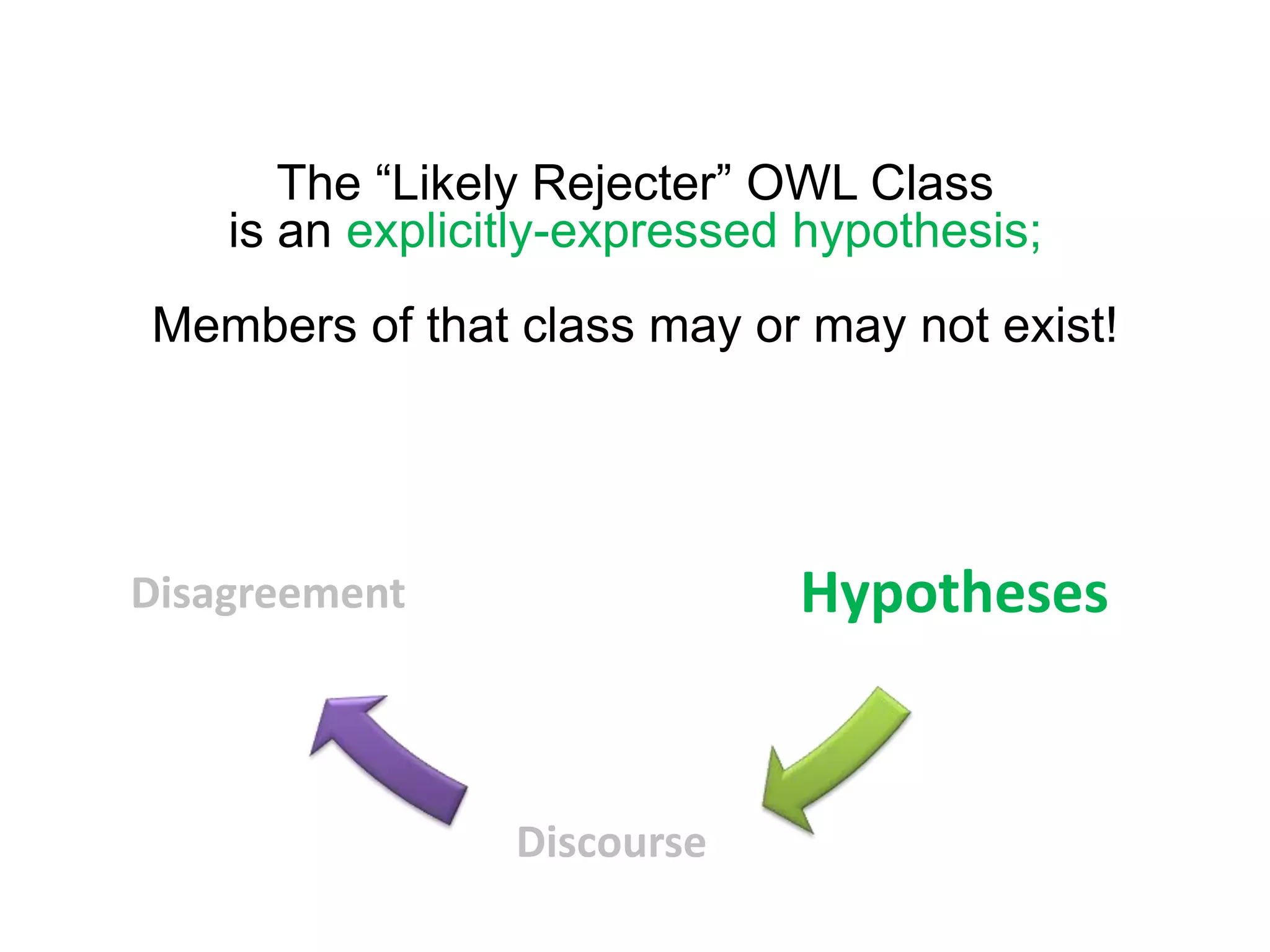
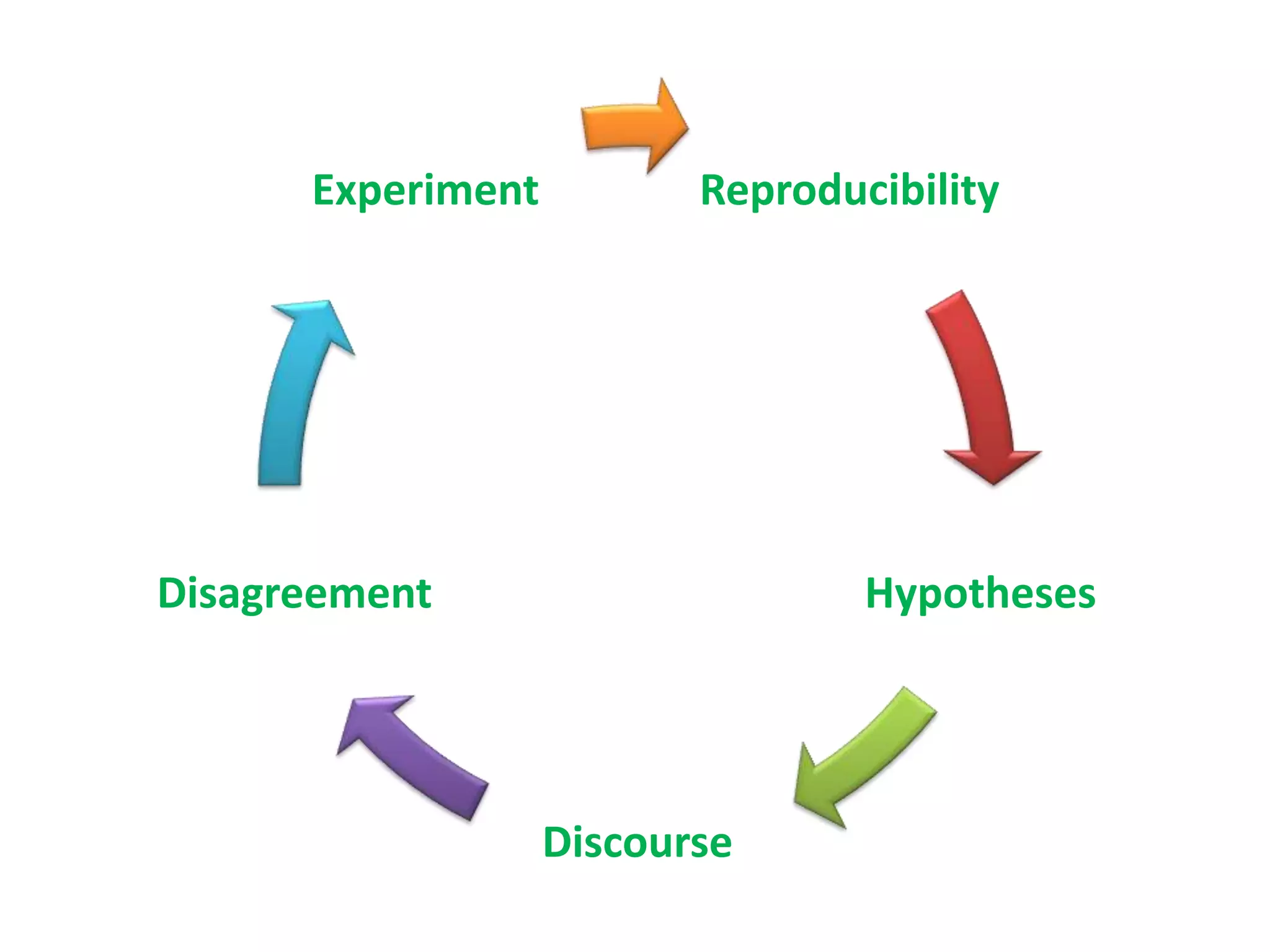
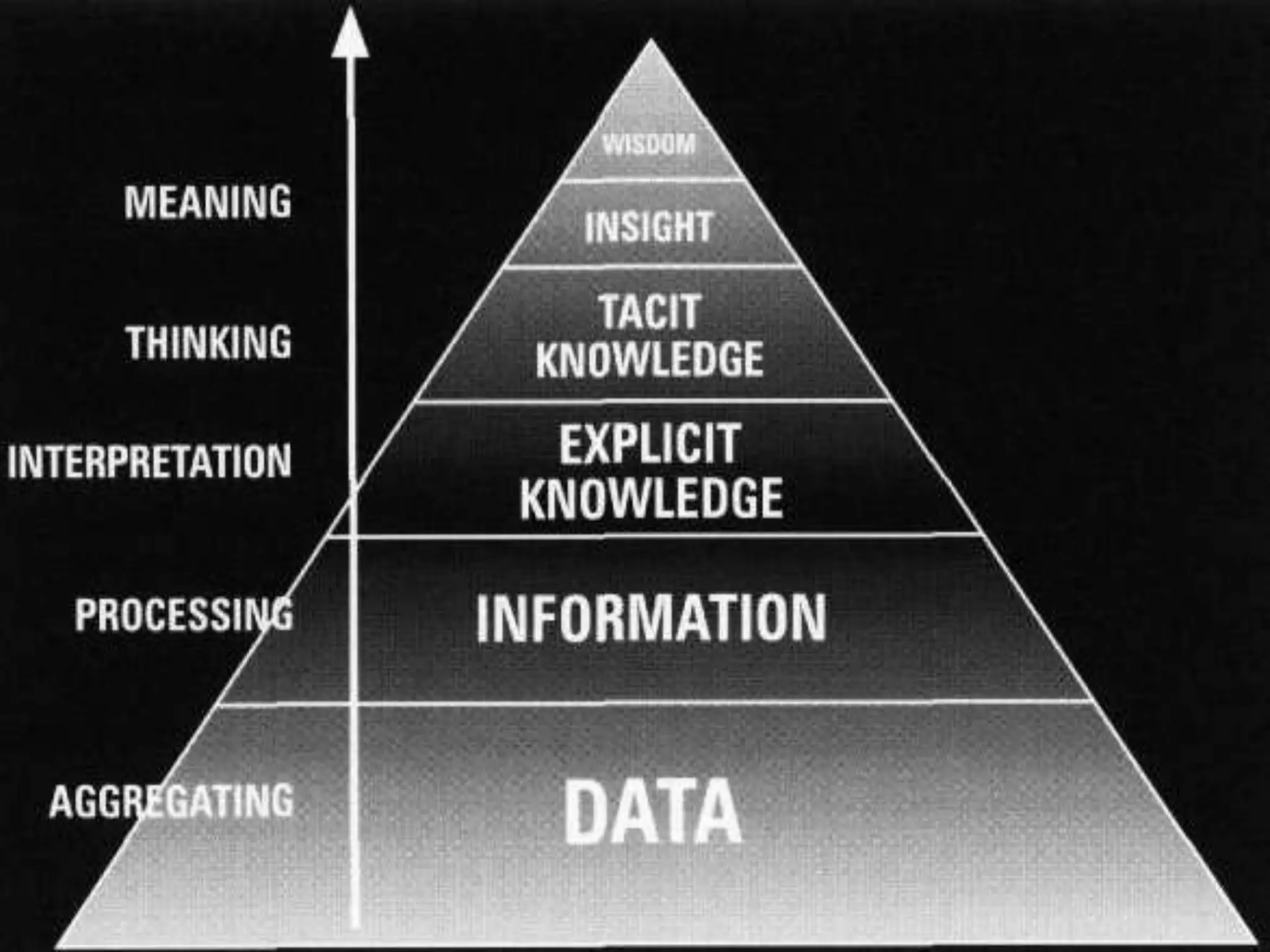
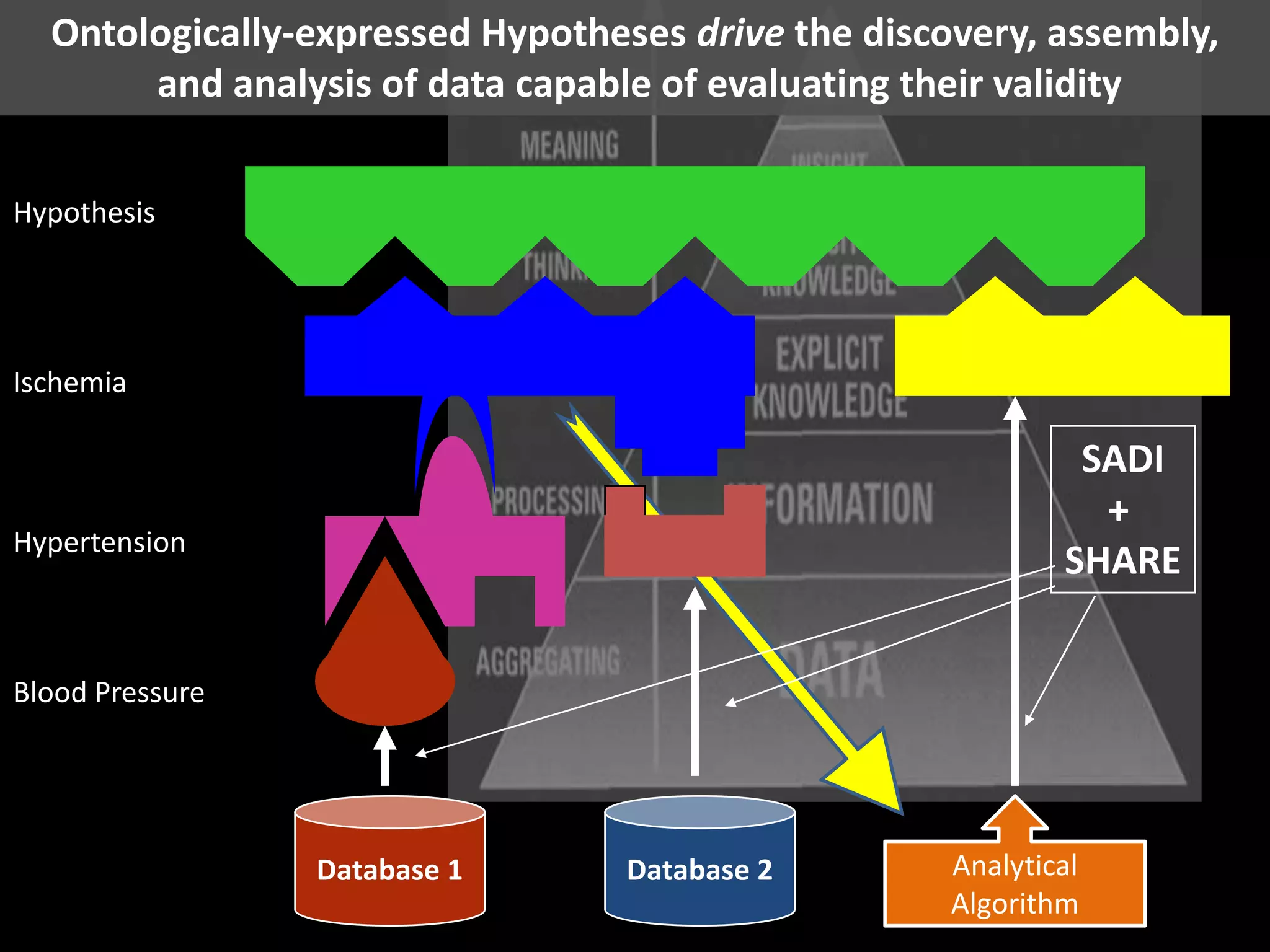
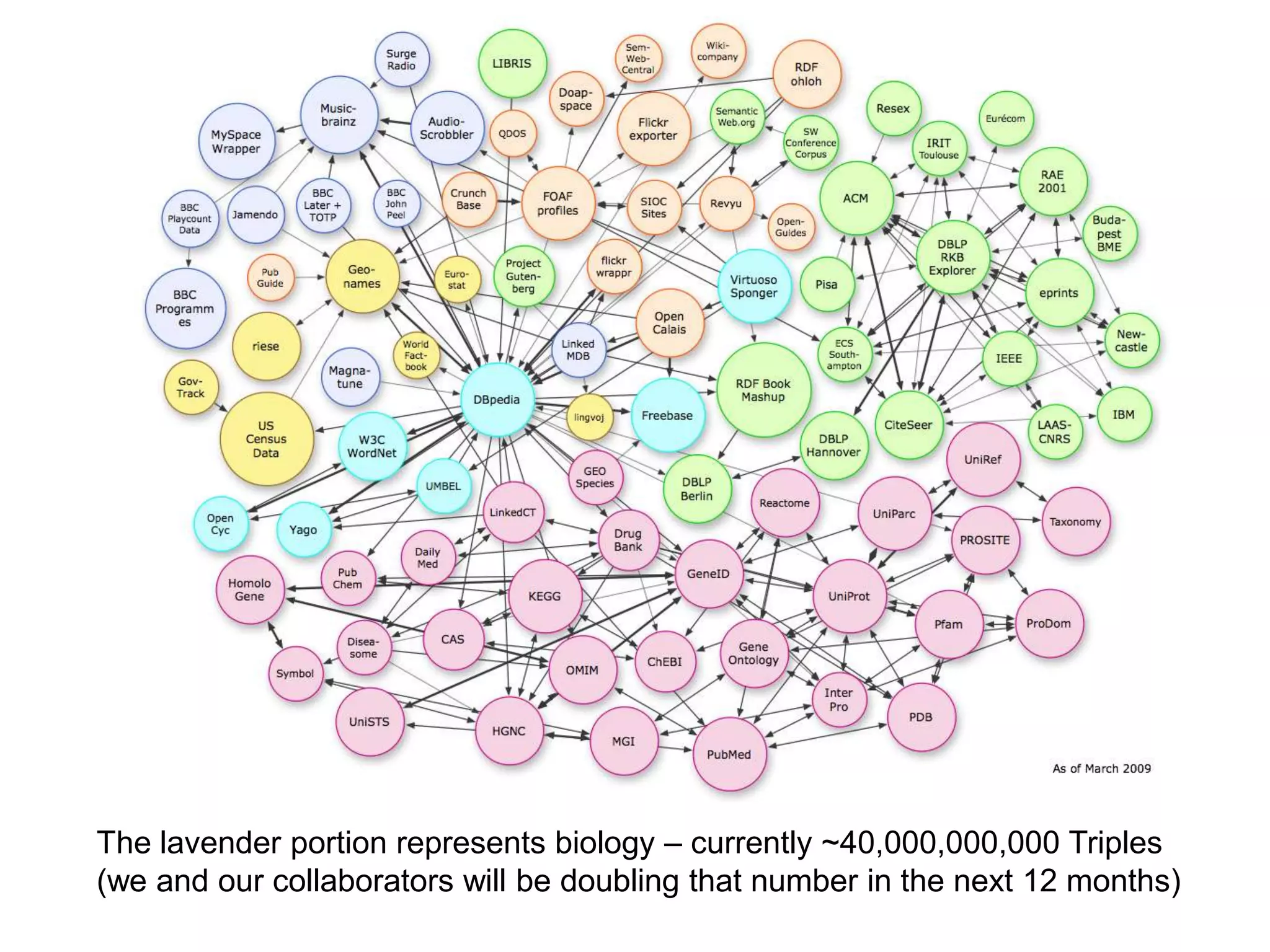
The document summarizes two demonstrations of the SADI and SHARE frameworks. The first demonstration shows how a SPARQL query is used to retrieve pathway information for a protein from multiple distributed data sources without using a database. The second demonstration shows clinical data for transplant patients being rejected being retrieved through analyzing definitions in an ontology and executing appropriate services.



![The Holy Grail:(this slide created circa 2002)Align the promoters of all serine threoninekinases involved exclusively in the regulation of cell sorting during wound healing in blood vessels.Retrieve and align 2000nt 5' from every serine/threonine kinase in Mus musculus expressed exclusively in the tunica [I | M |A] whose expression increases 5X or more within 5 hours of wounding but is not activated during the normal development of blood vessels, and is <40% homologous in the active site to kinases known to be involved in cell-cycle regulation in any other species.](https://image.slidesharecdn.com/icapture-101010193247-phpapp01/75/The-Scientific-Method-on-the-Semantic-Web-4-2048.jpg)







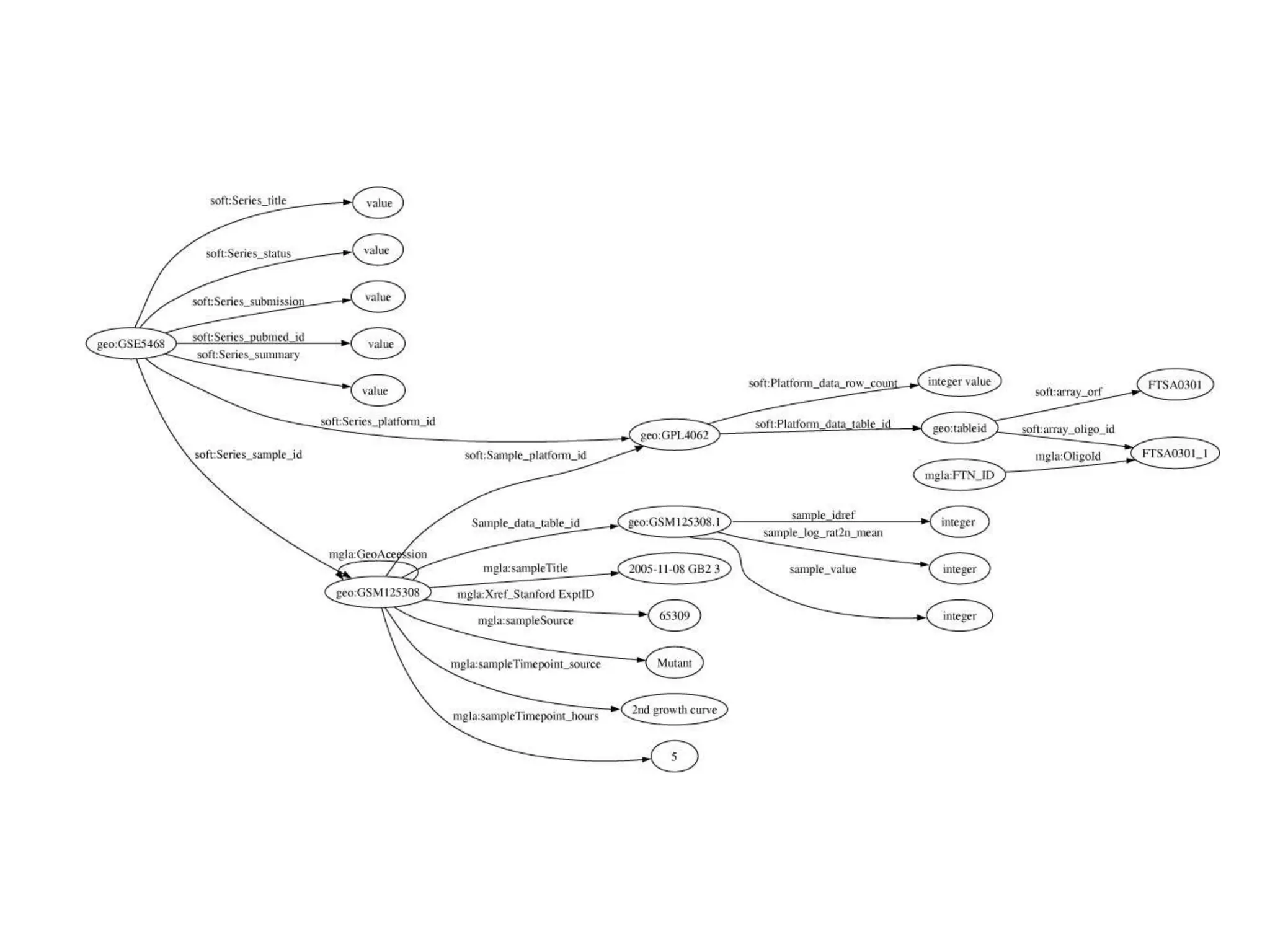



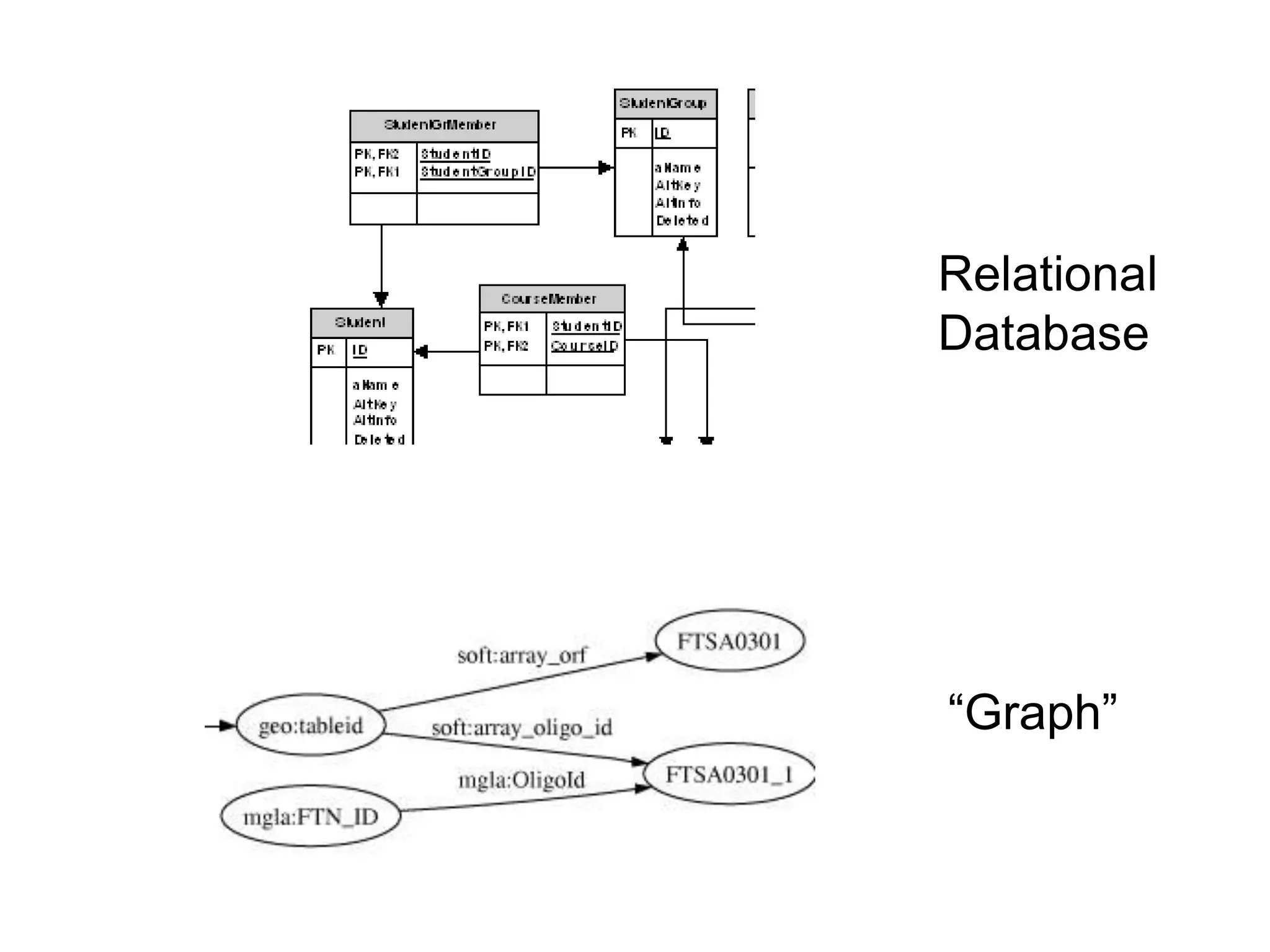
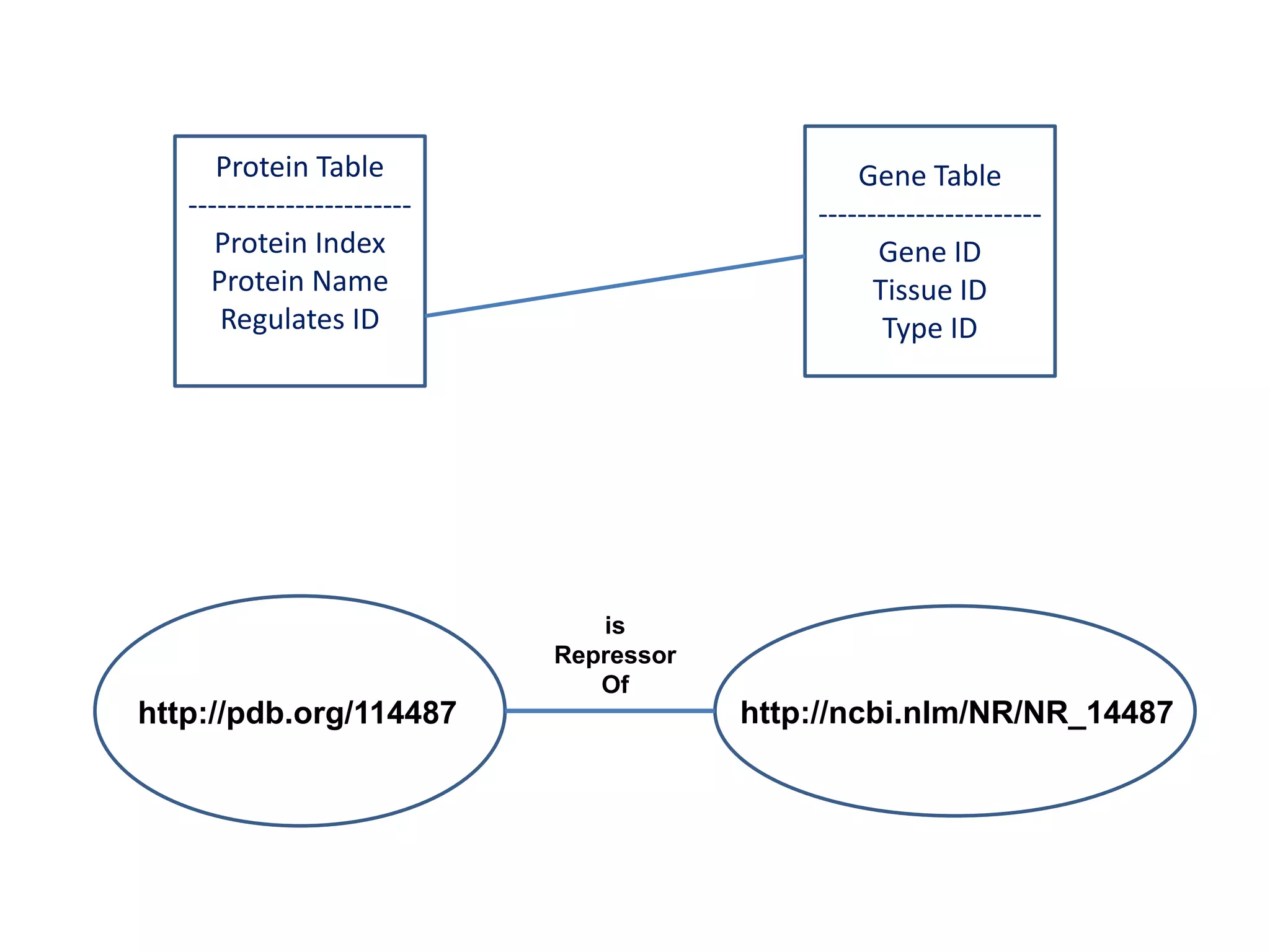
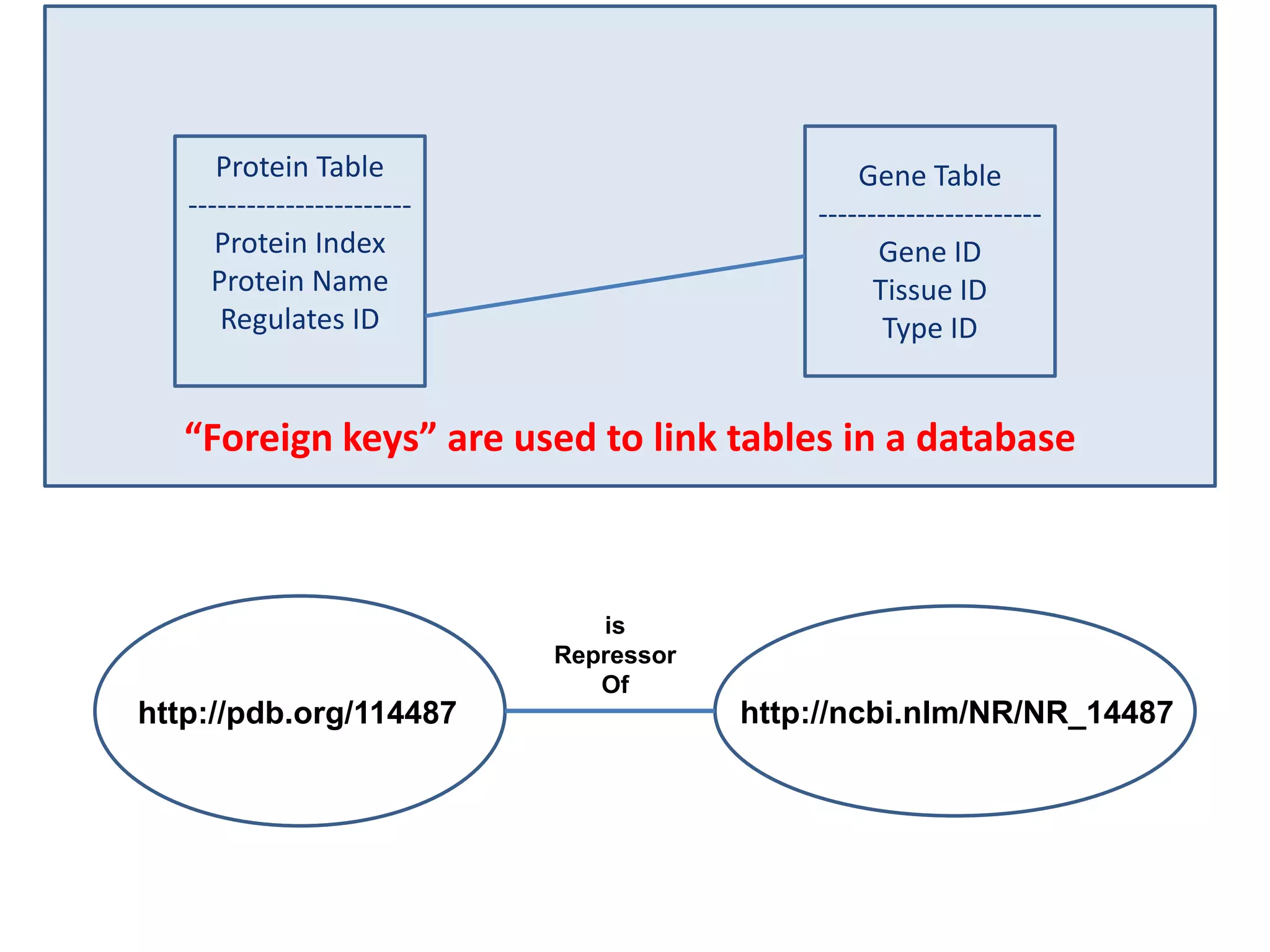
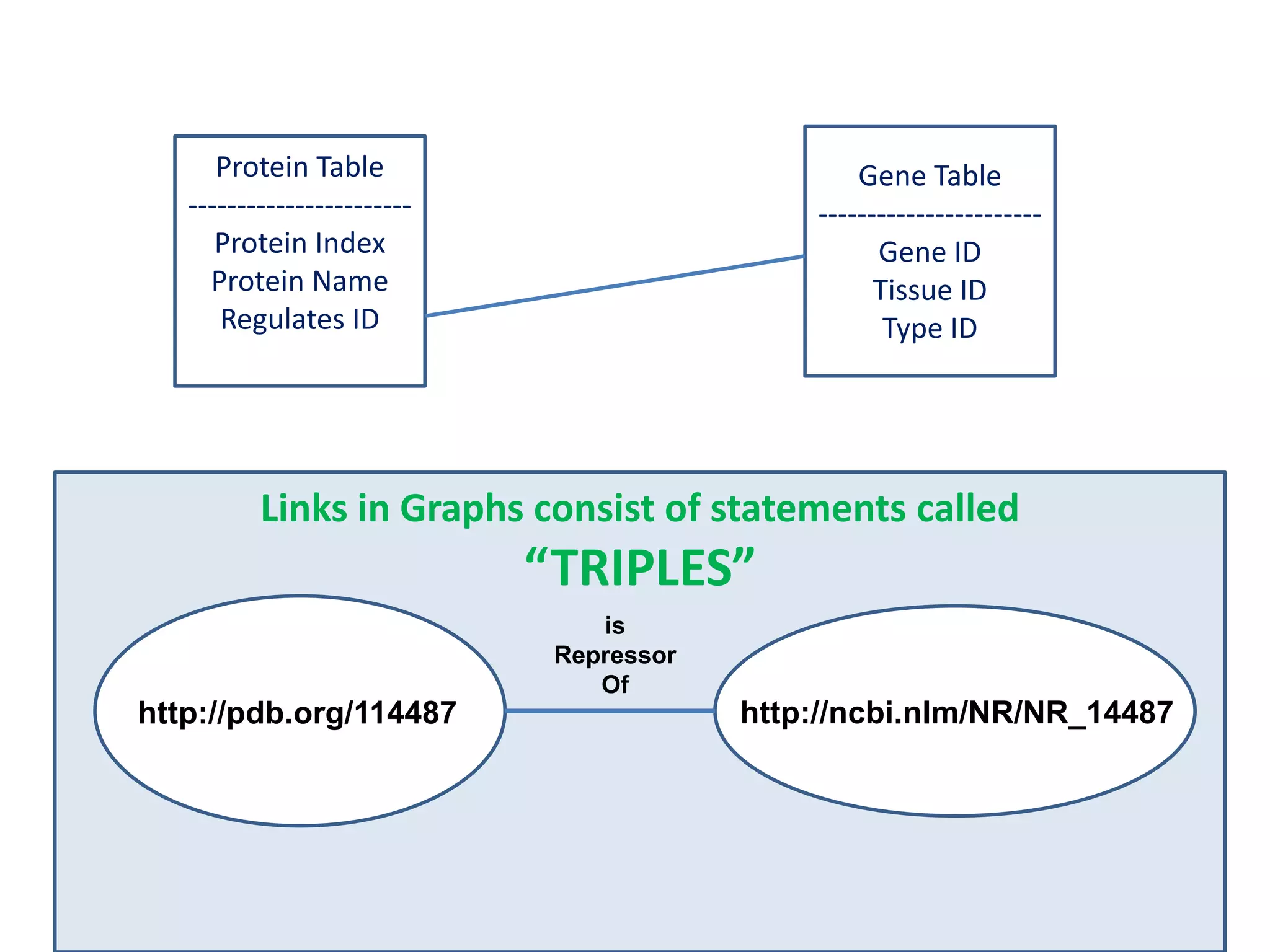
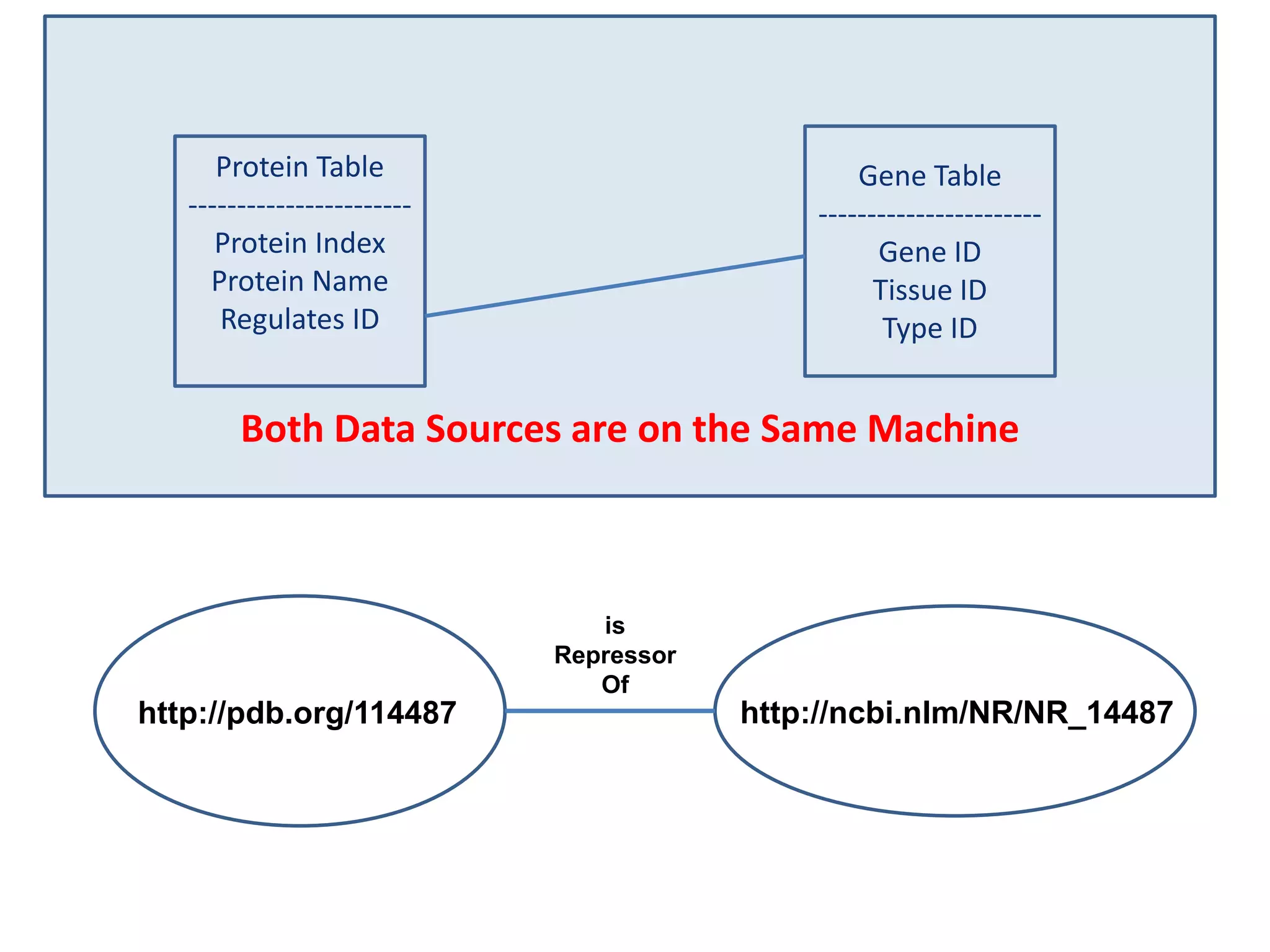
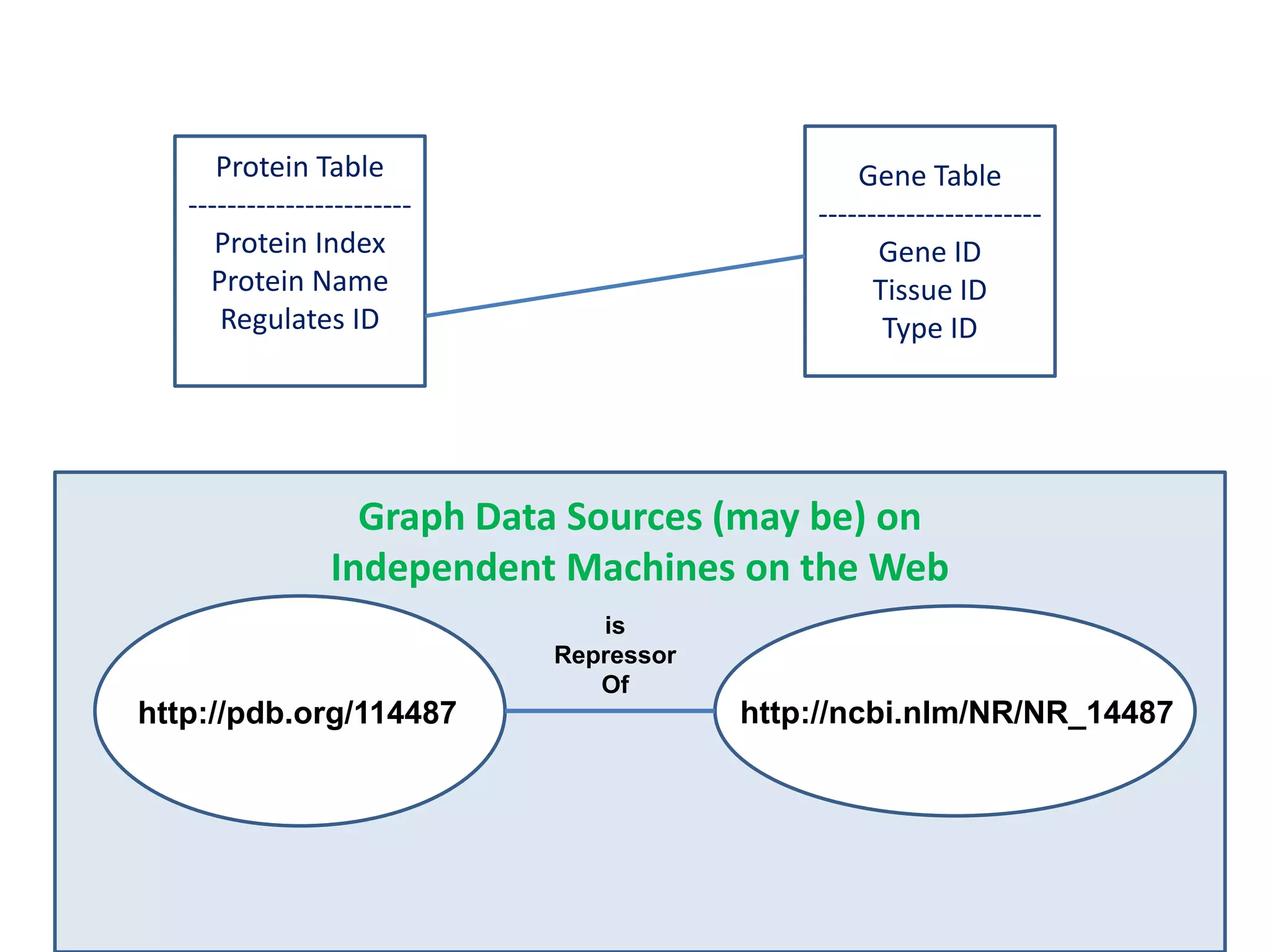
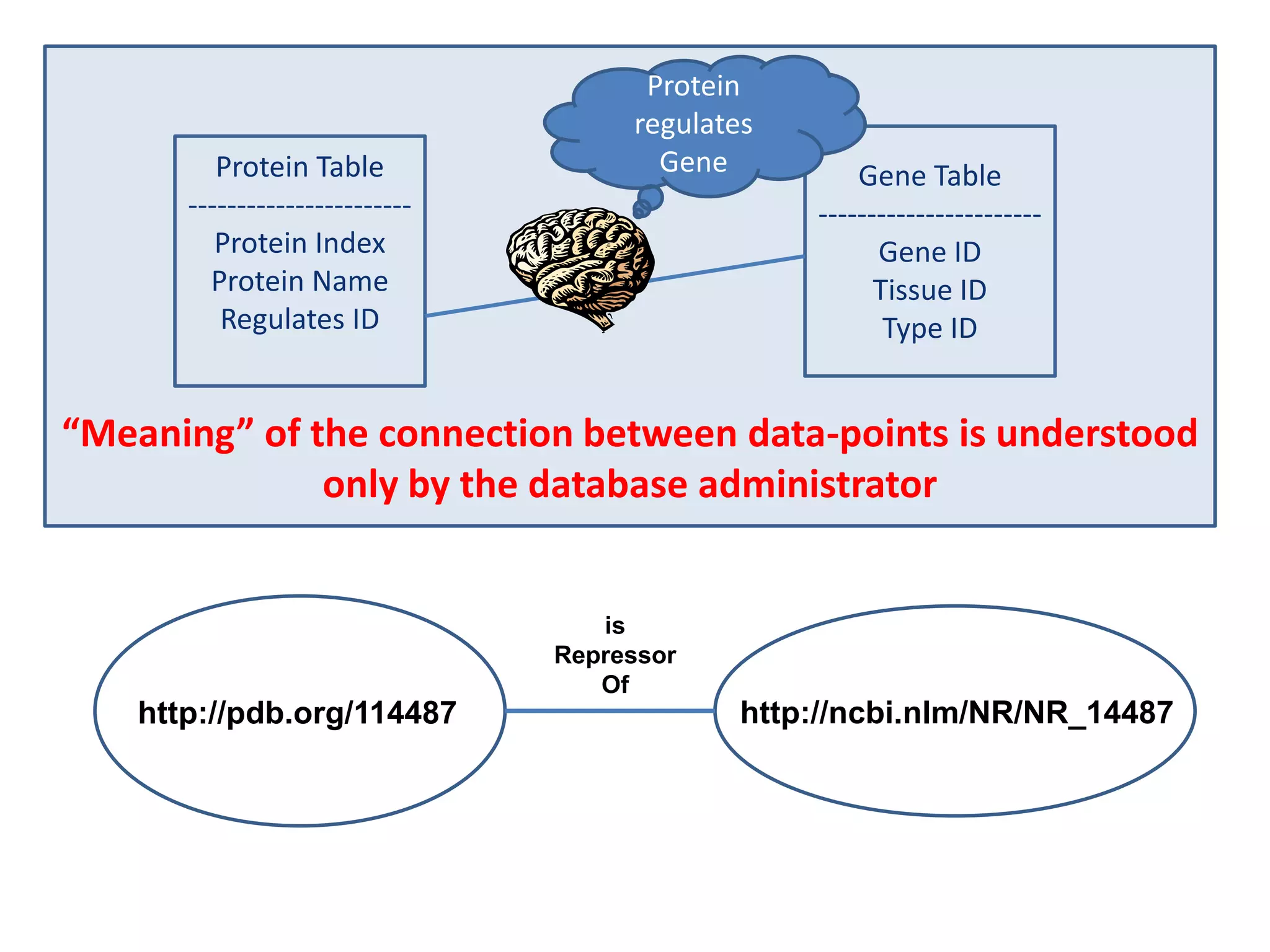
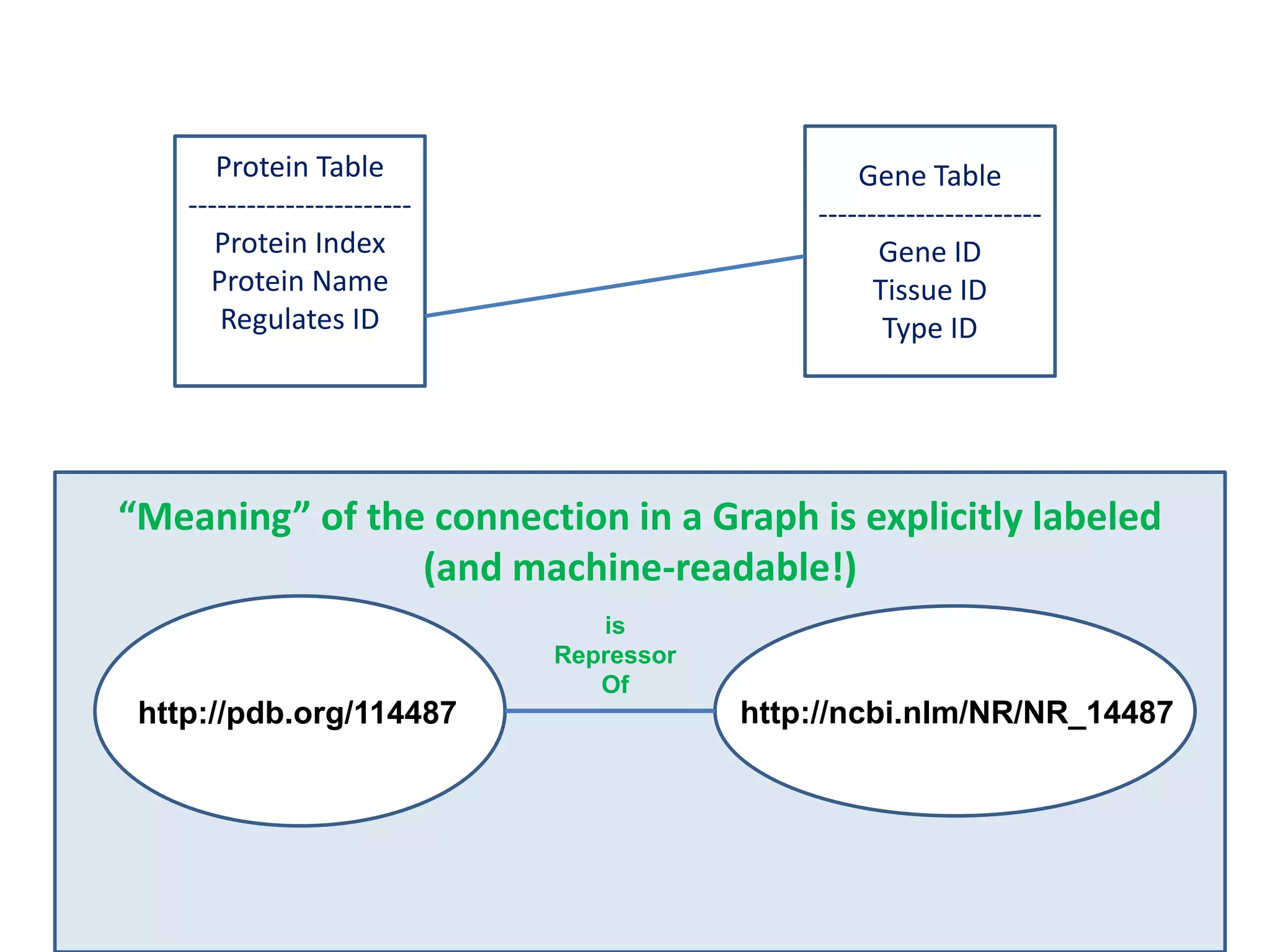



















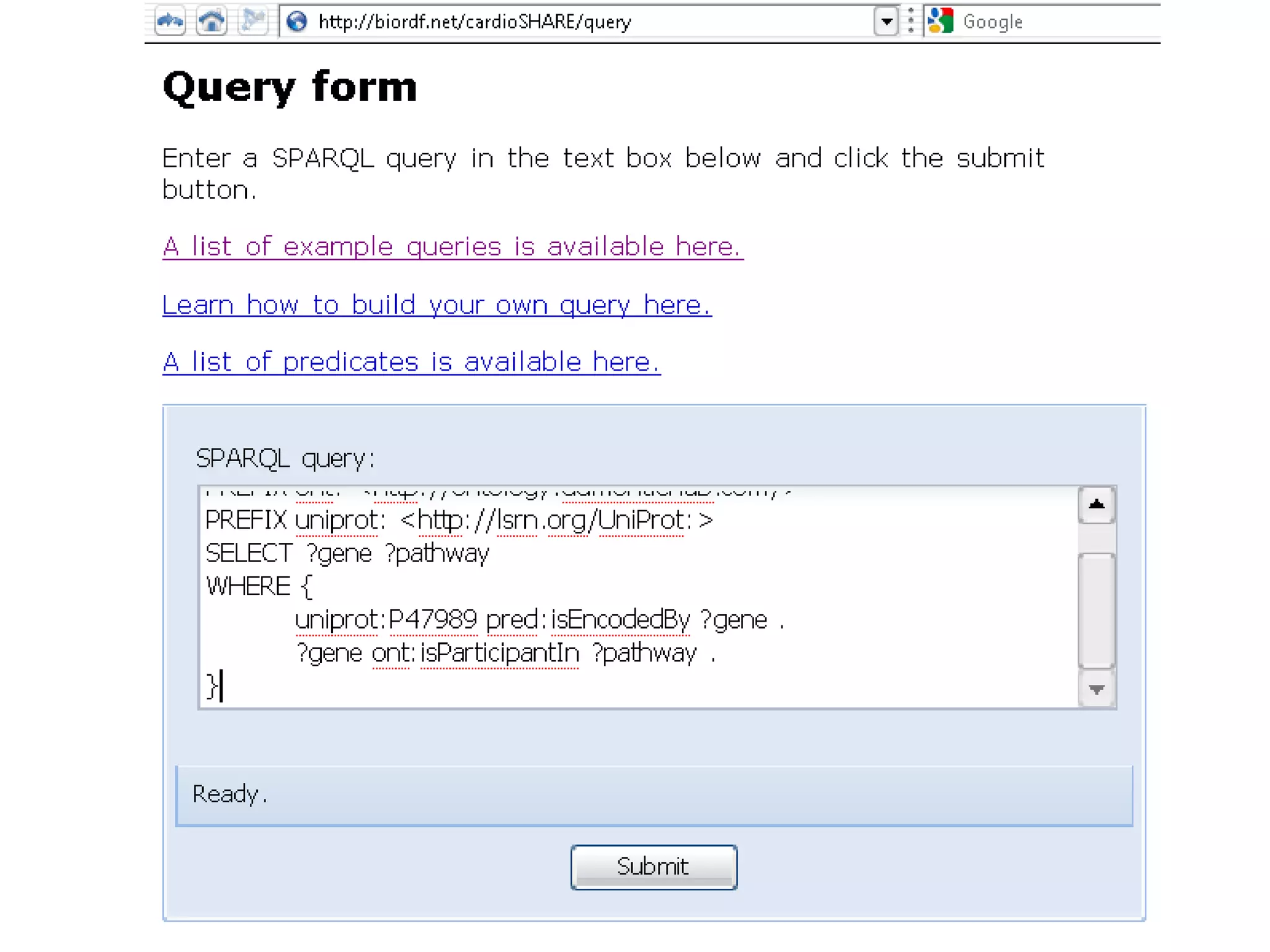
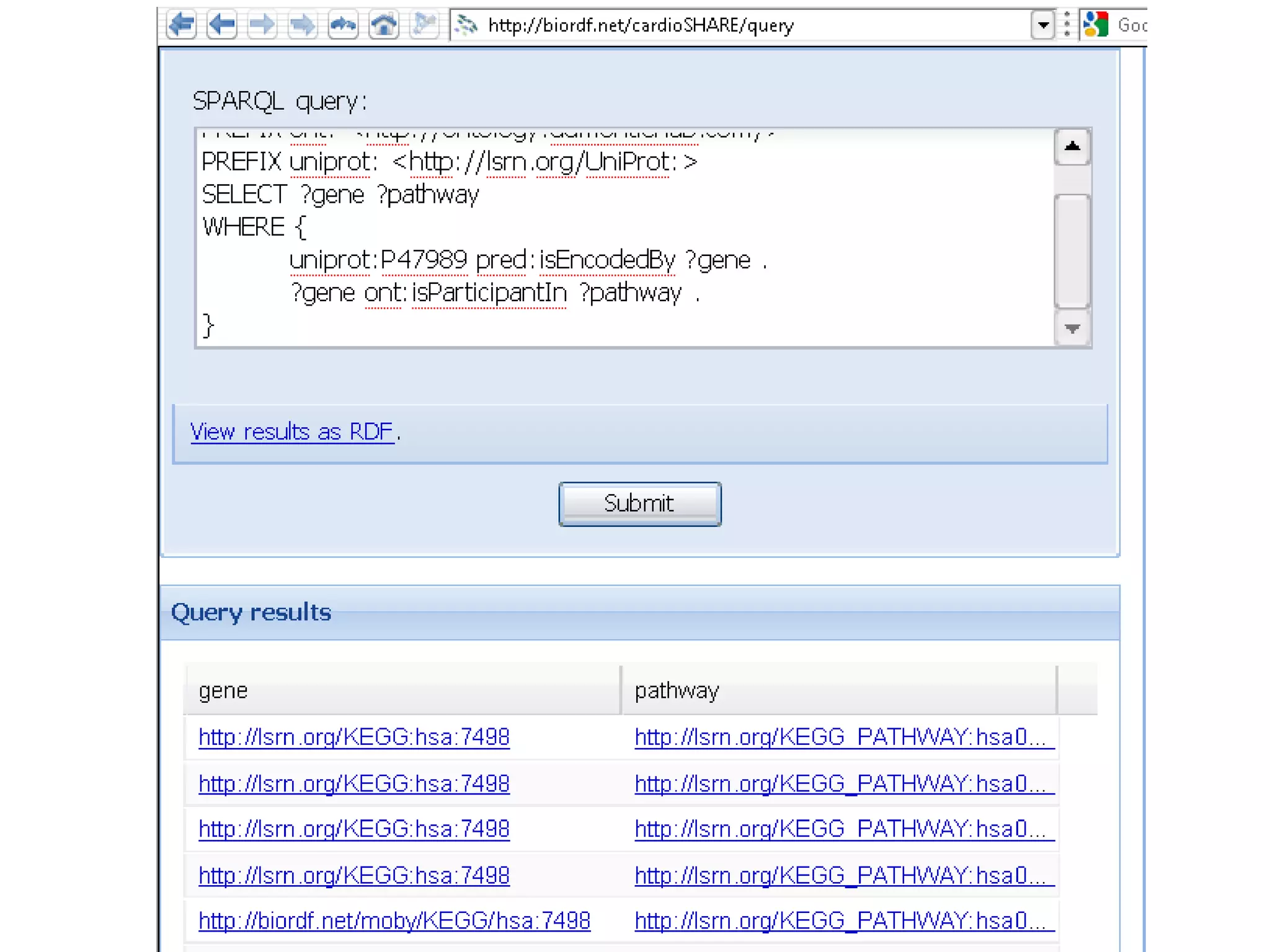





![Holy Grail Demo #1Align the promoters of all serine threonine kinases involved exclusively in the regulation of cell sorting during wound healing in blood vessels.Retrieve and align 2000nt 5' from every serine/threonine kinase in Mus musculus expressed exclusively in the tunica [I | M |A] whose expression increases 5X or more within 5 hours of wounding but is not activated during the normal development of blood vessels, and is <40% homologous in the active site to kinases known to be involved in cell-cycle regulation in any other species.](https://image.slidesharecdn.com/icapture-101010193247-phpapp01/75/The-Scientific-Method-on-the-Semantic-Web-52-2048.jpg)





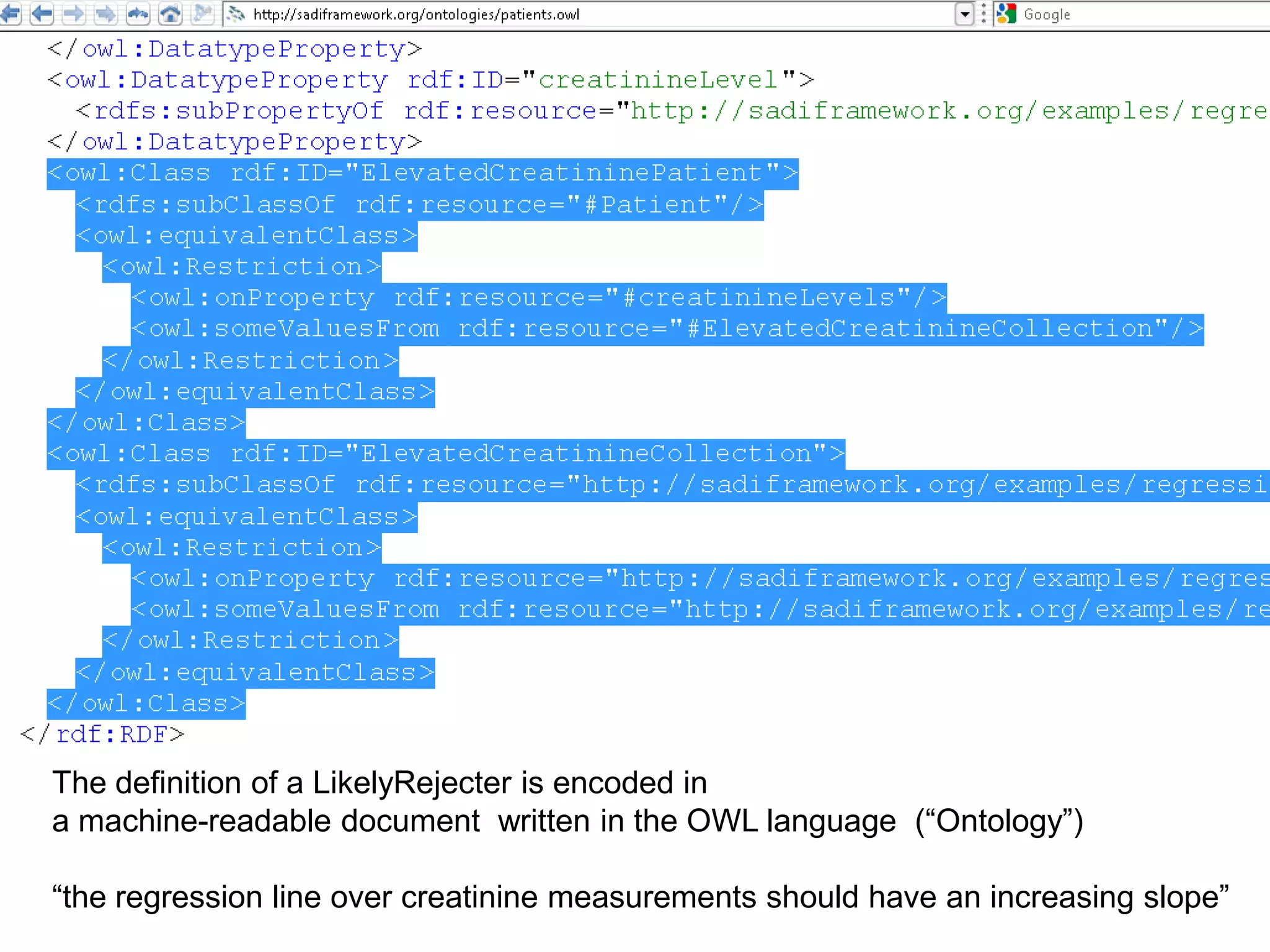
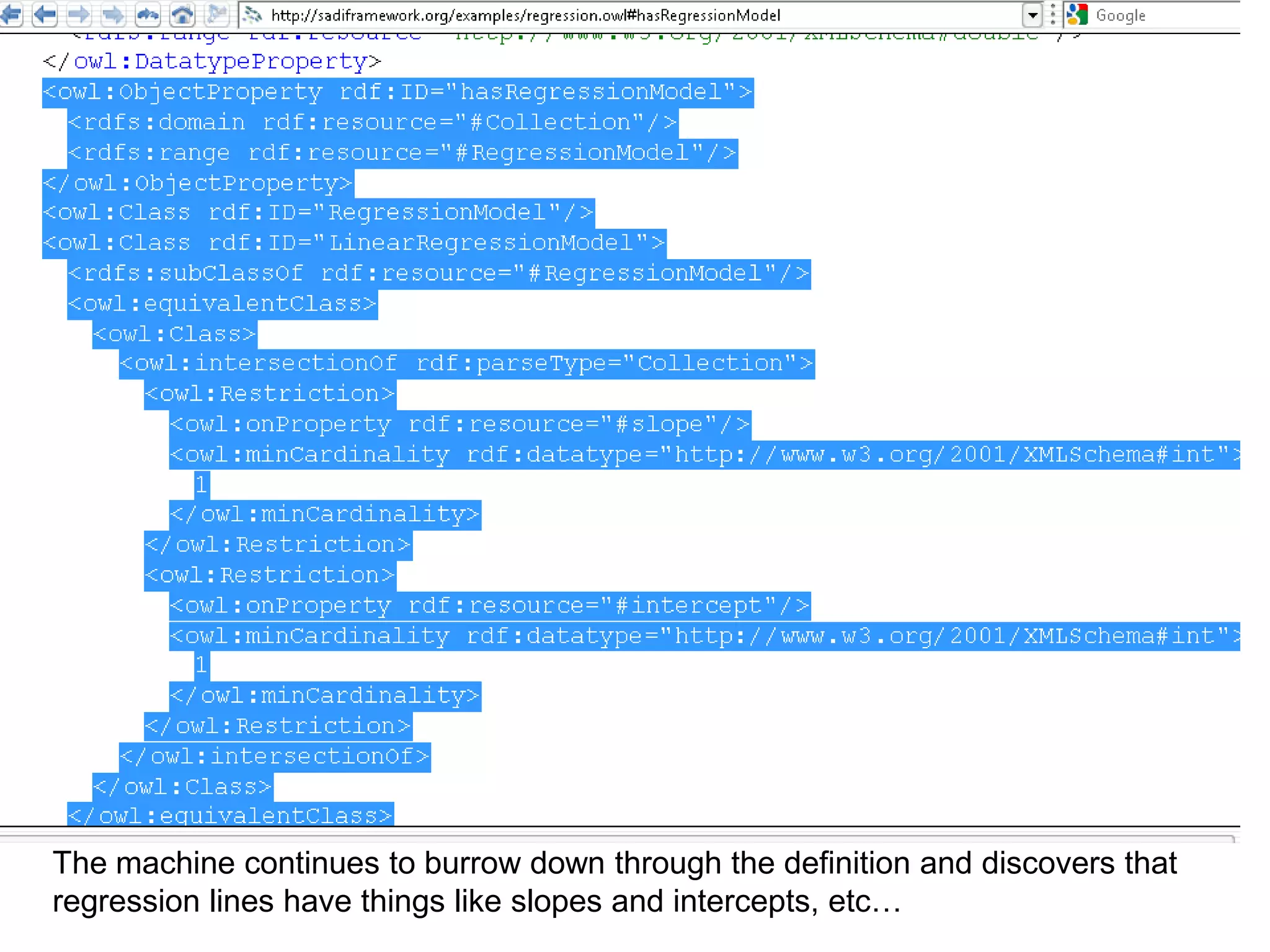


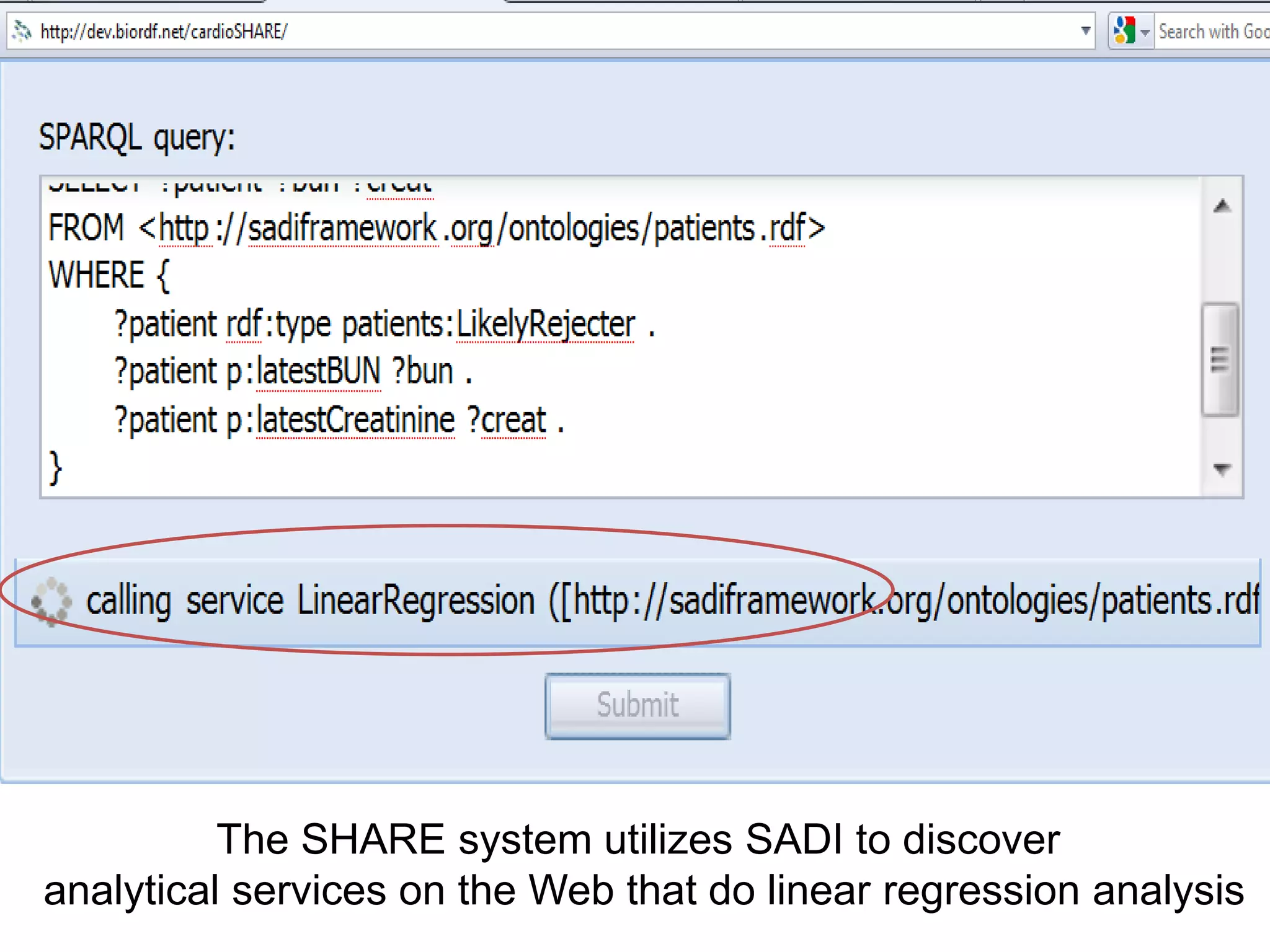
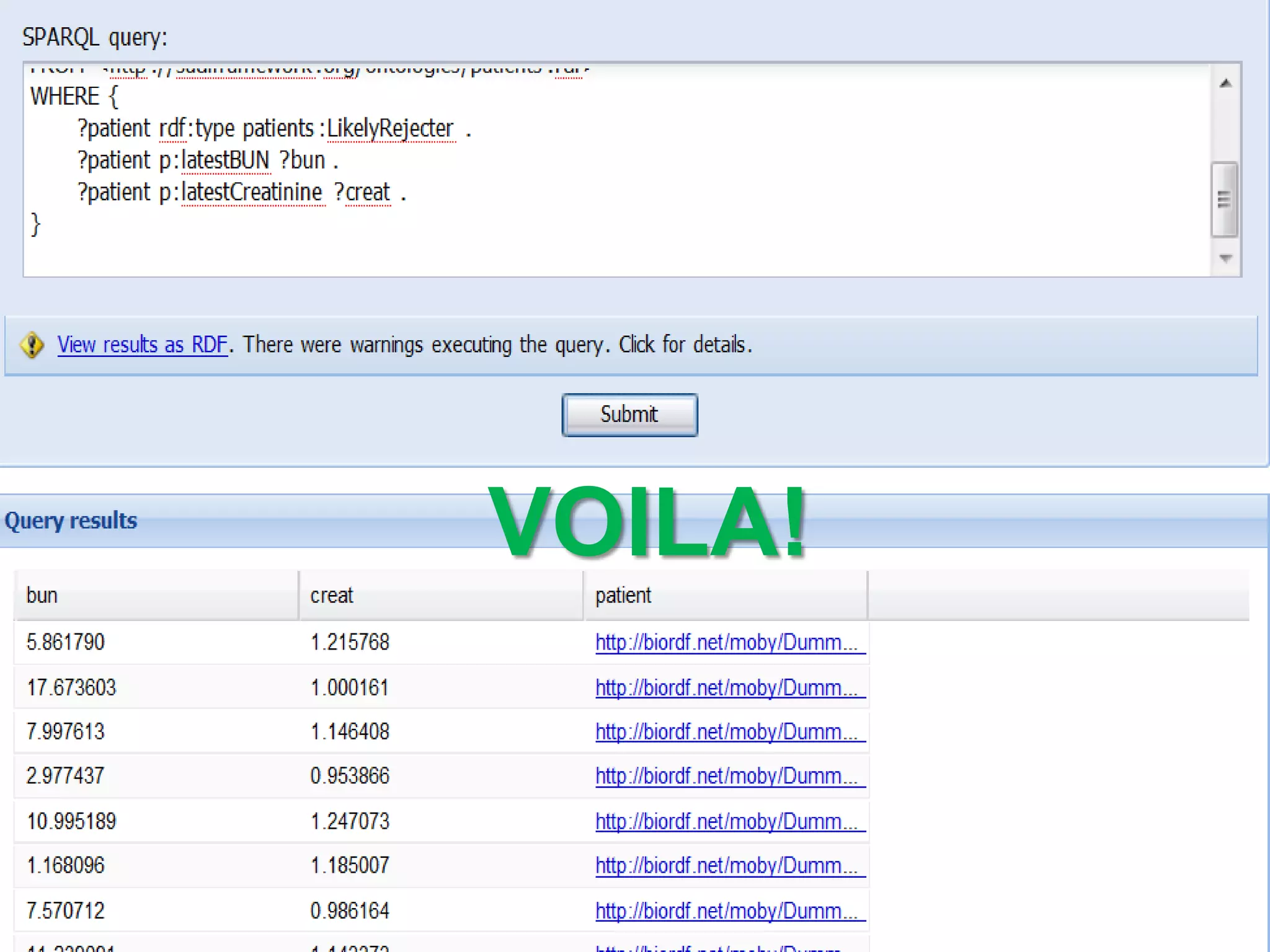








![Holy Grail Demo #2Align the promoters of all serine threonine kinases involved exclusively in the regulation of cell sorting during wound healing in blood vessels.Retrieve and align 2000nt 5' from every serine/threonine kinase in Mus musculus expressed exclusively in the tunica [I | M |A] whose expression increases 5X or more within 5 hours of wounding but is not activated during the normal development of blood vessels, and is <40% homologous in the active site to kinases known to be involved in cell-cycle regulation in any other species.](https://image.slidesharecdn.com/icapture-101010193247-phpapp01/75/The-Scientific-Method-on-the-Semantic-Web-75-2048.jpg)