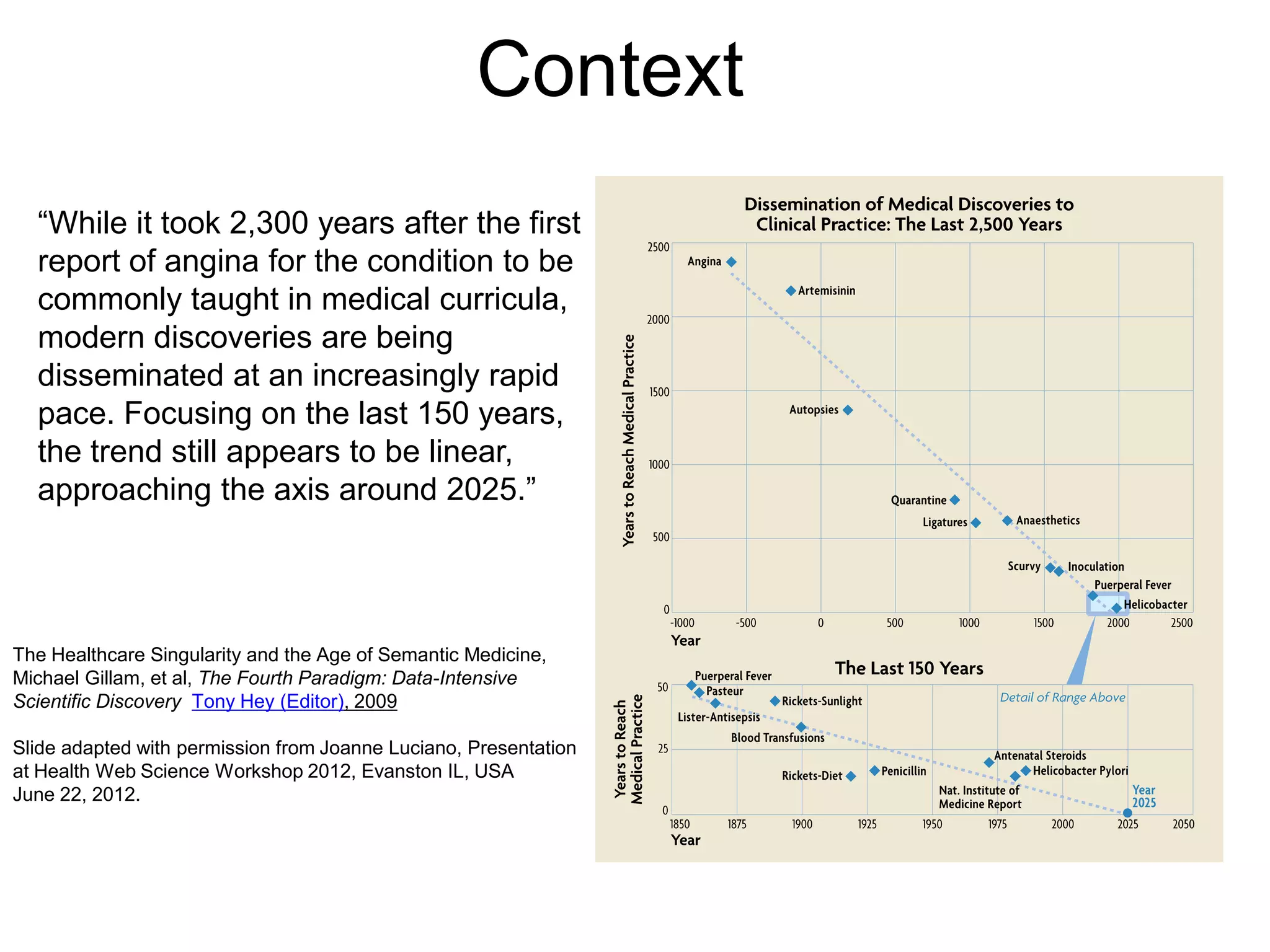
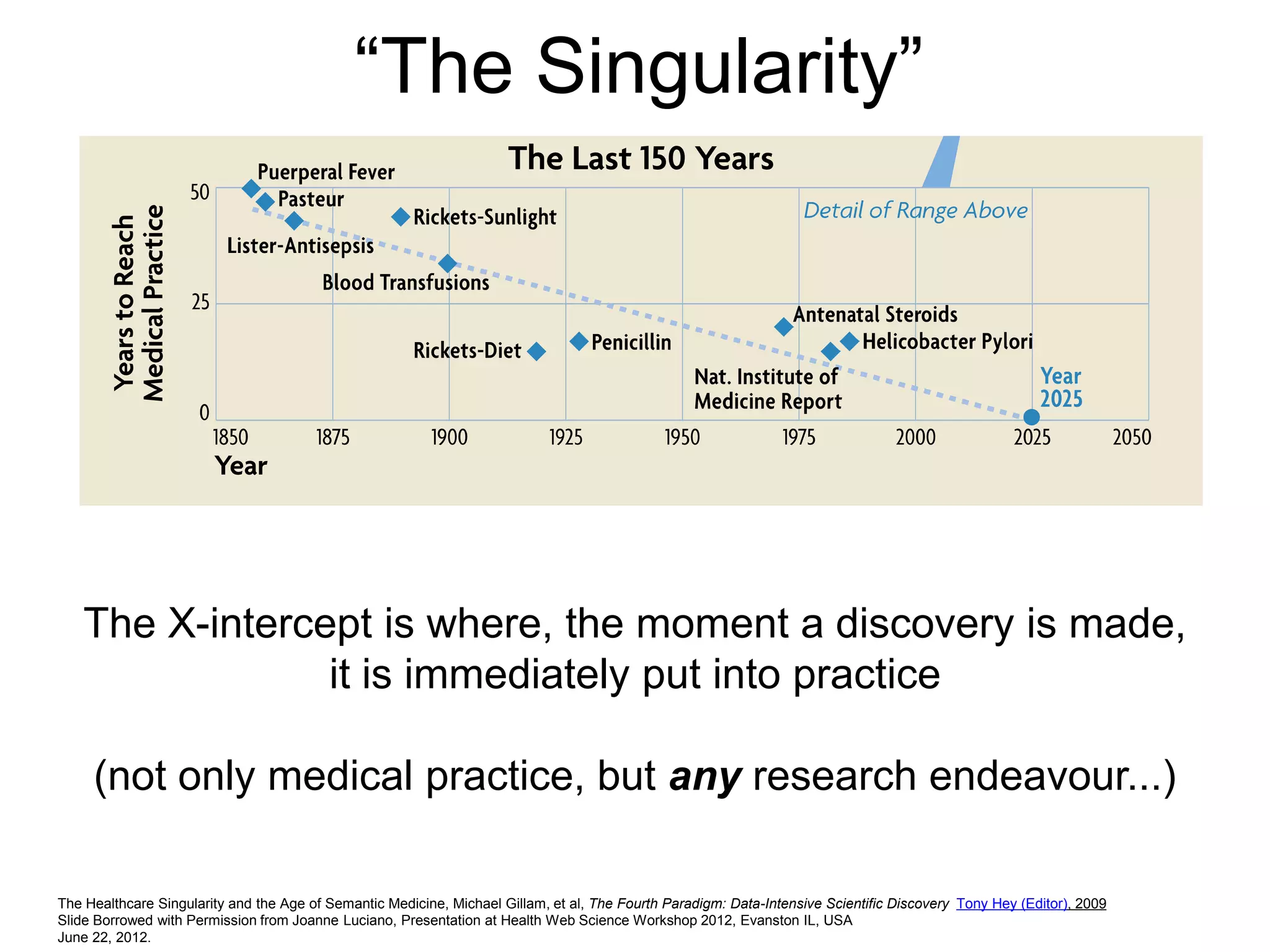
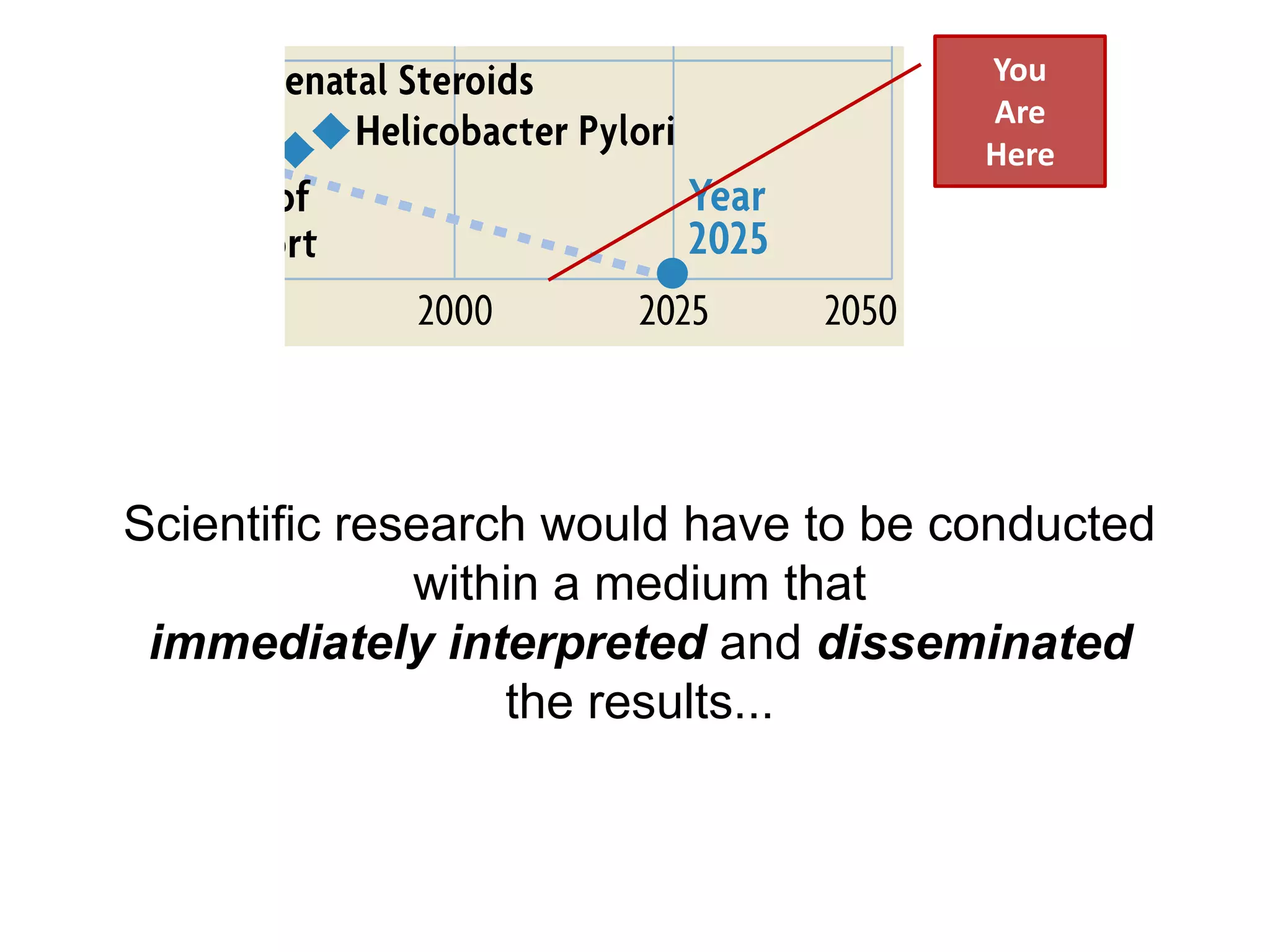
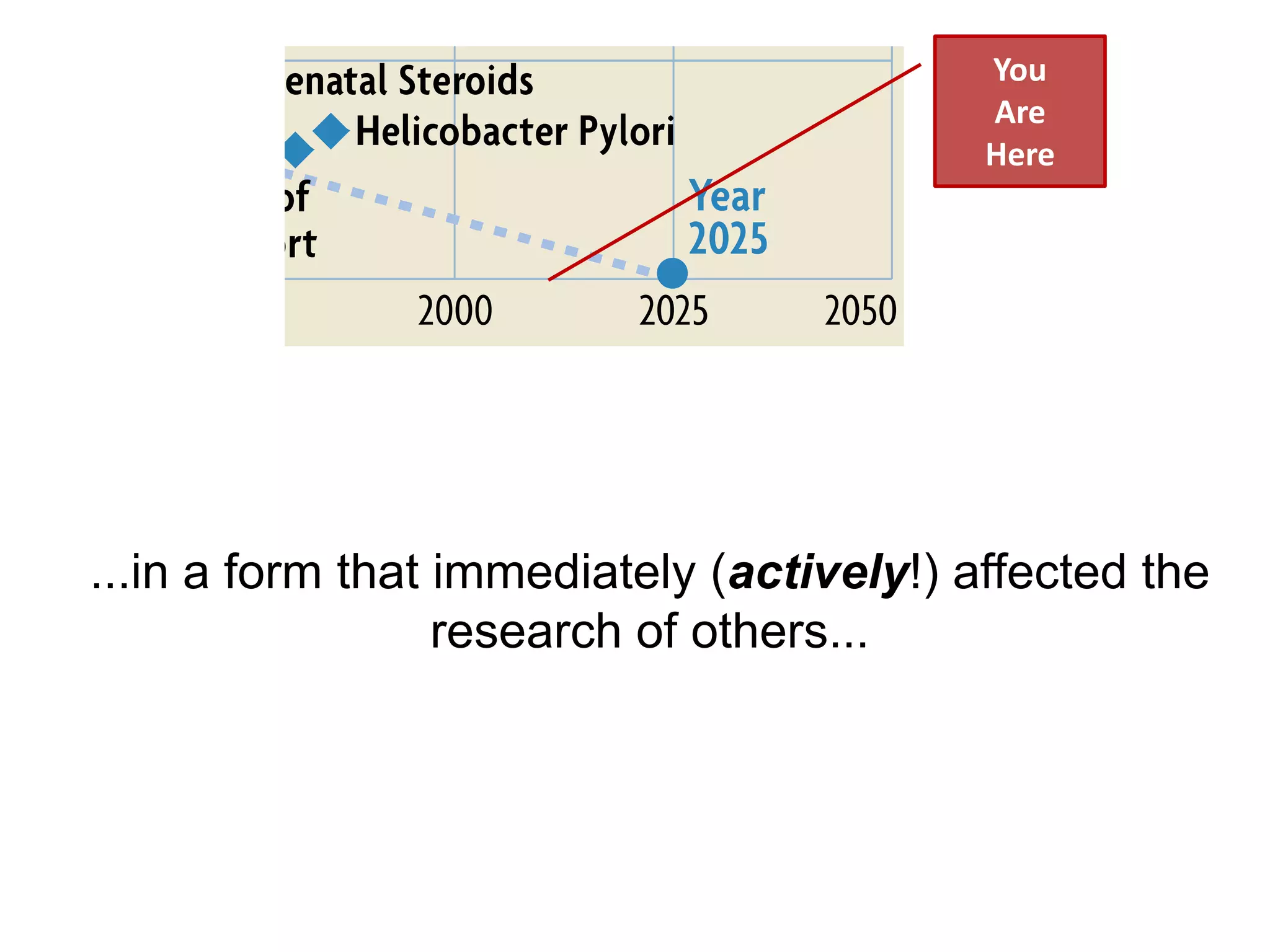
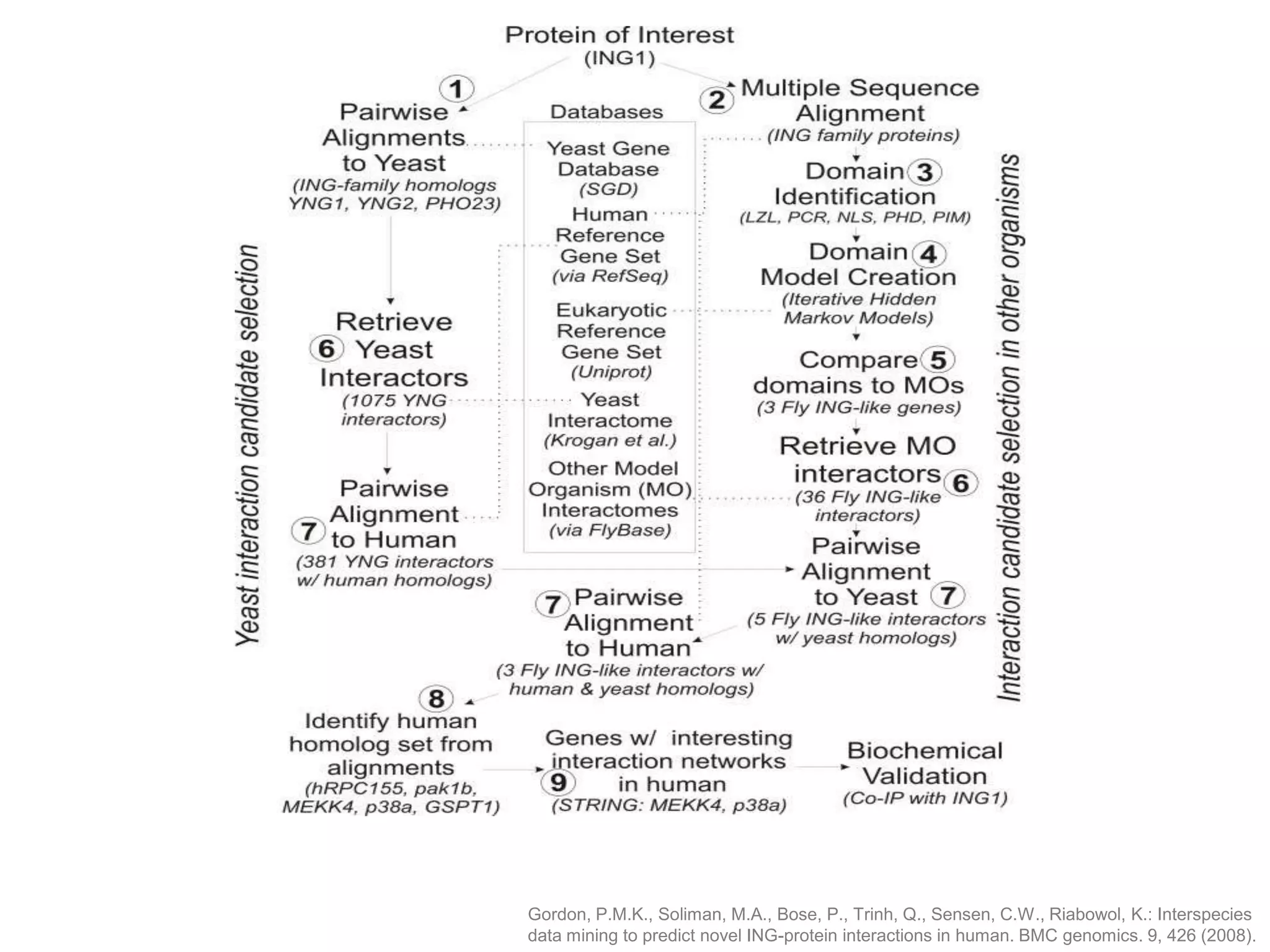
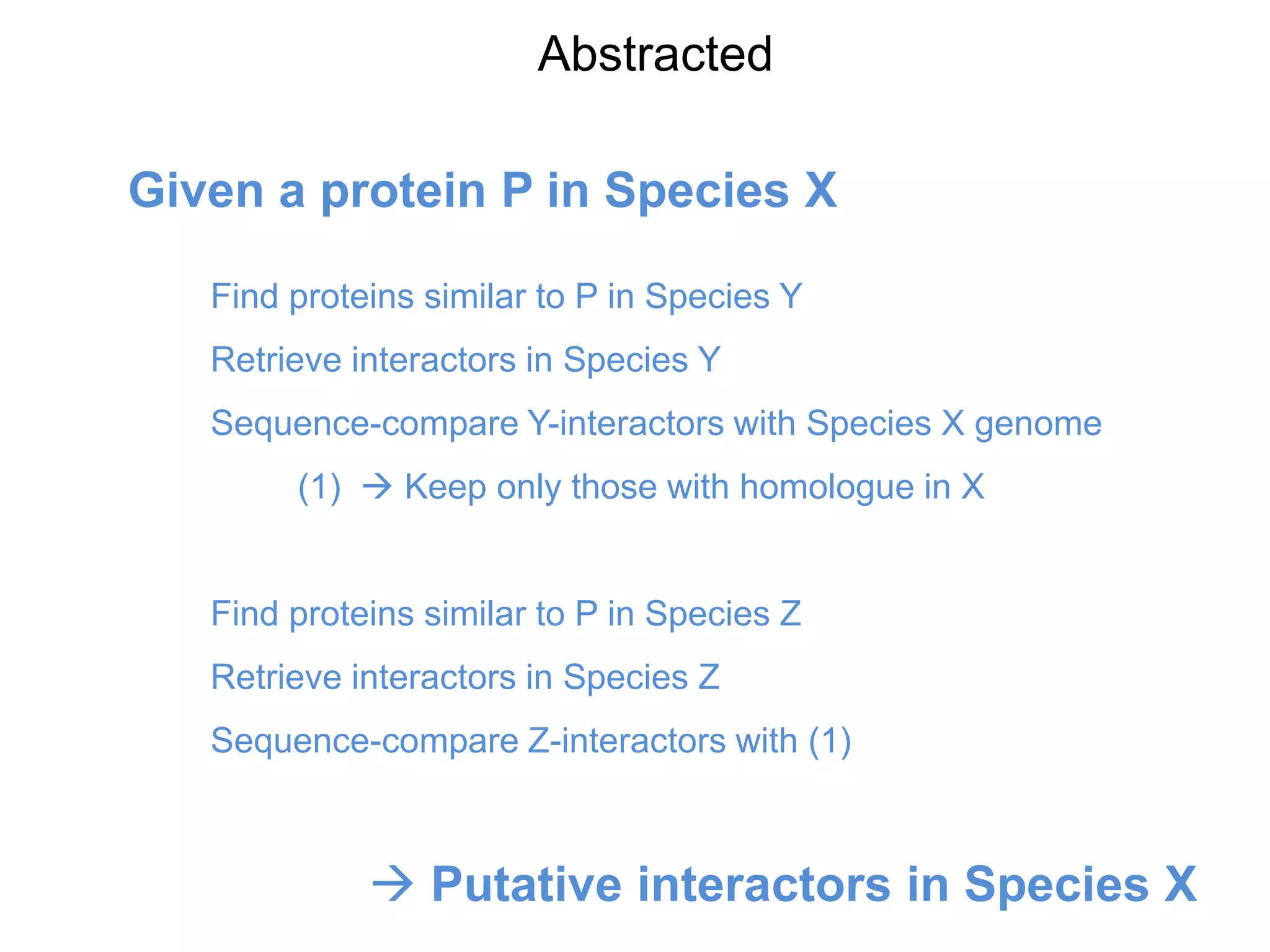
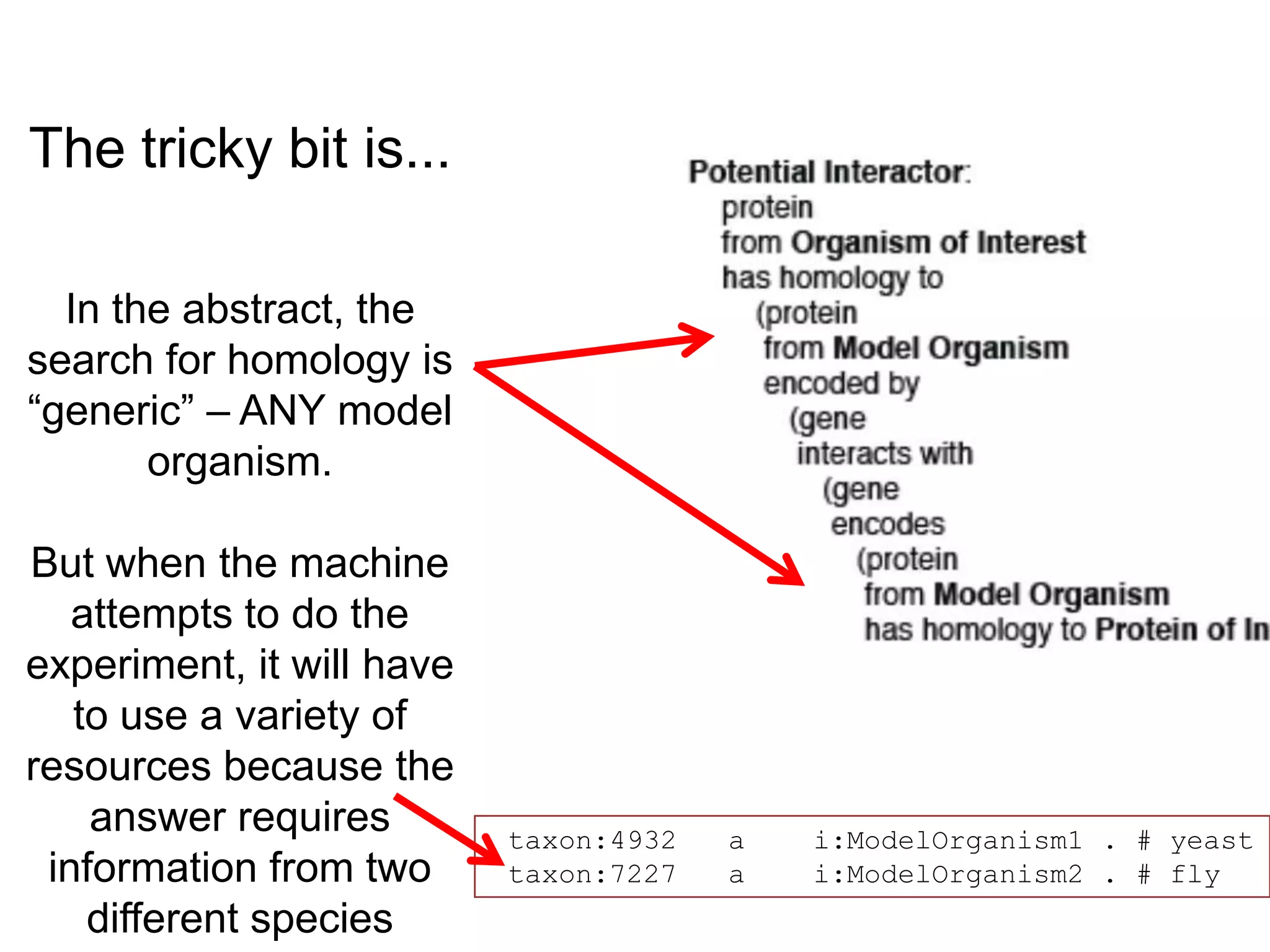
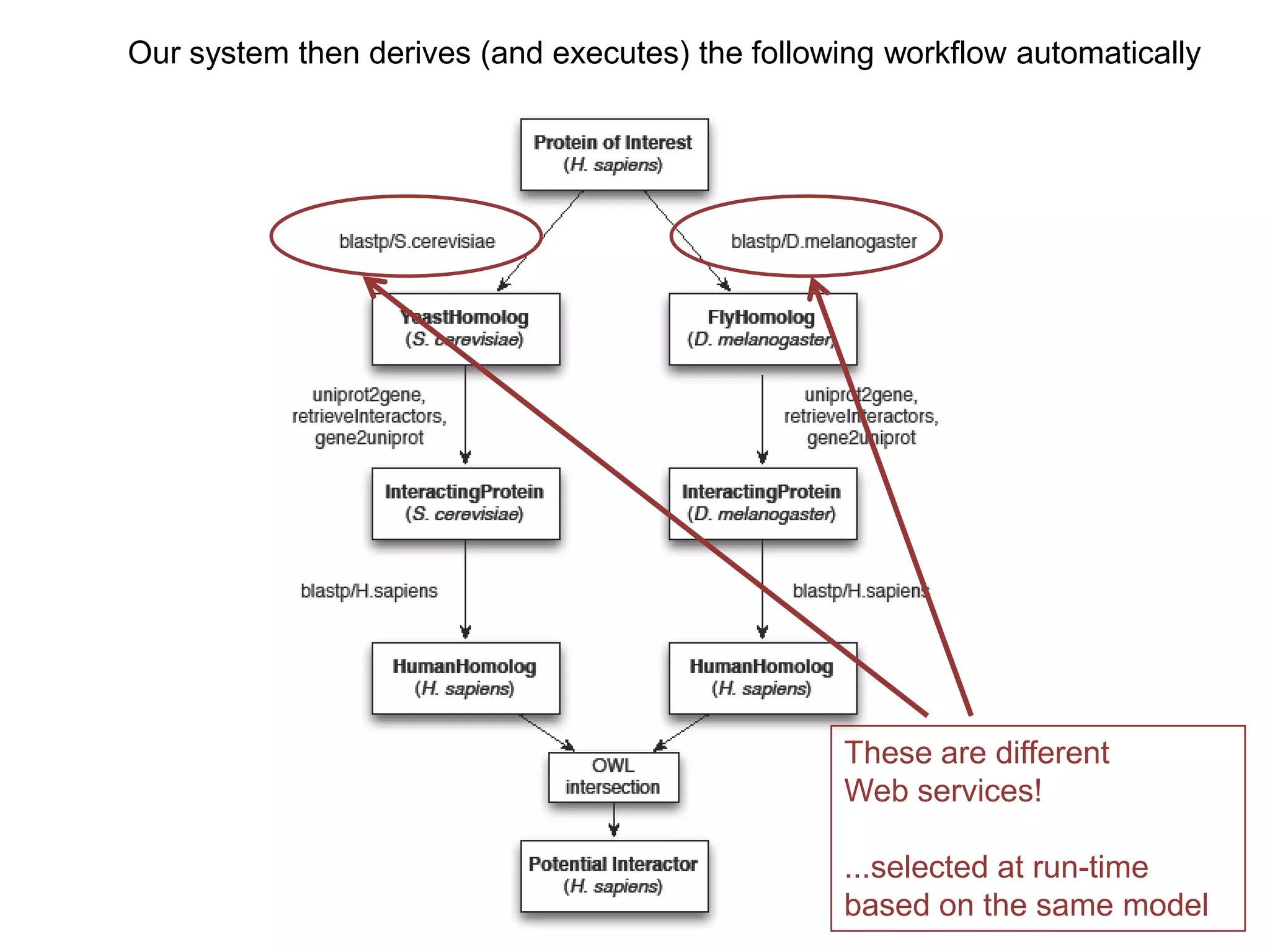
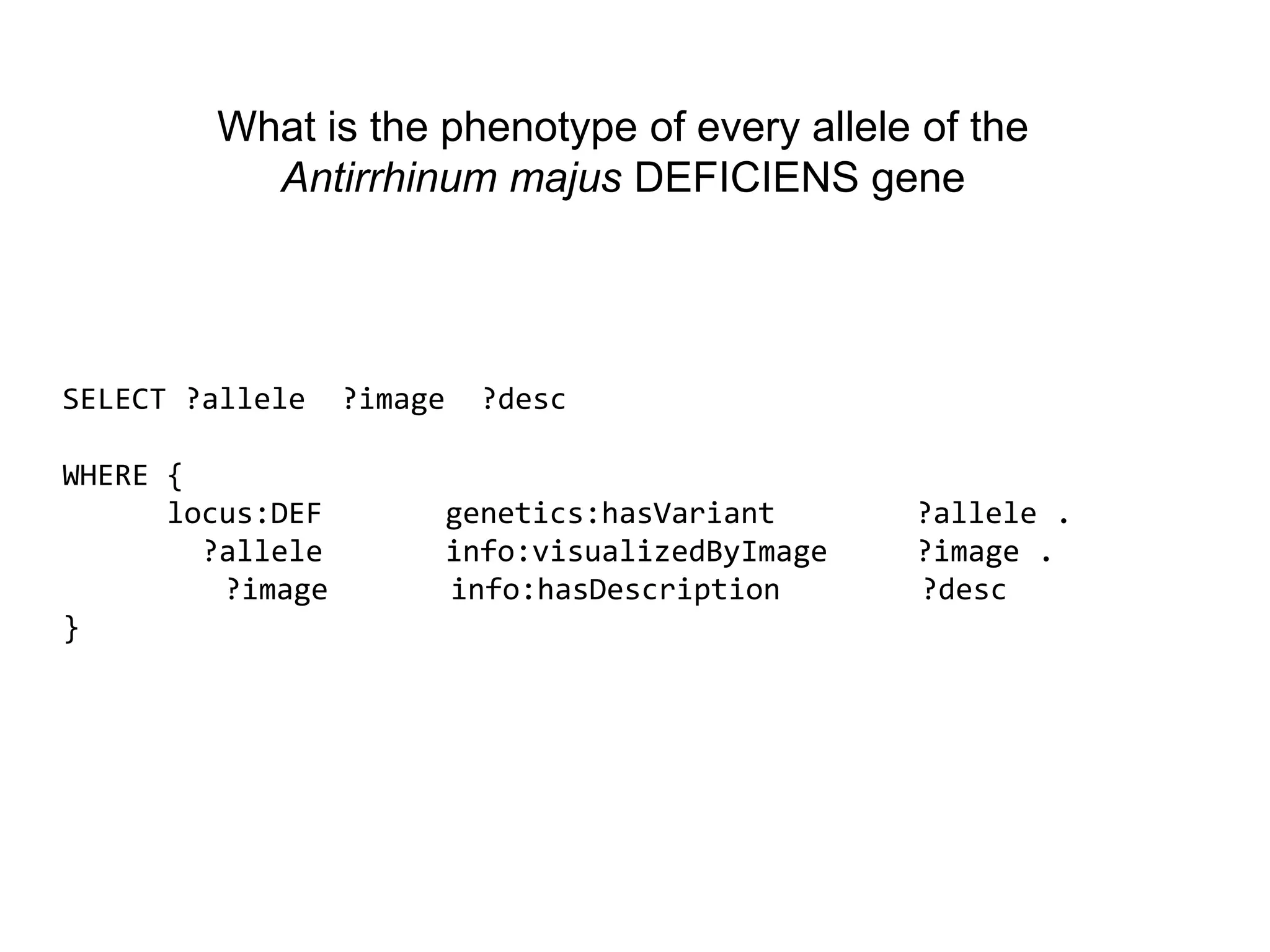
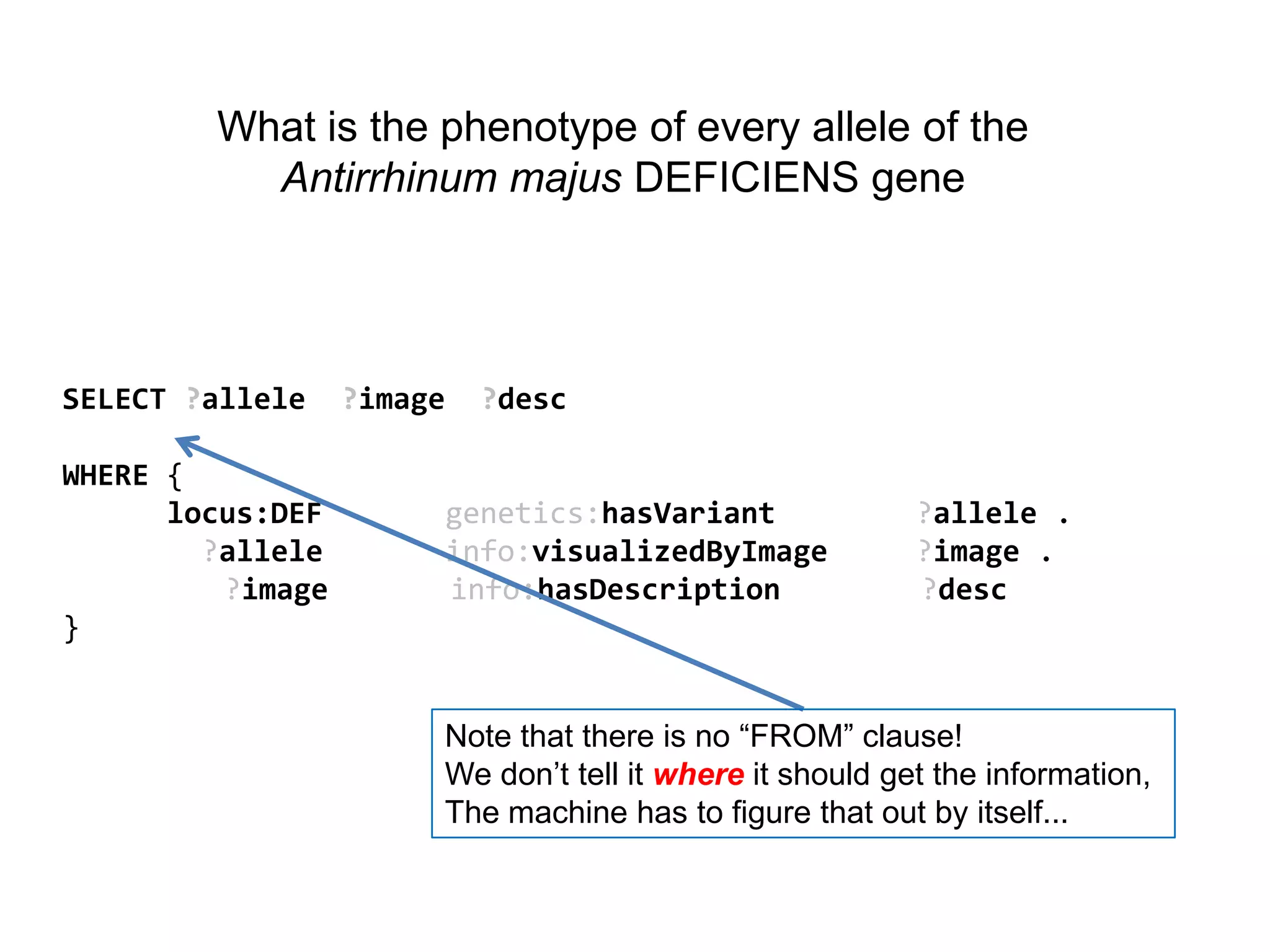
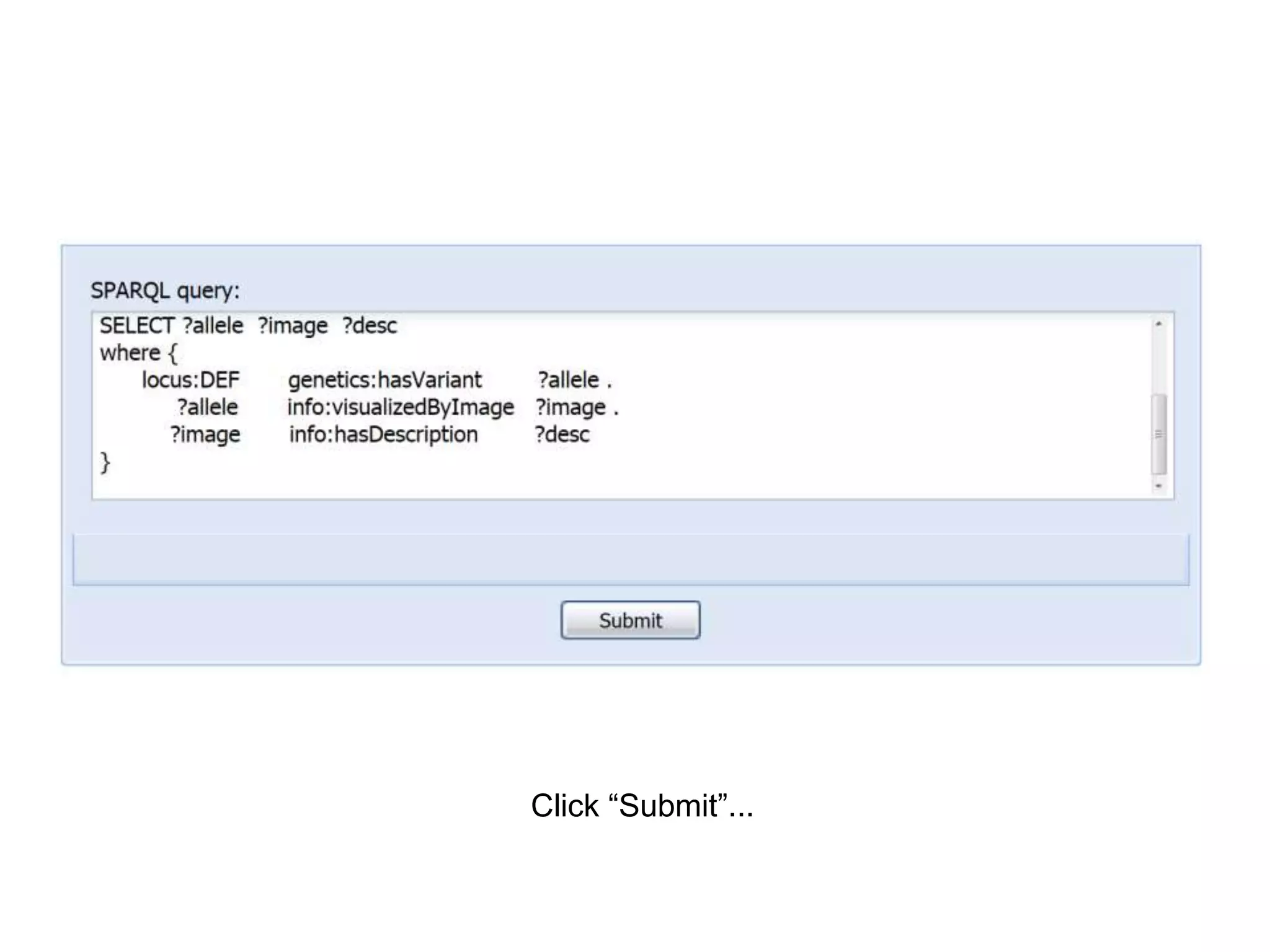
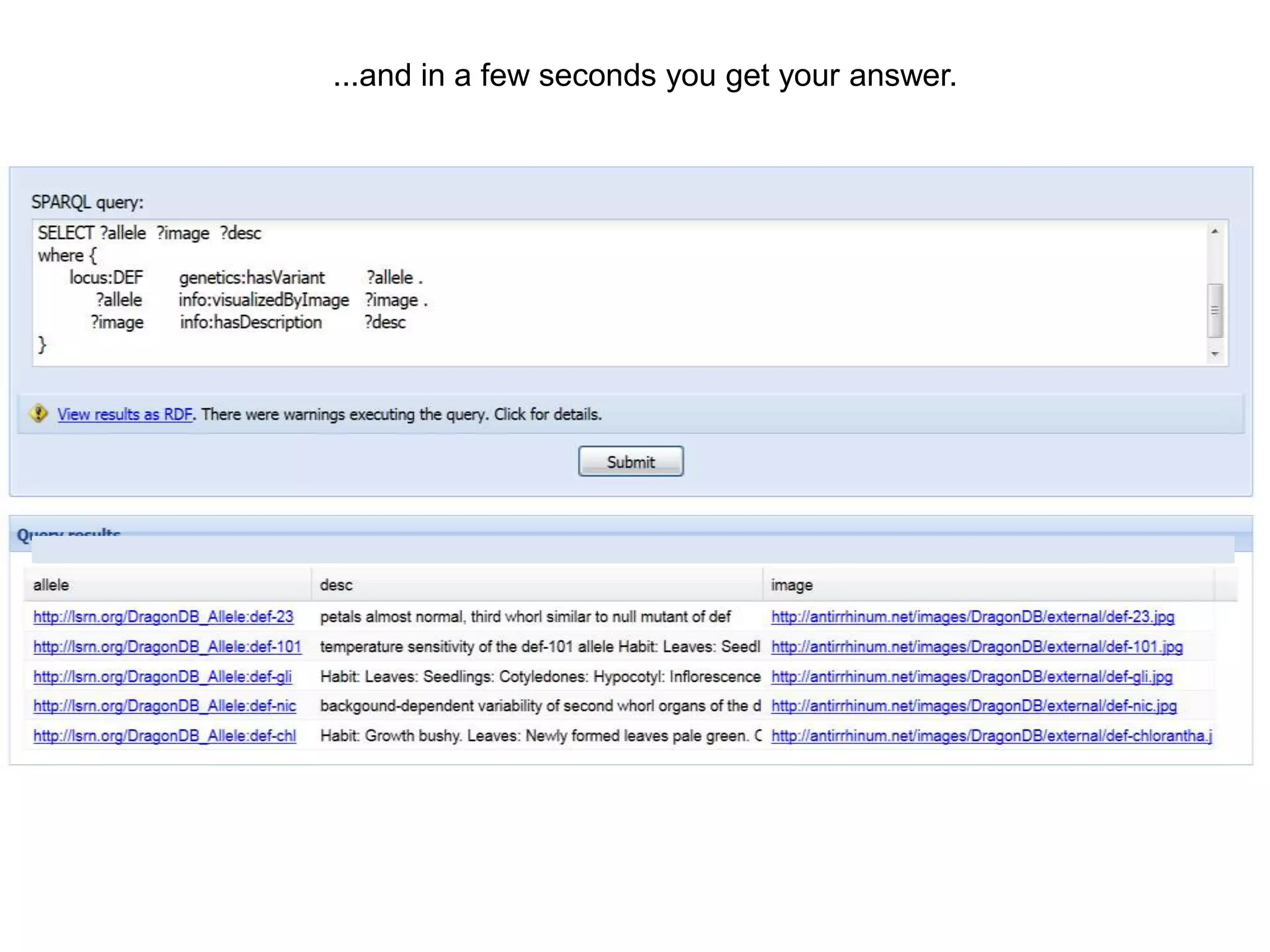
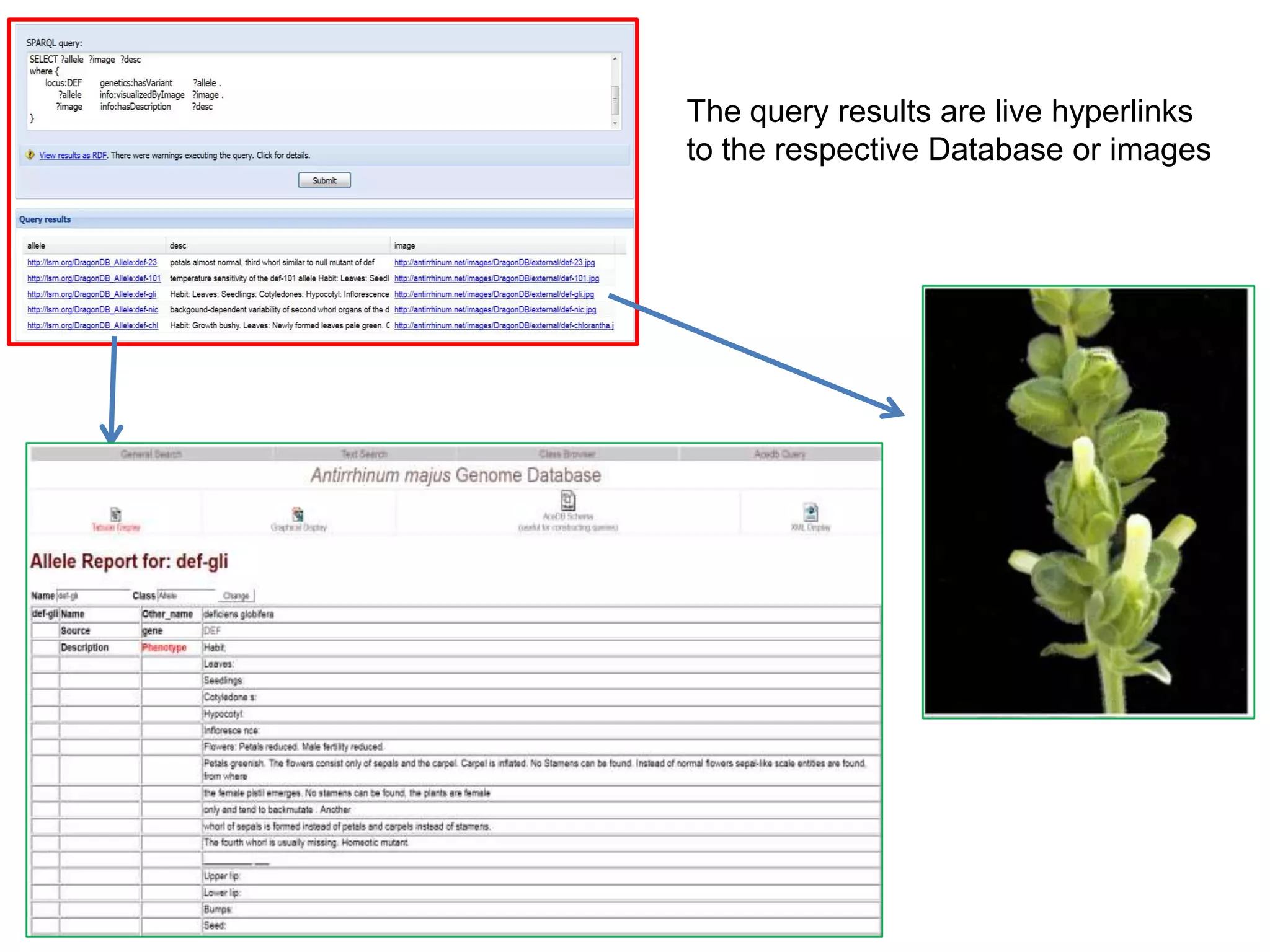
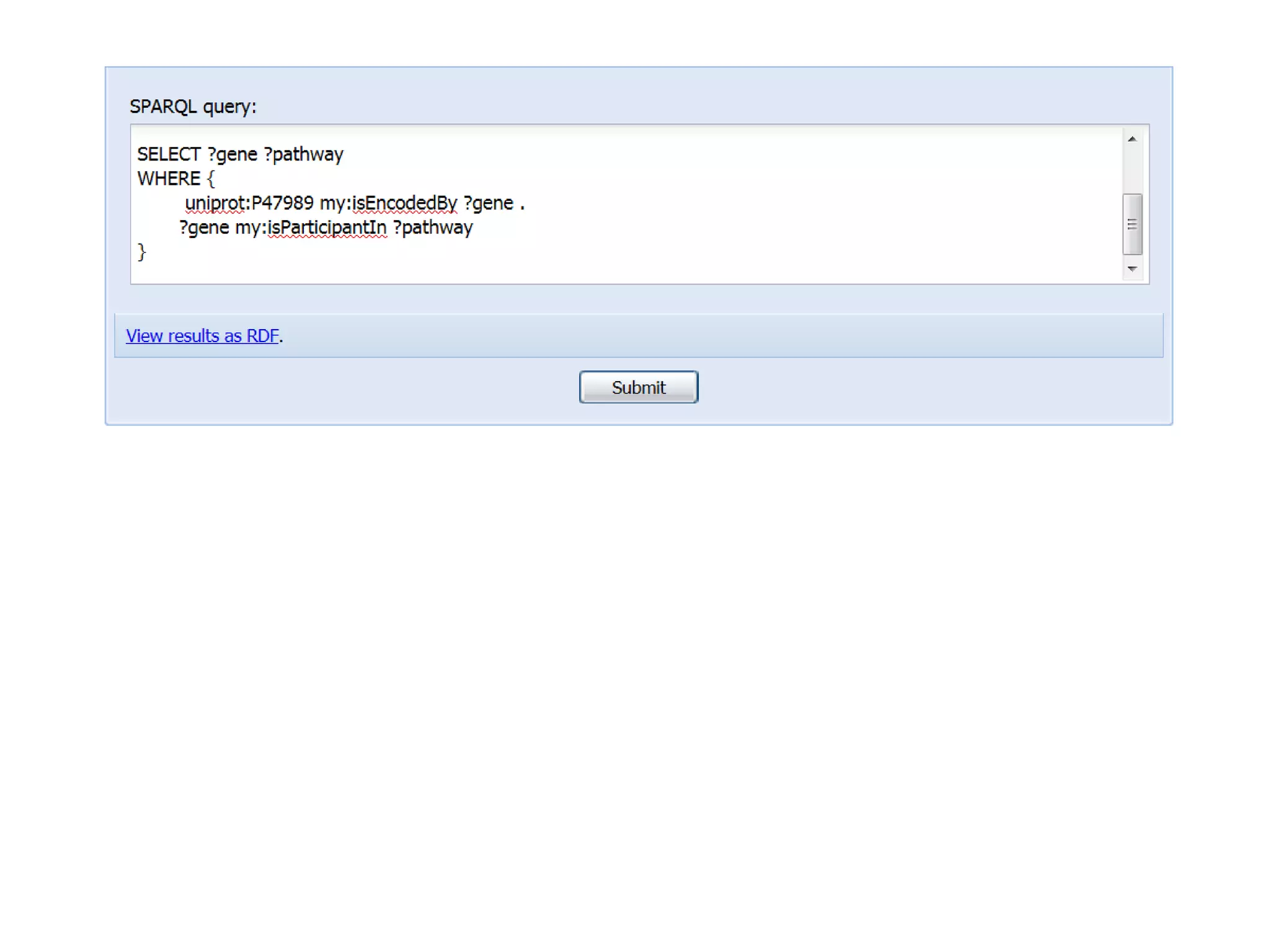
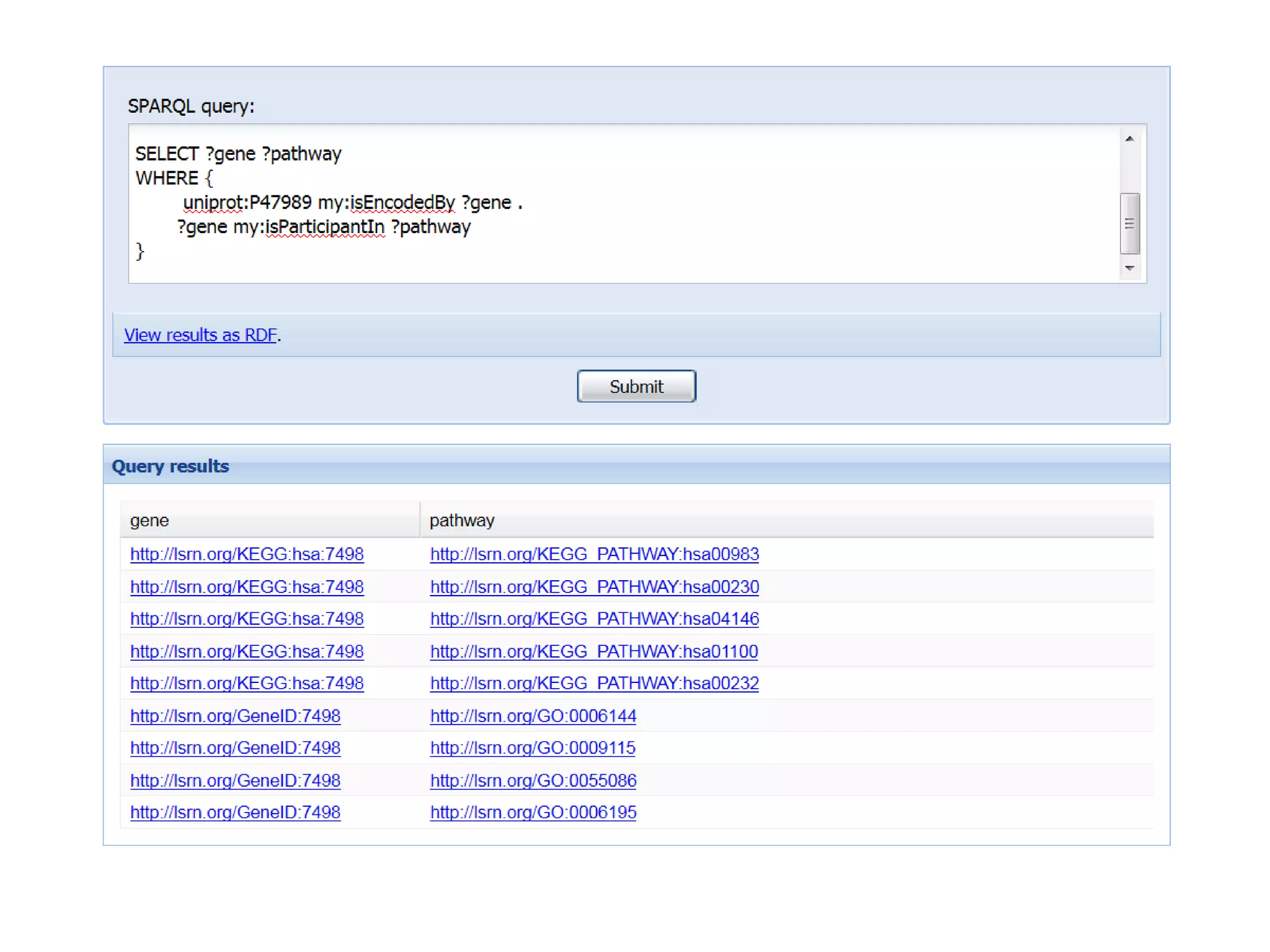
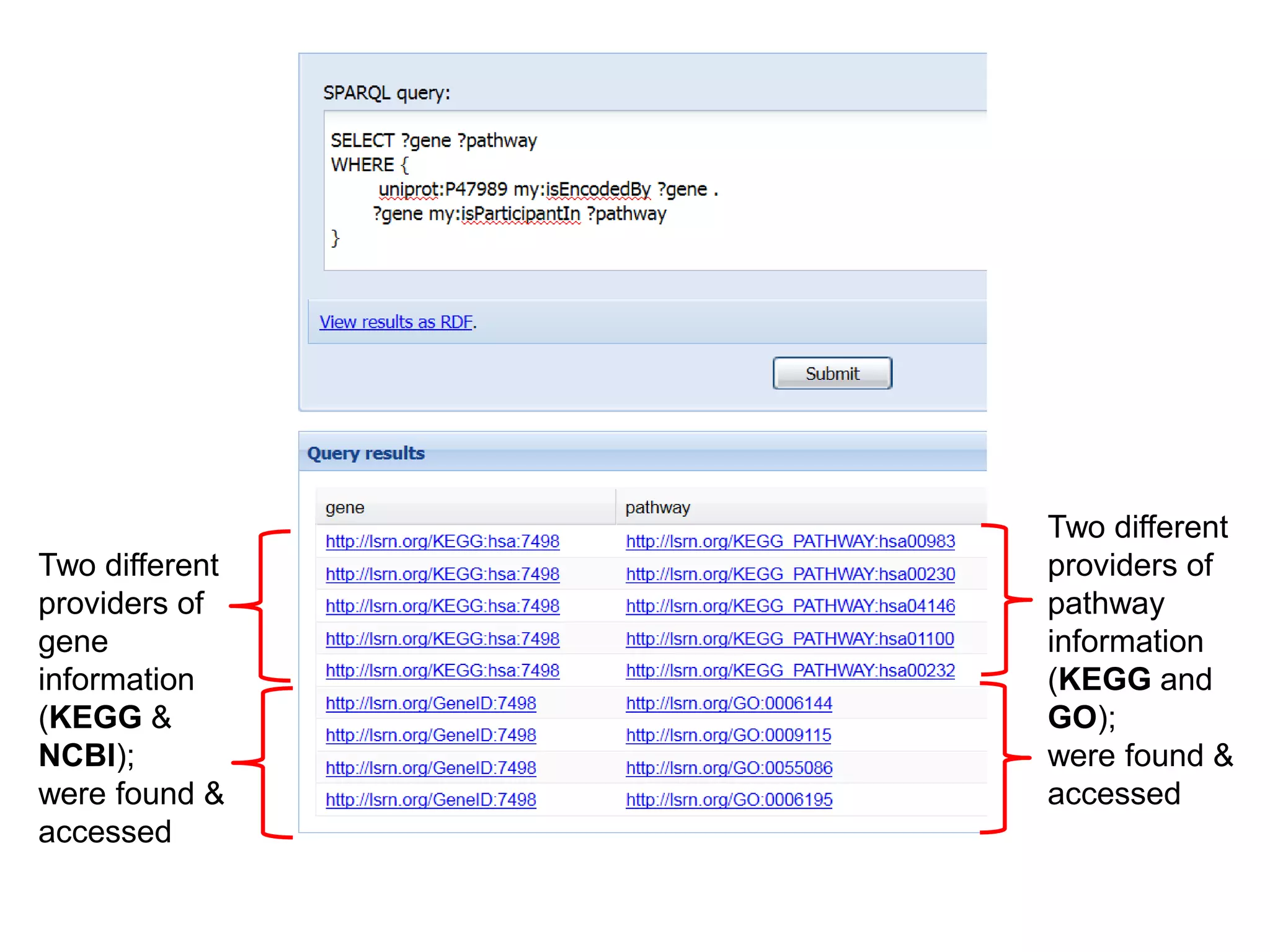
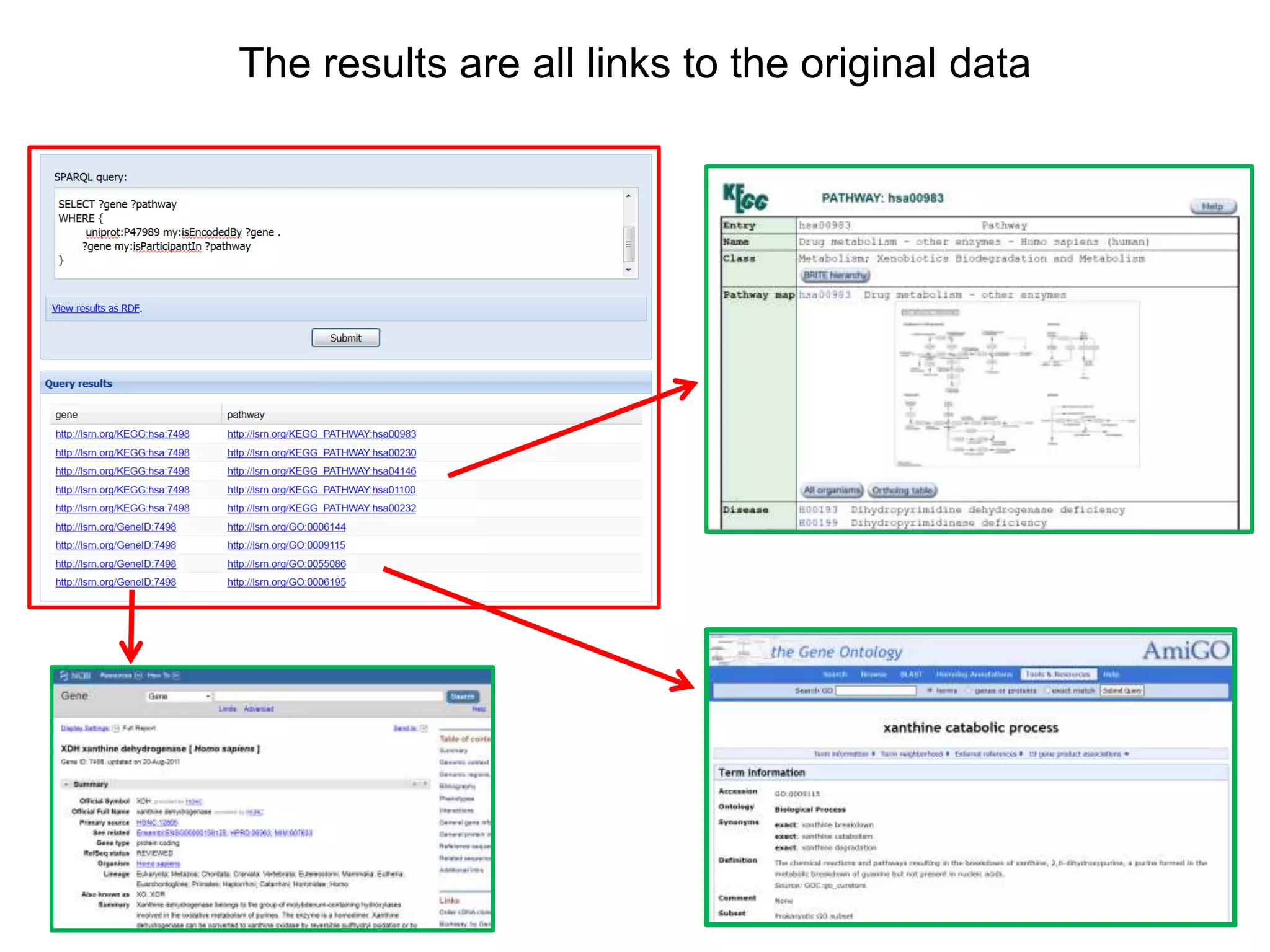
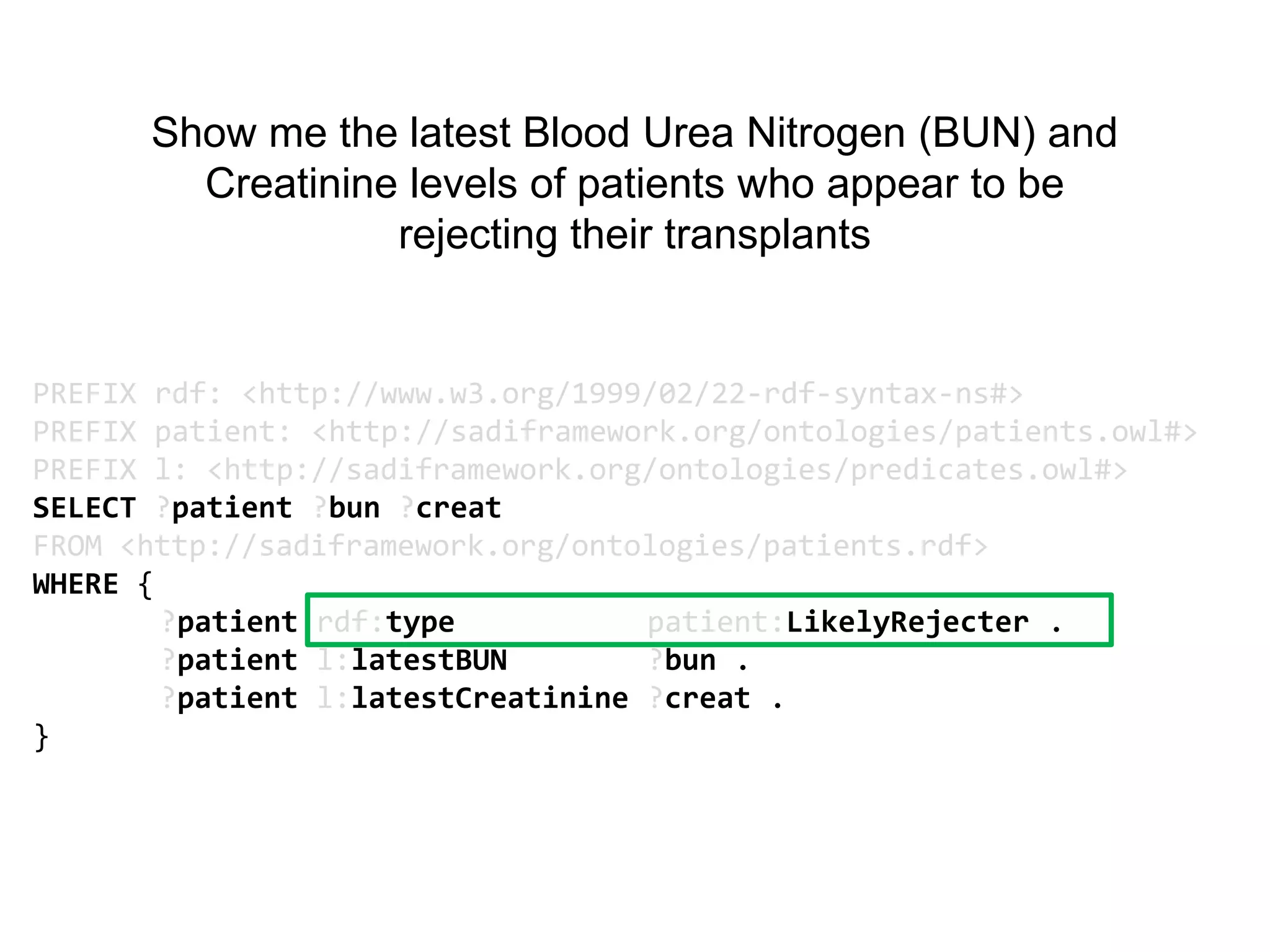
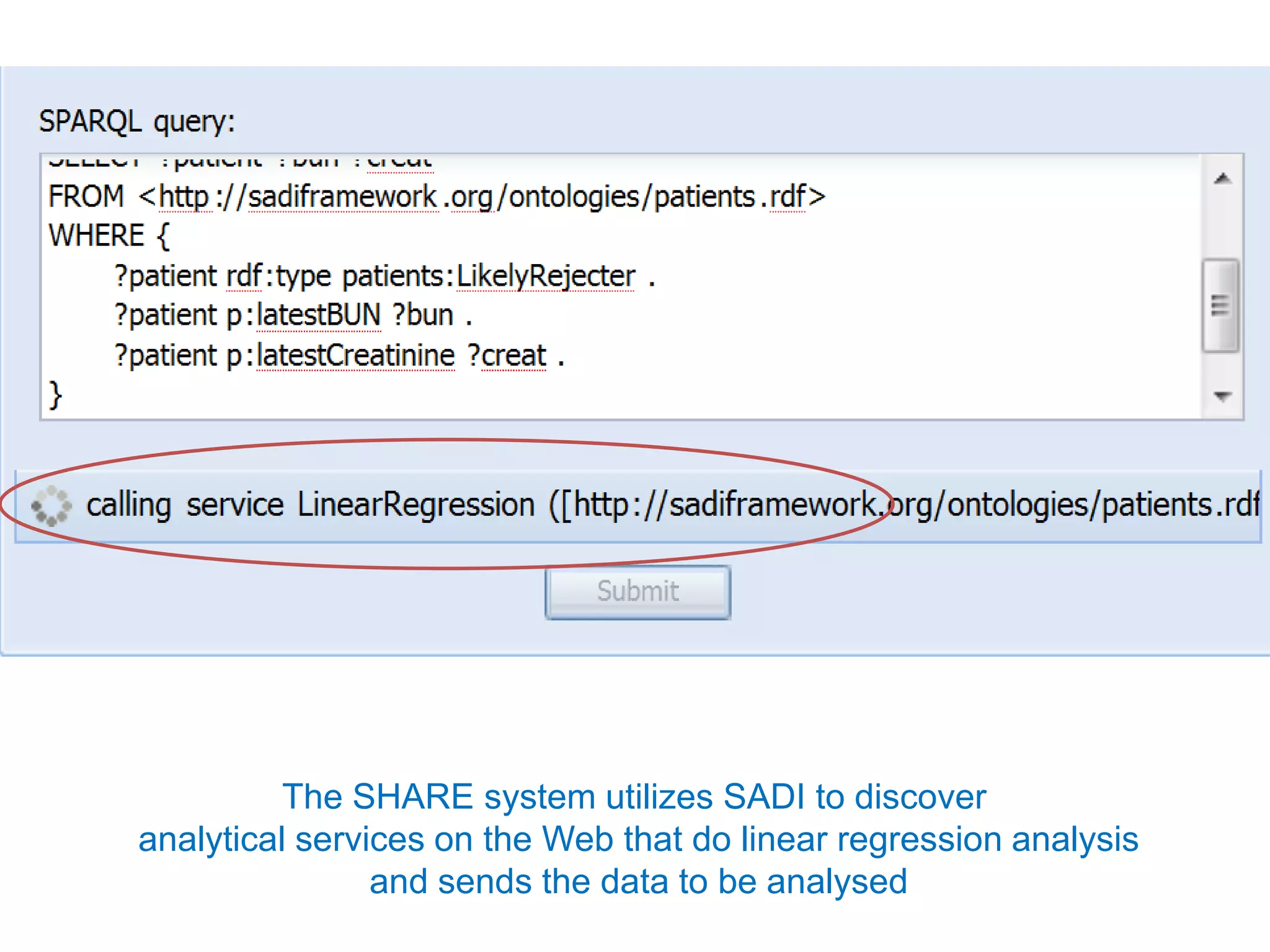
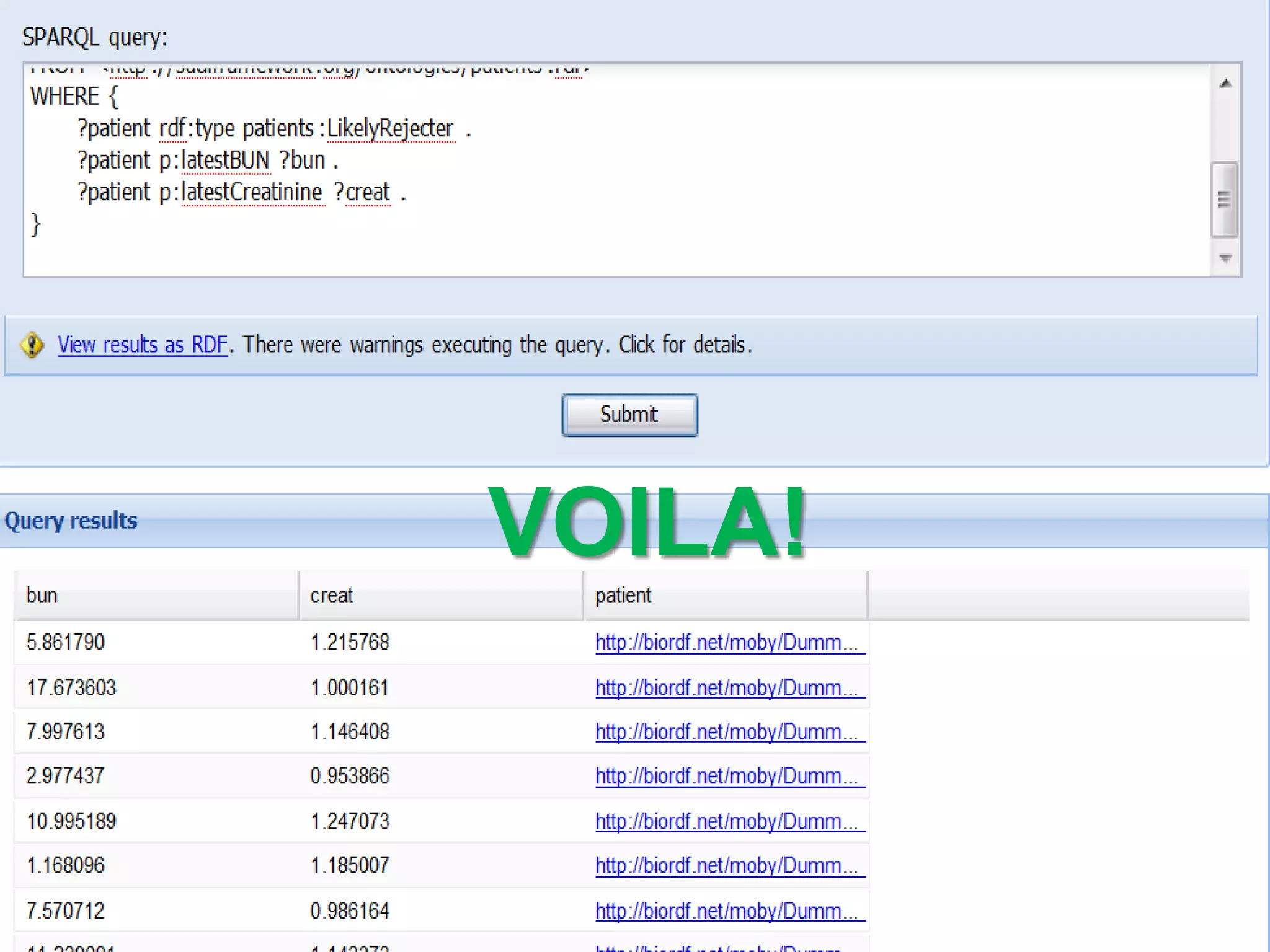
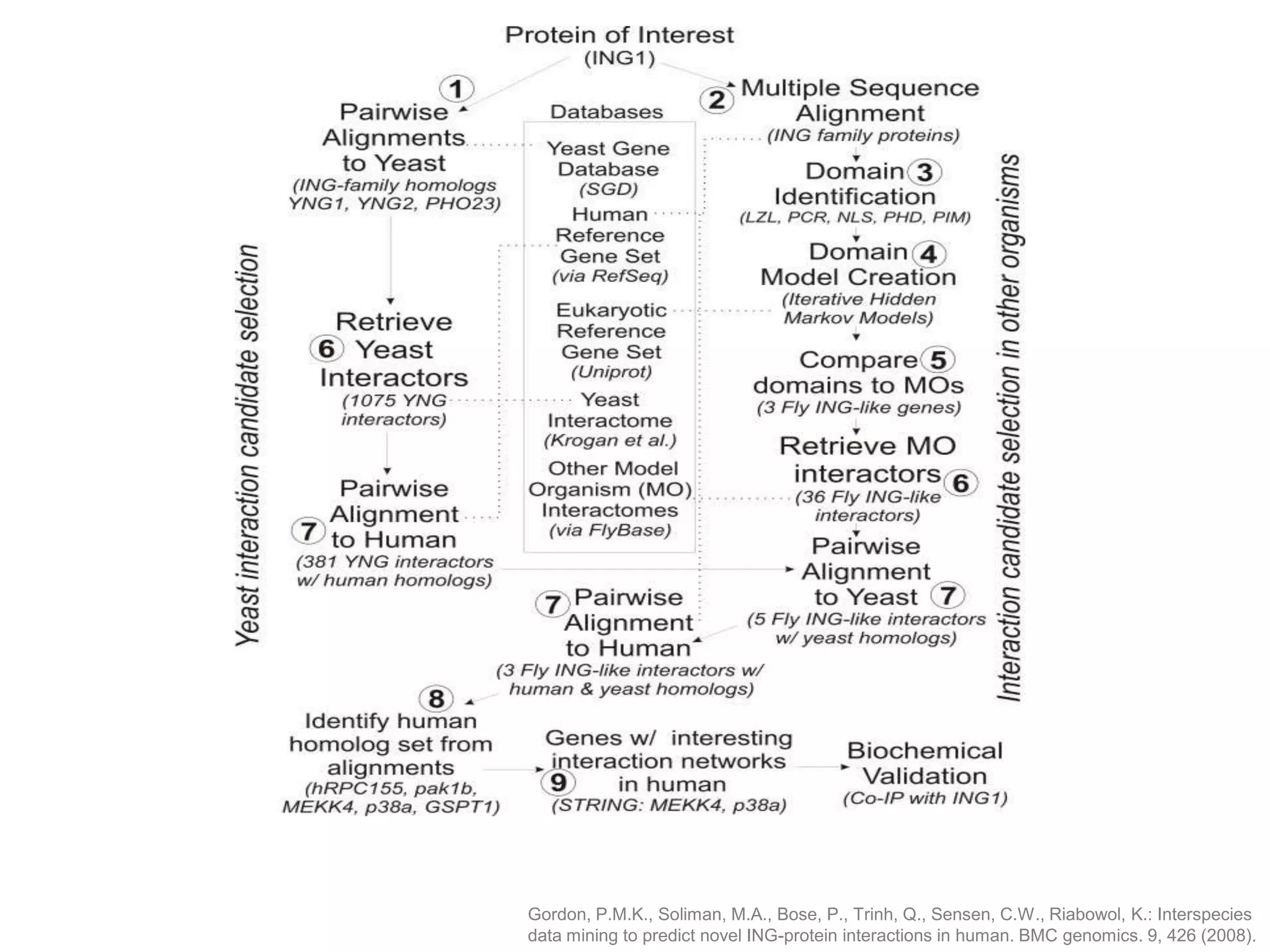
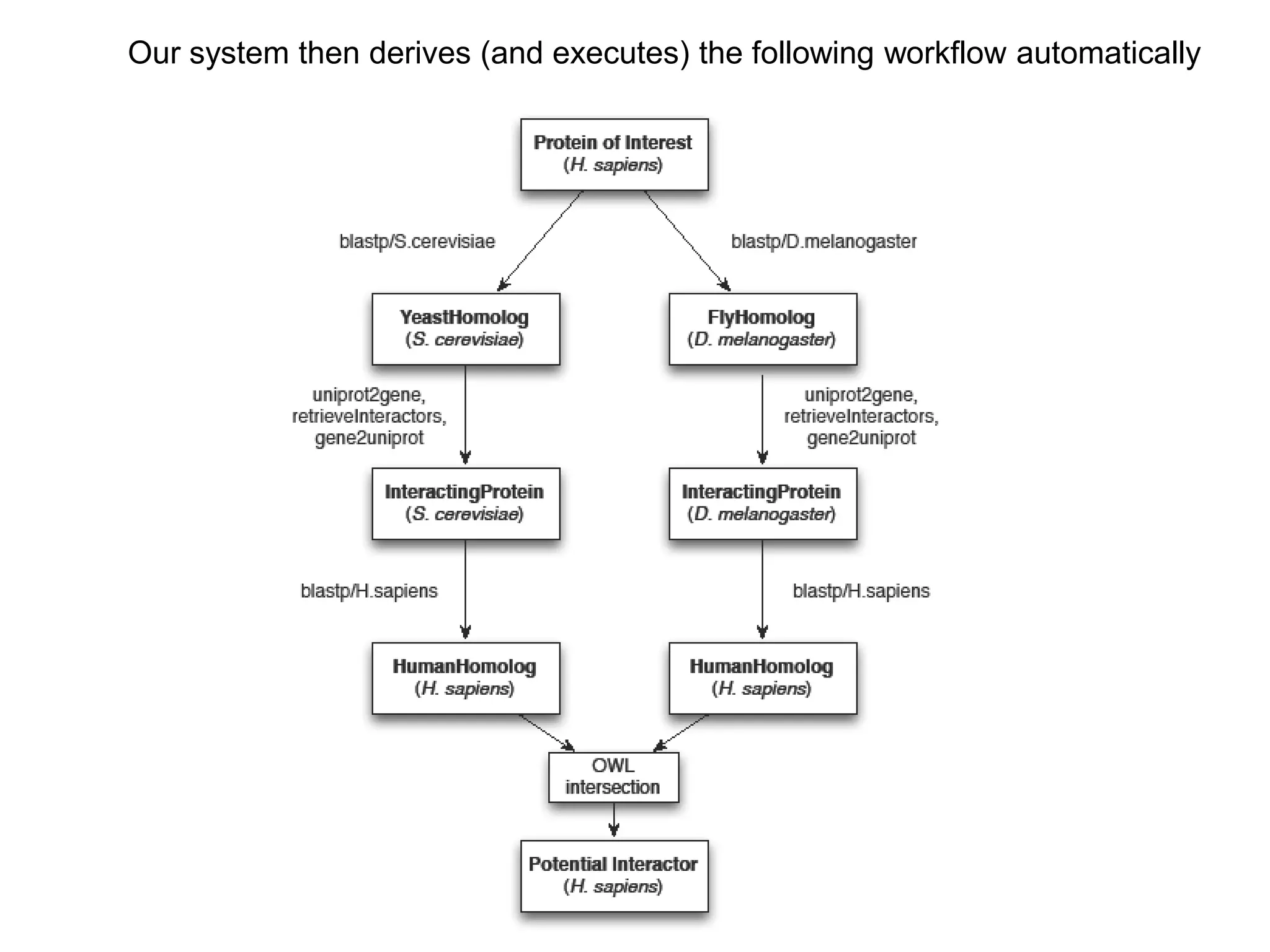
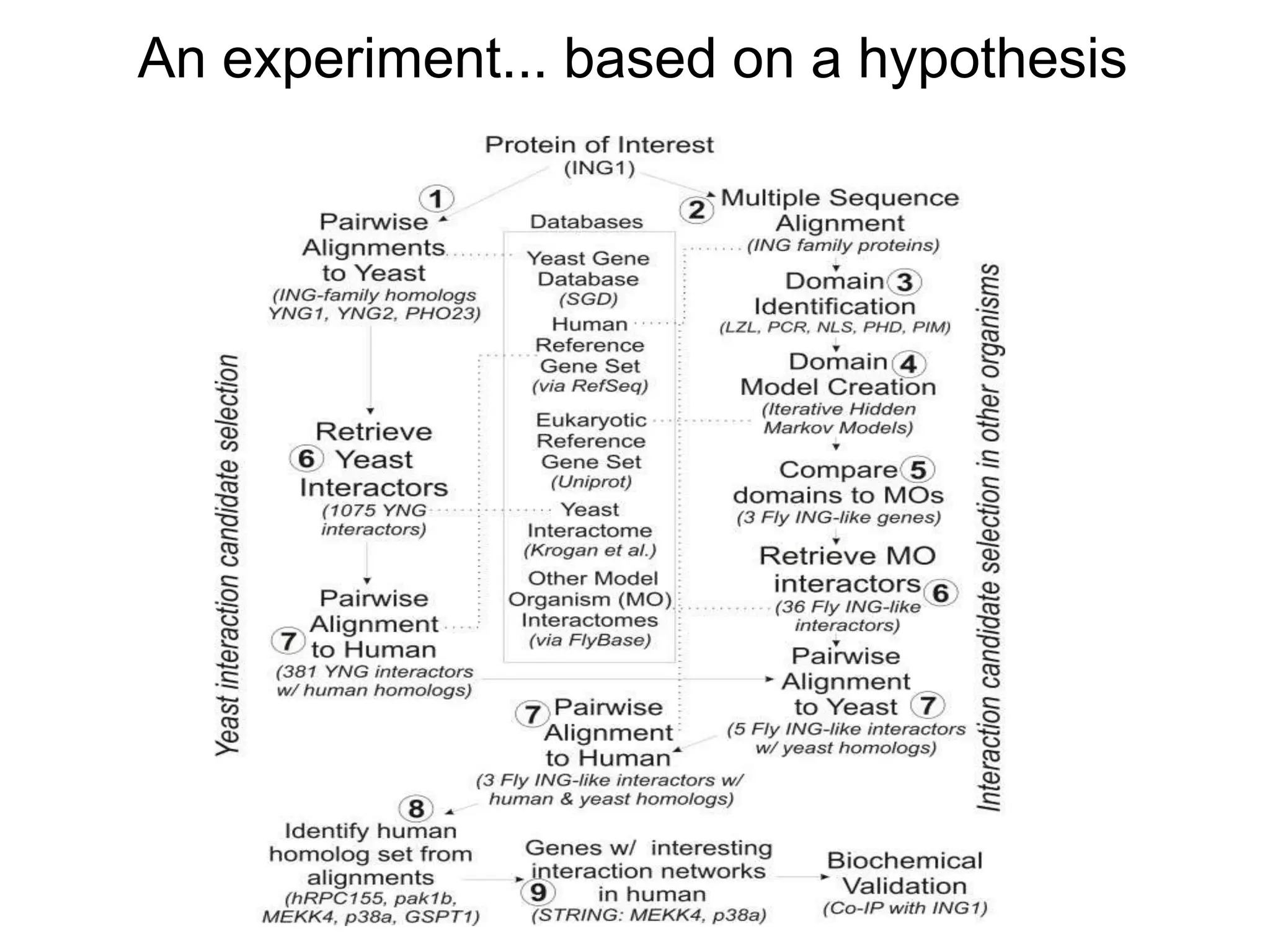
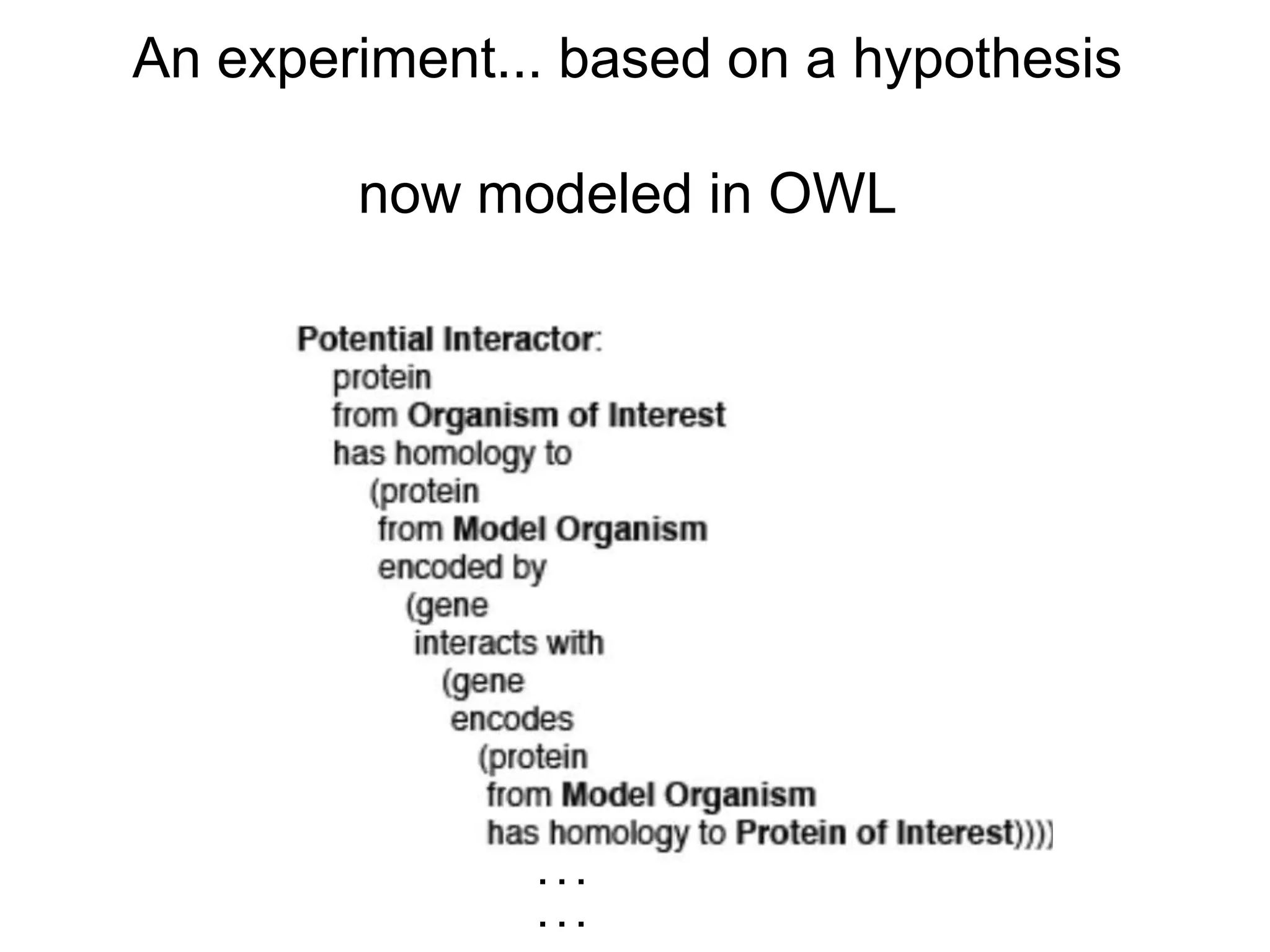
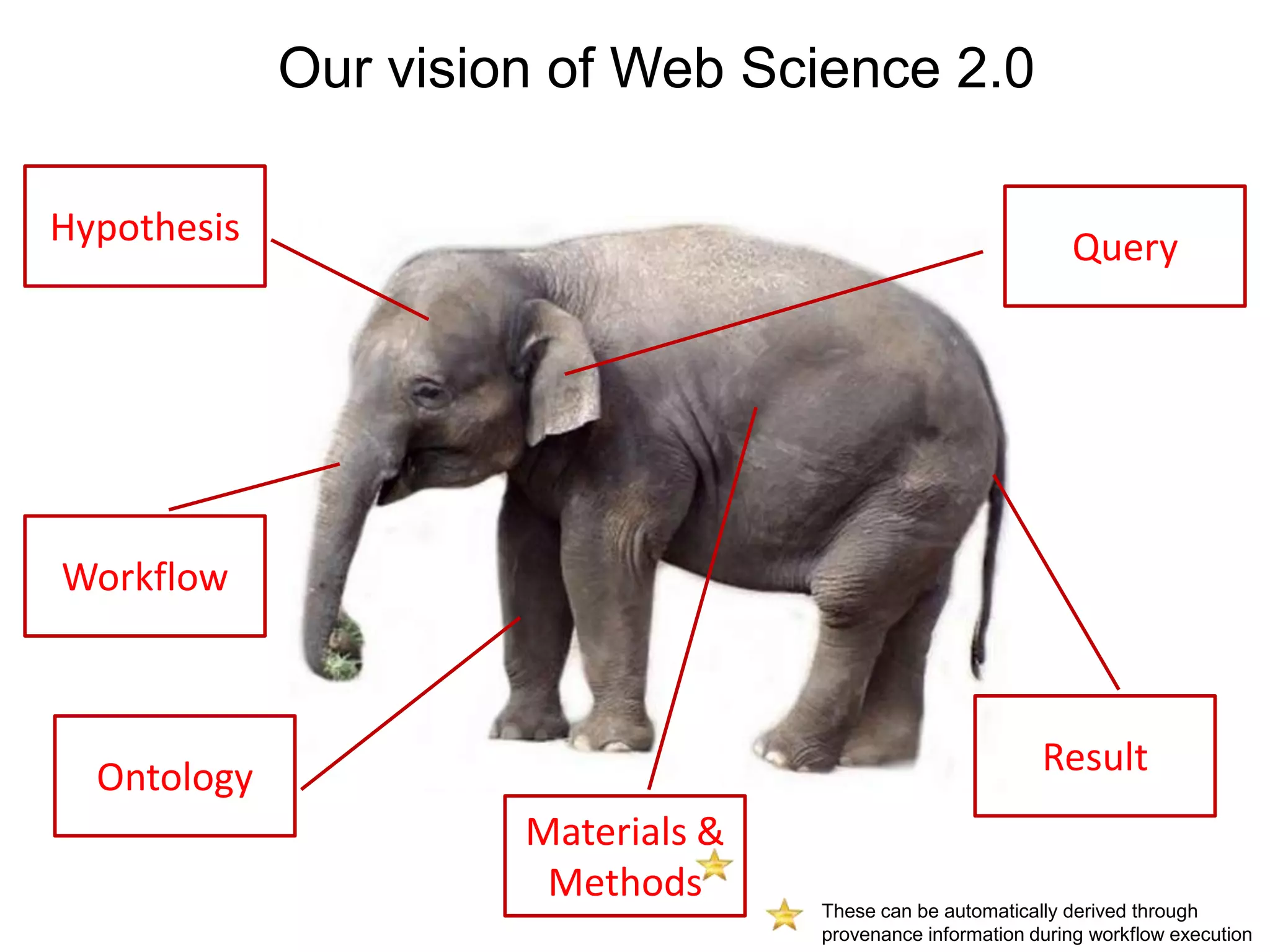
The document discusses the concept of Web Science 2.0, highlighting advancements in conducting in silico research that allow for immediate application of discoveries in scientific practice. It emphasizes the use of ontologies, particularly the Web Ontology Language (OWL), to automate workflows in bioinformatics, enabling machines to interpret and interact with data without needing human intervention. The paper illustrates this through examples of complex multi-database queries and the adaptability of published models for re-use in various research contexts.