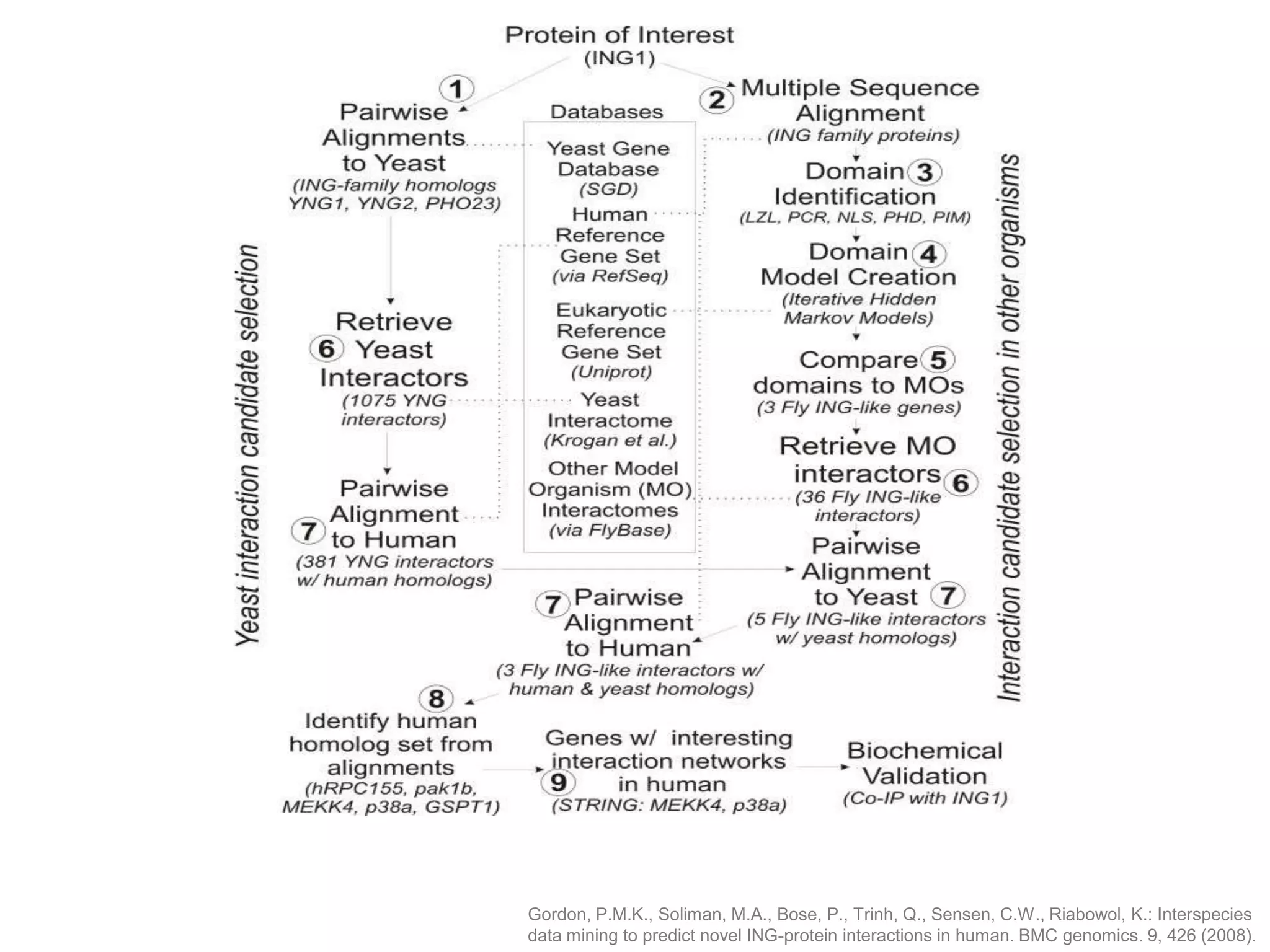
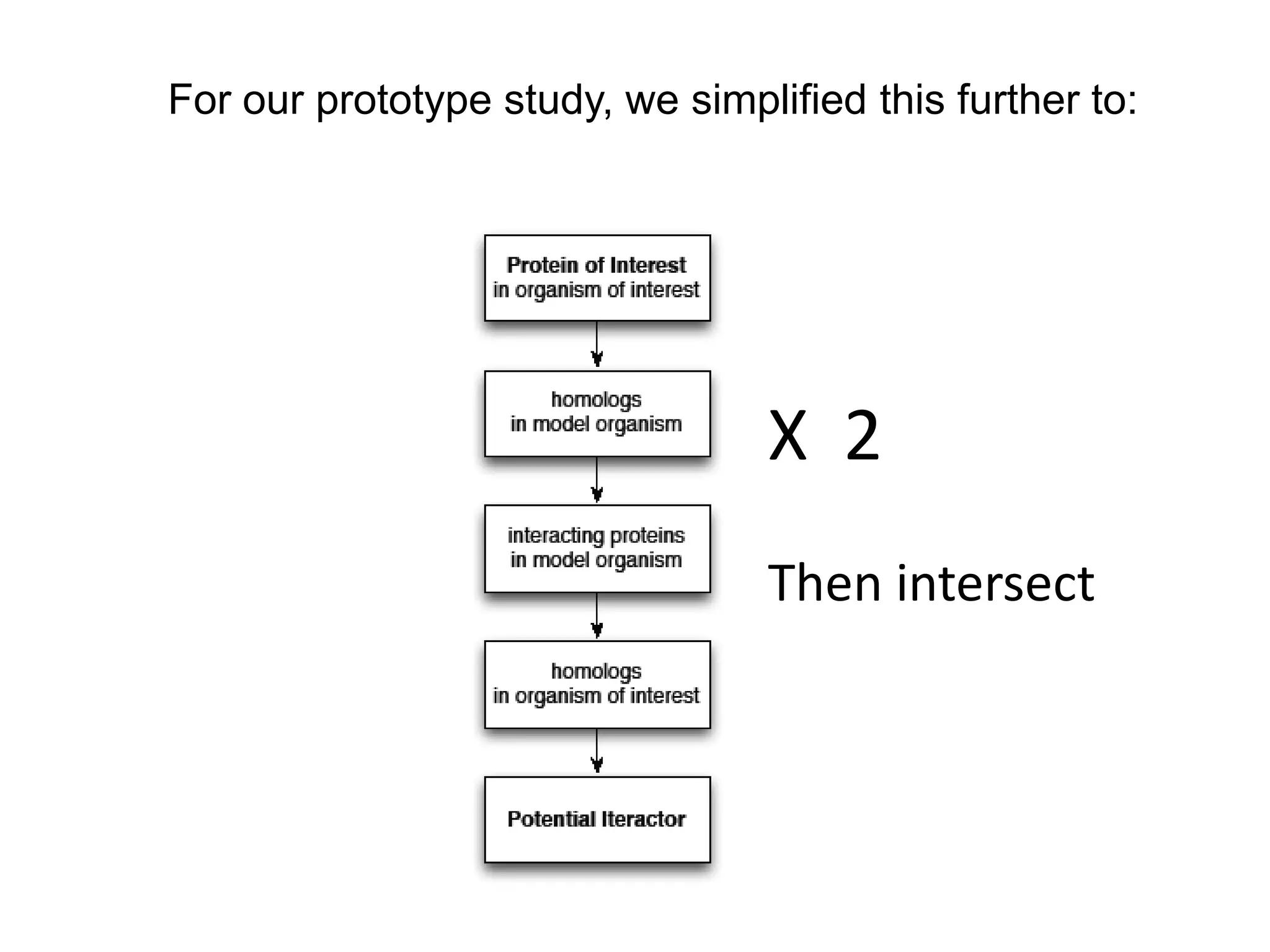
The document discusses the use of SPARQL-DL as an abstract workflow language to automate the evaluation of hypotheses in bioinformatics through semantic web services. It explores how a model can be created to predict protein interactions across species using ontology representation and highlights the flexibility of generating workflows dynamically based on the semantic relationships established in the model. Additionally, it addresses the challenges in re-purposing workflows and advocates for a user-friendly, personalized interface to enhance data-driven research in medical contexts.








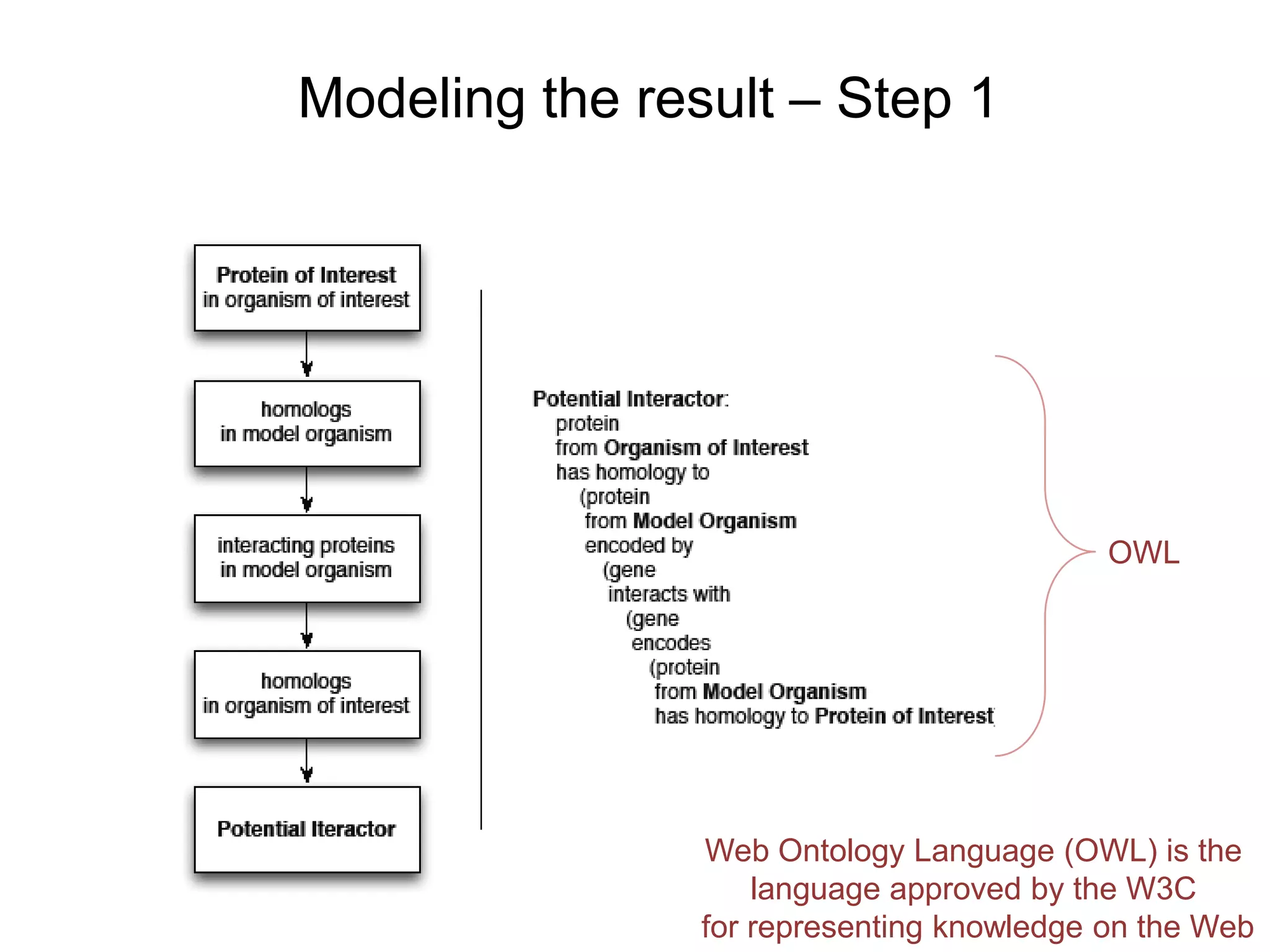
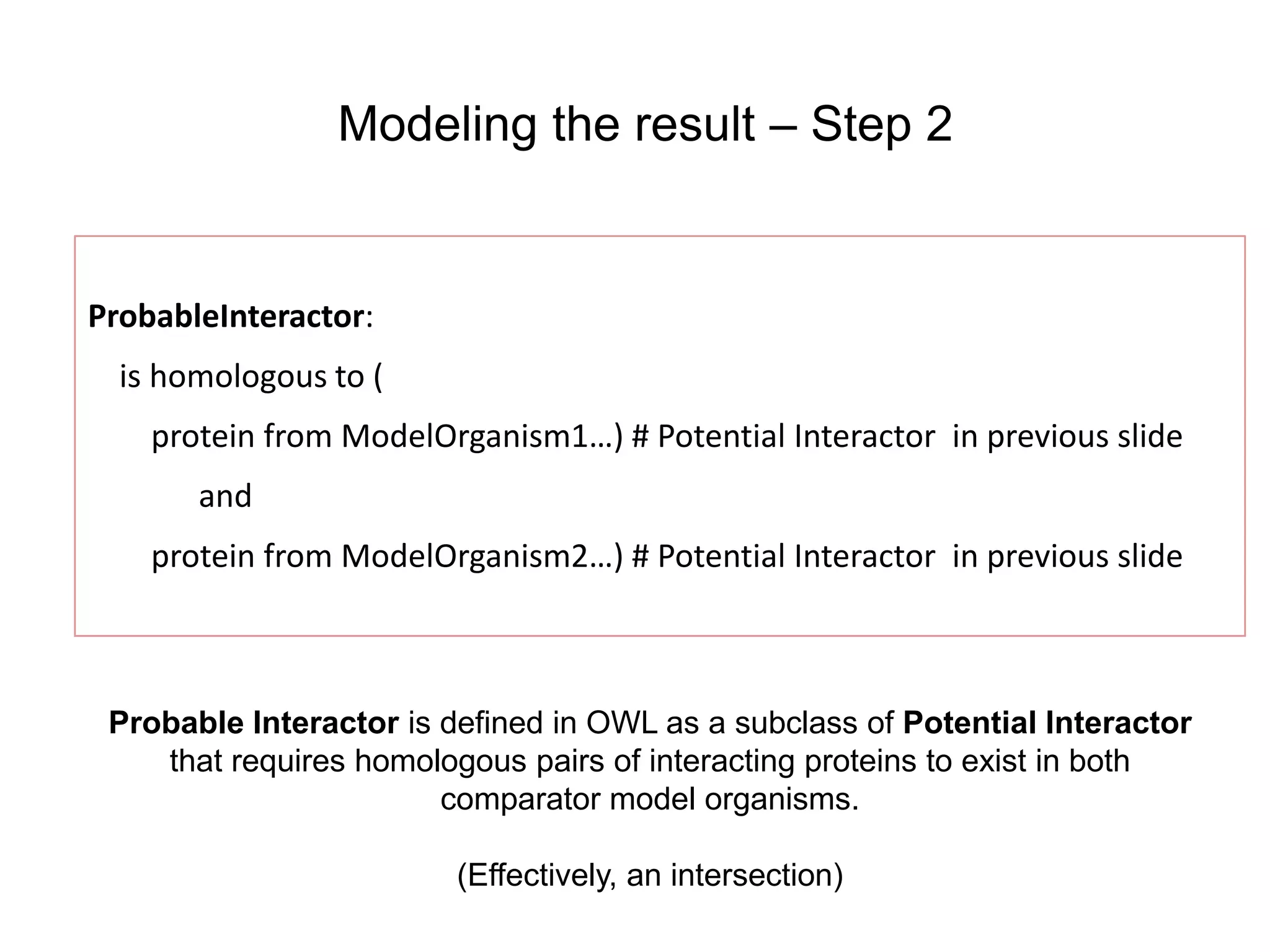





























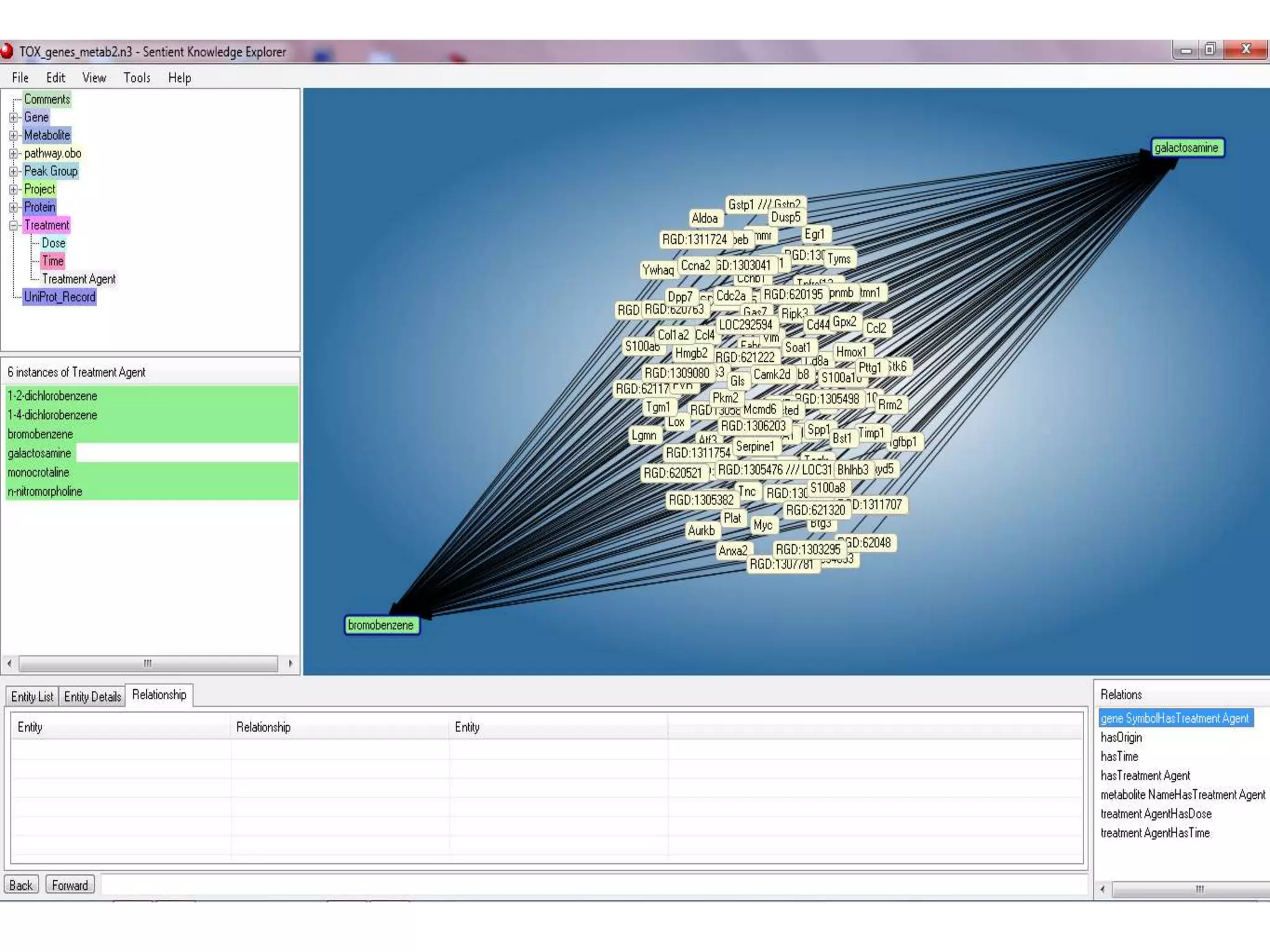
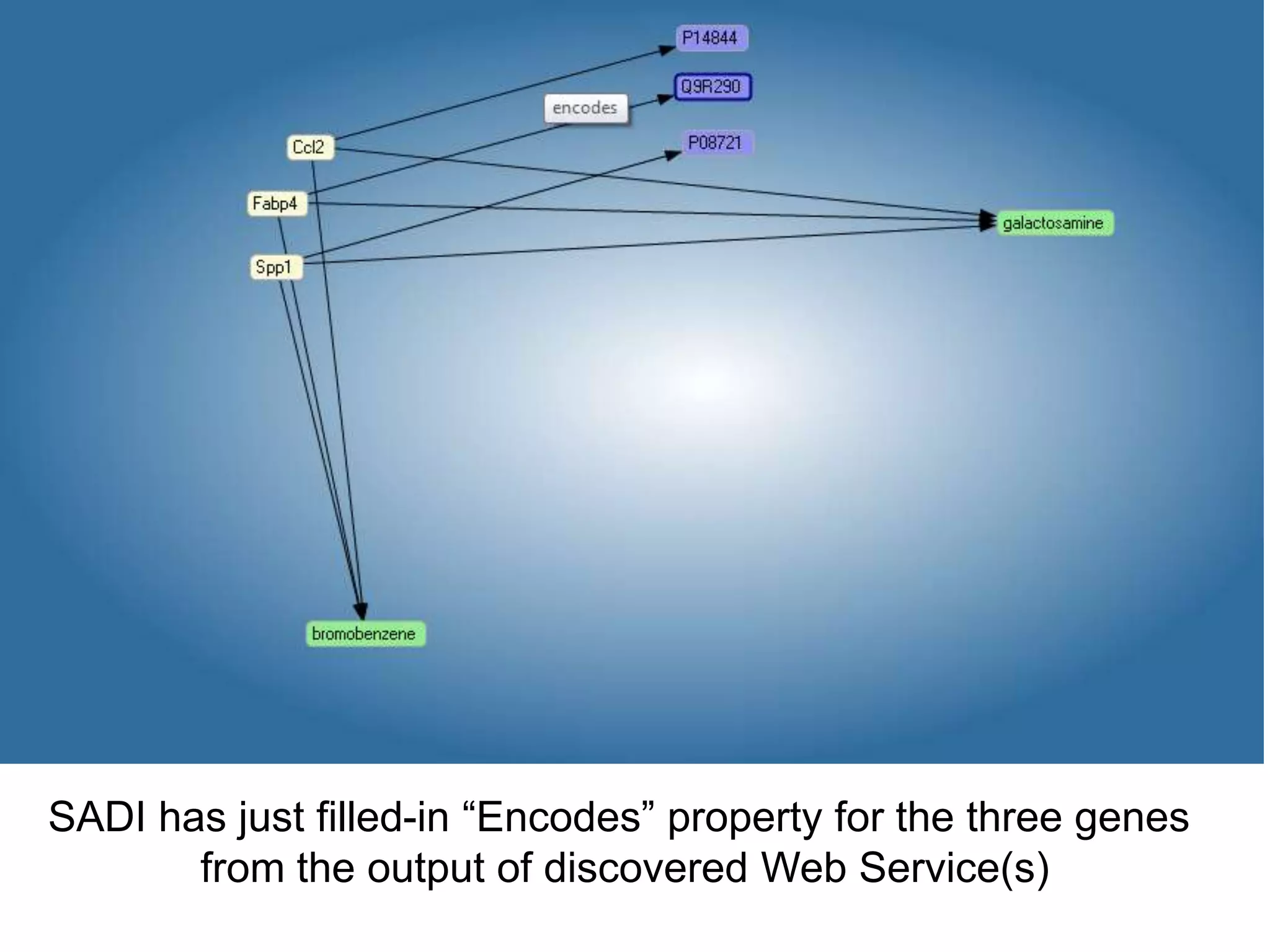
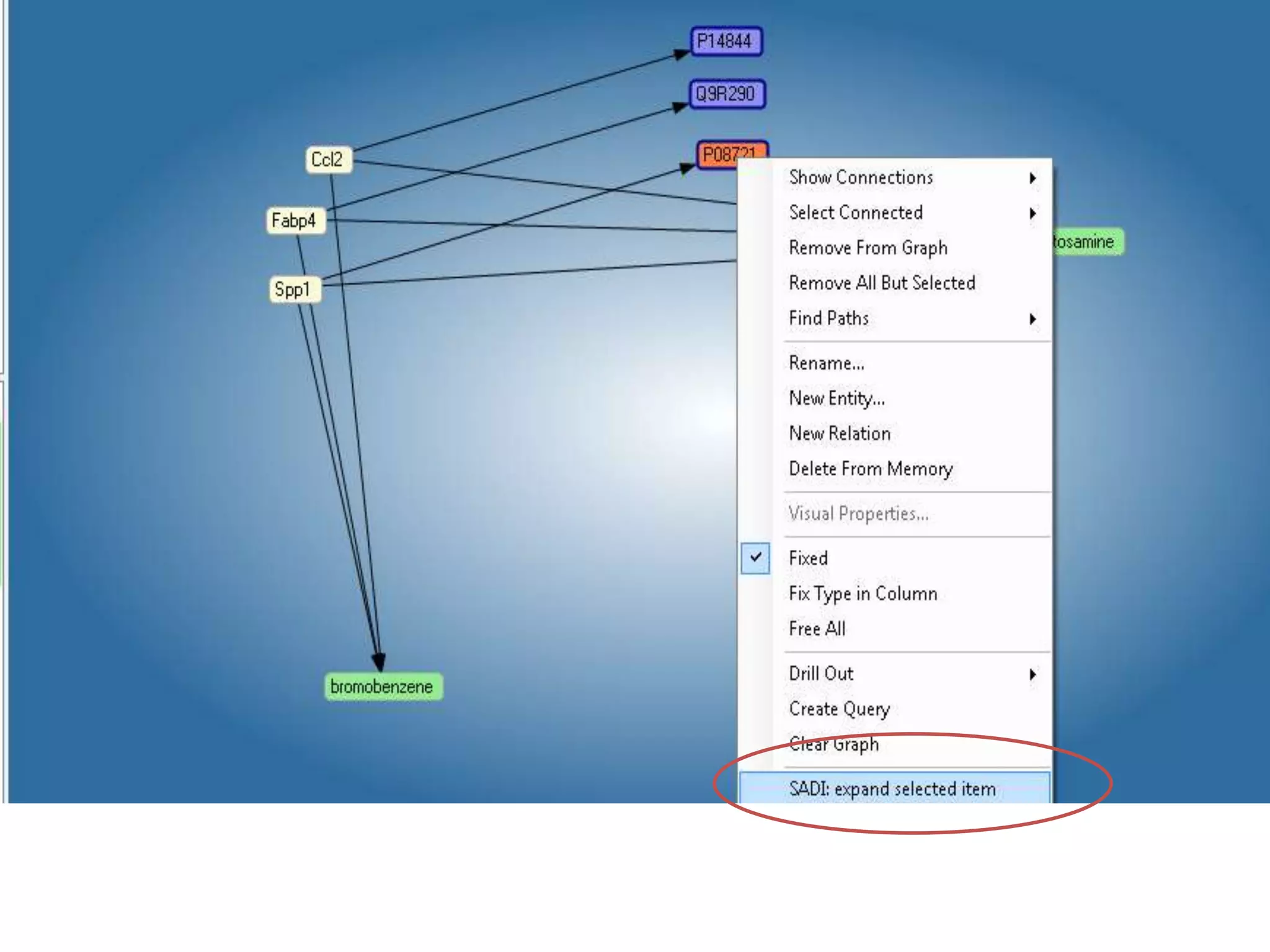
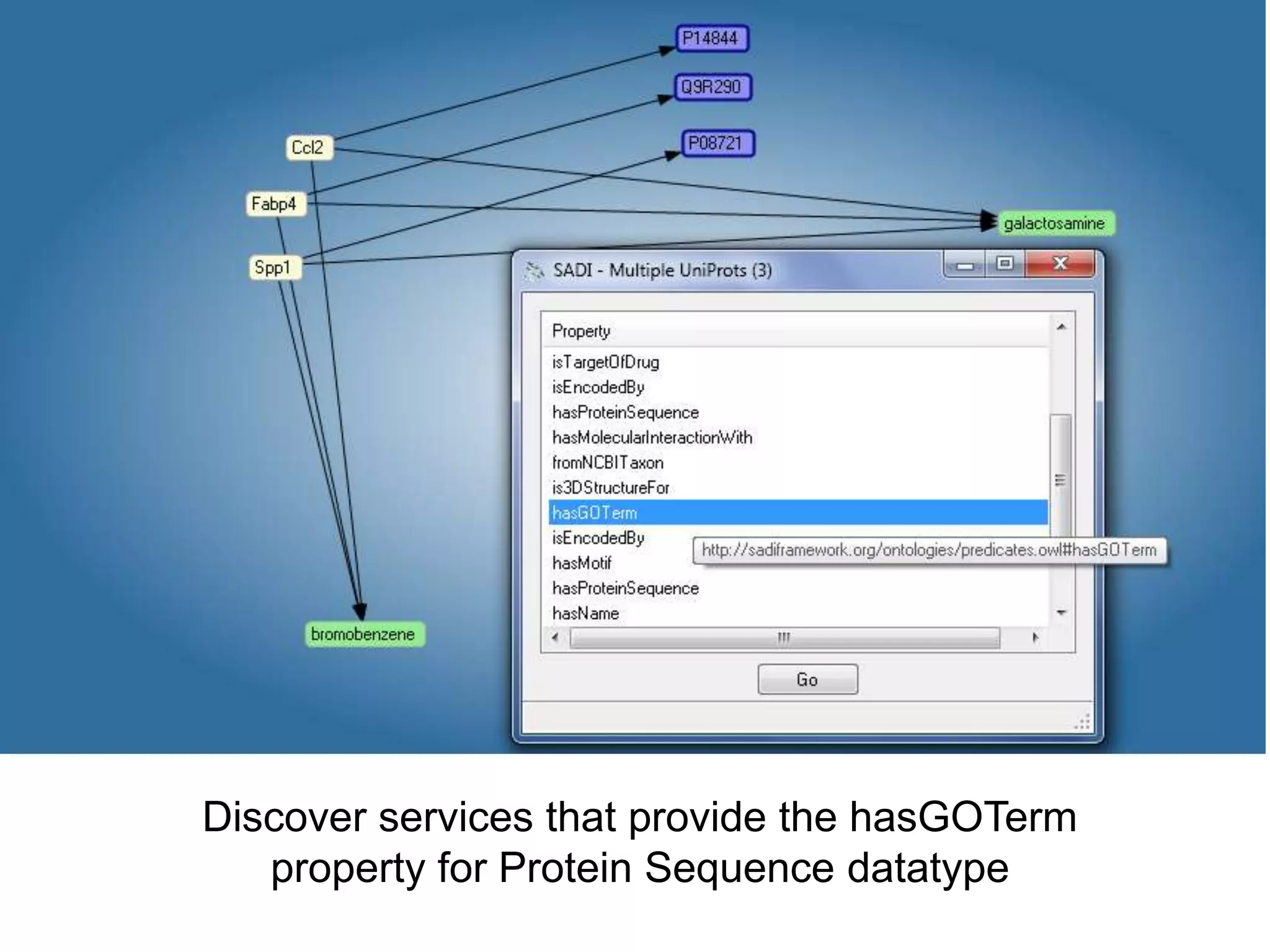



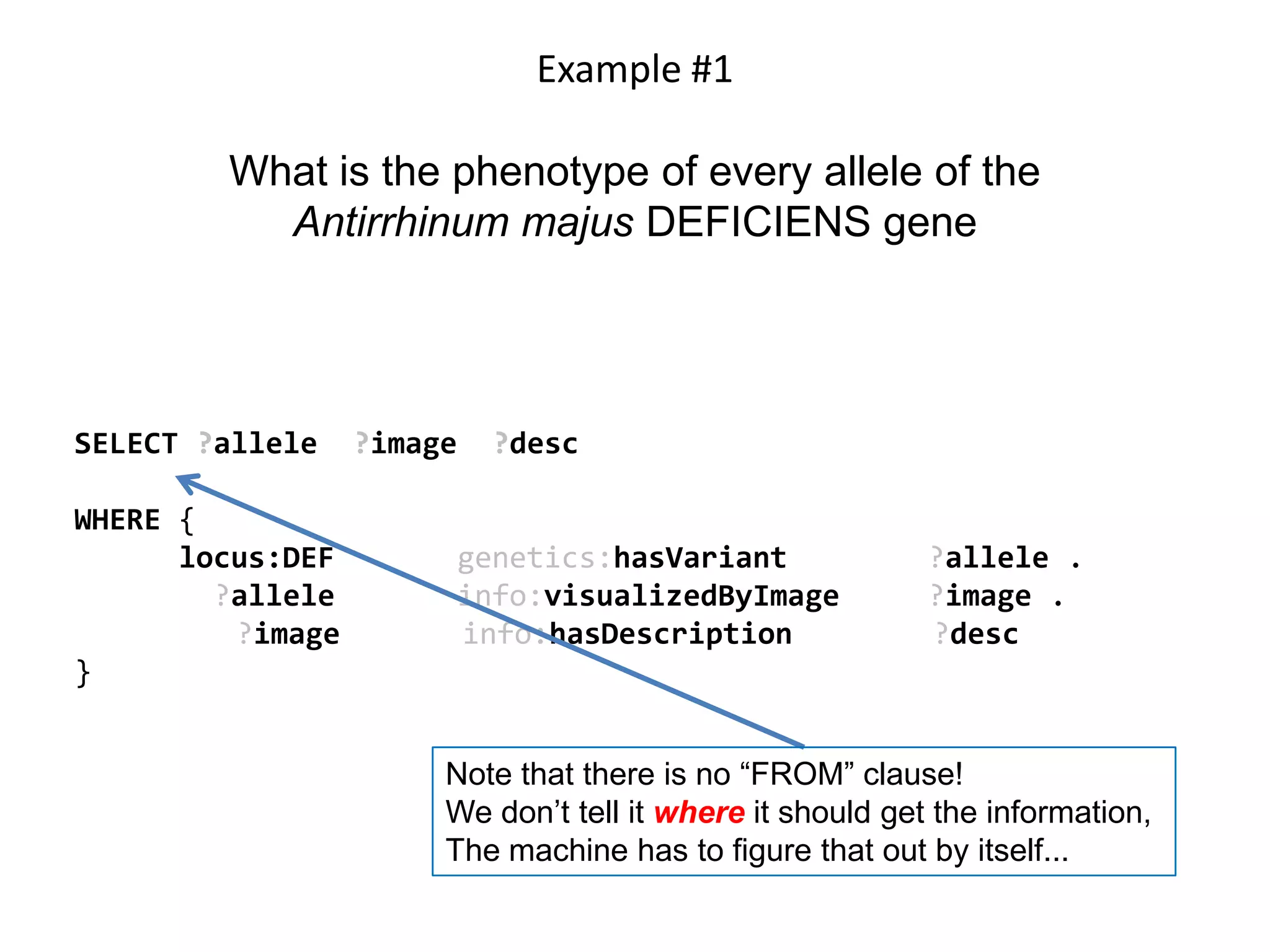

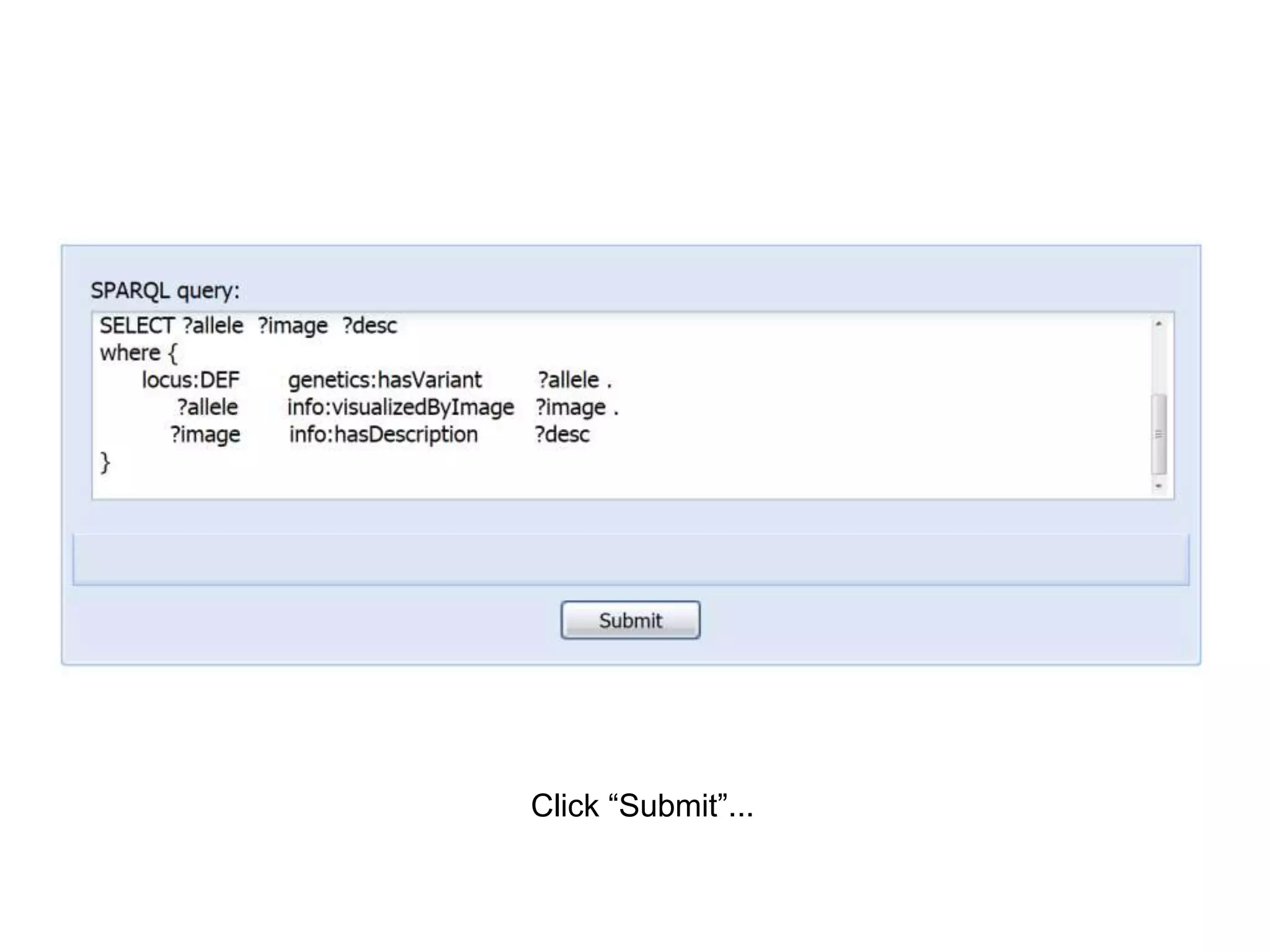
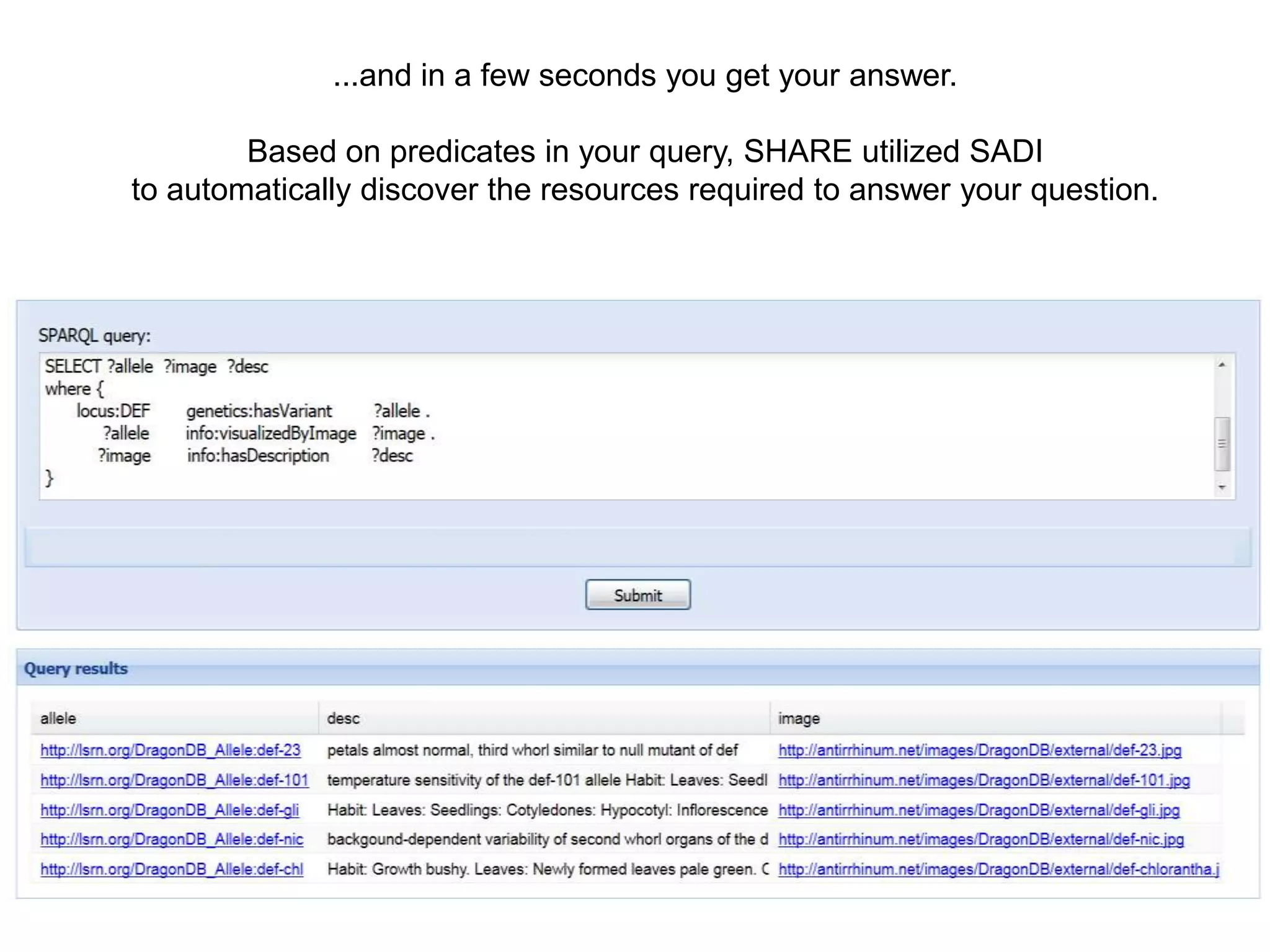






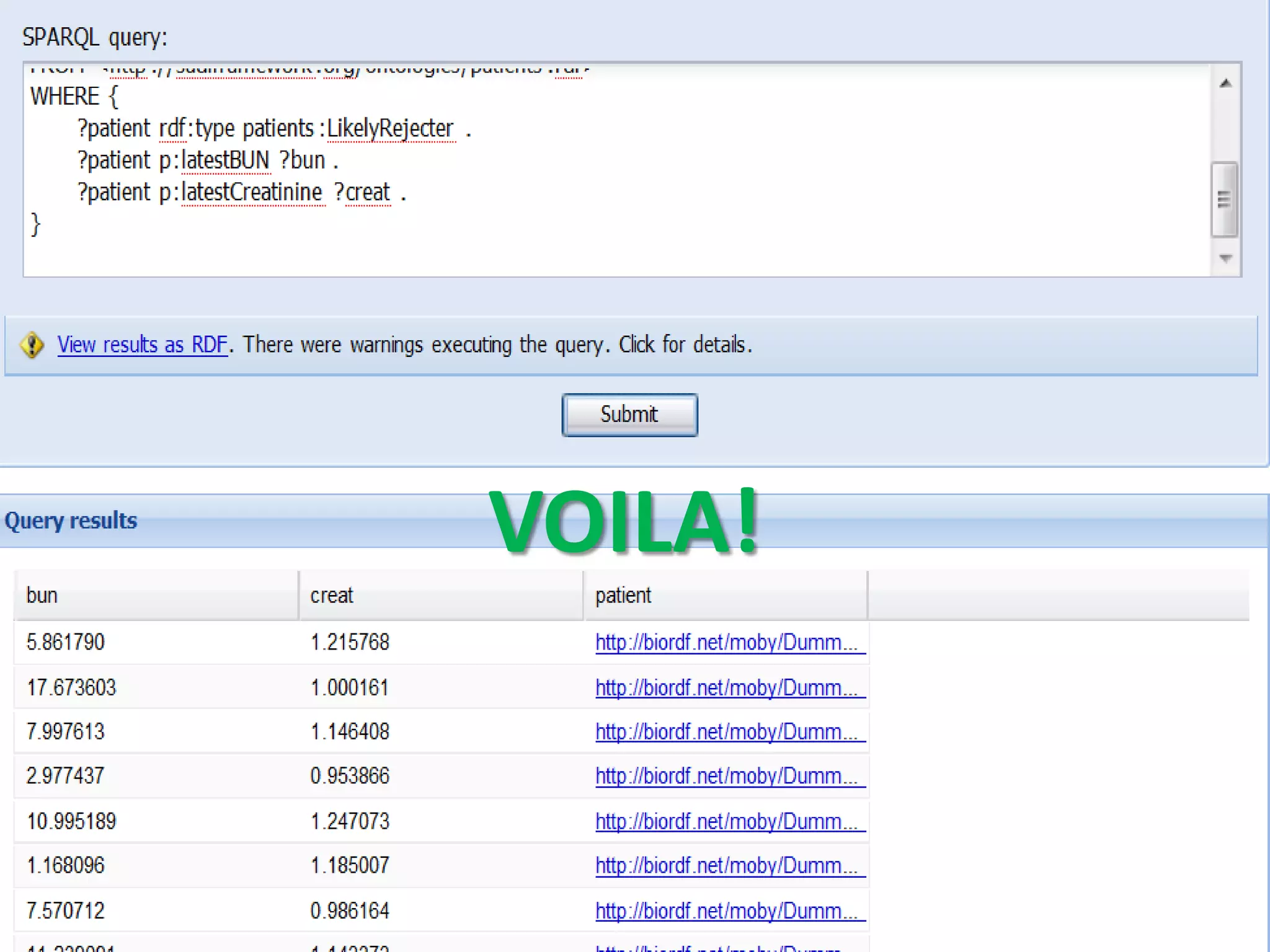


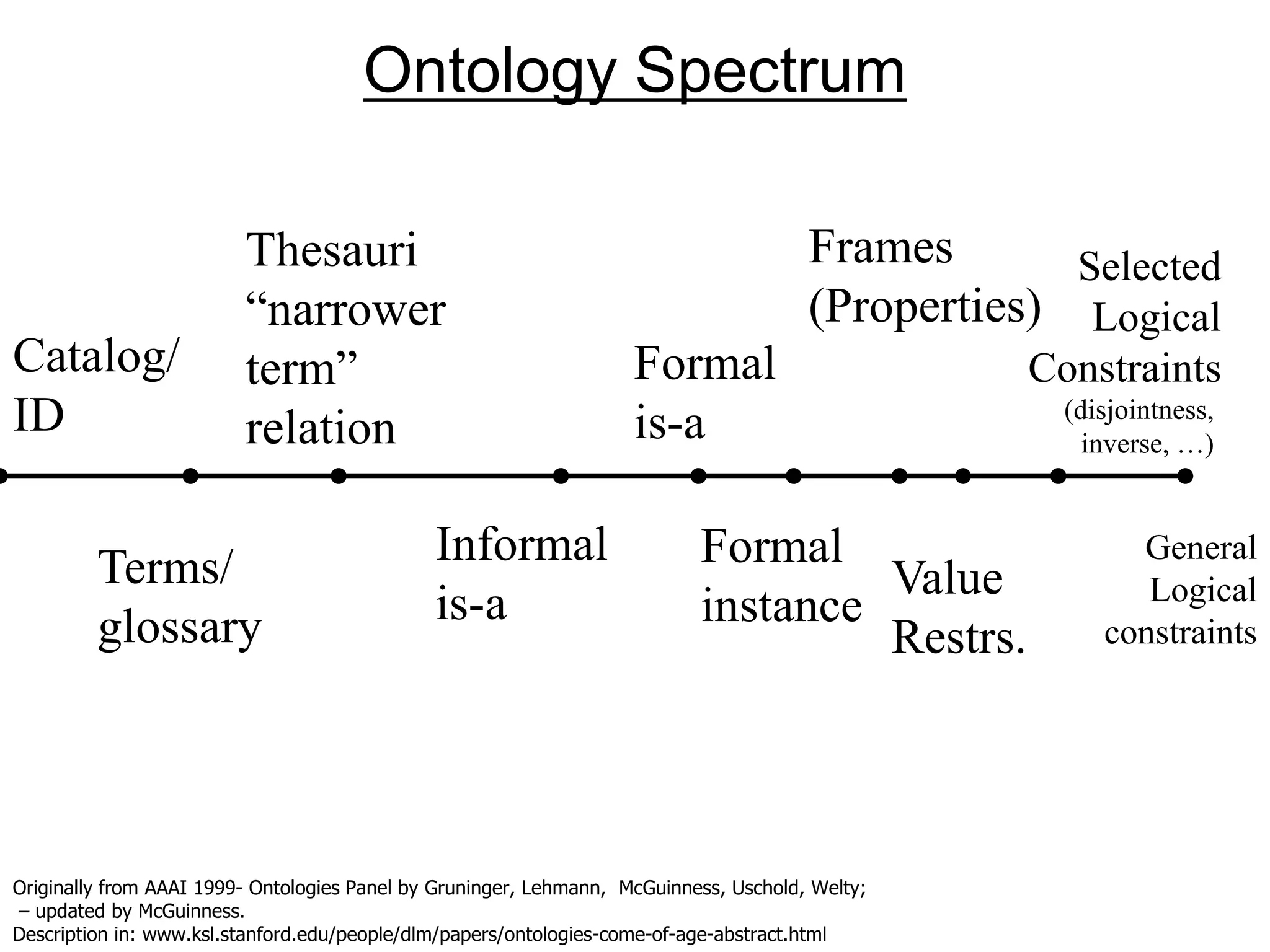
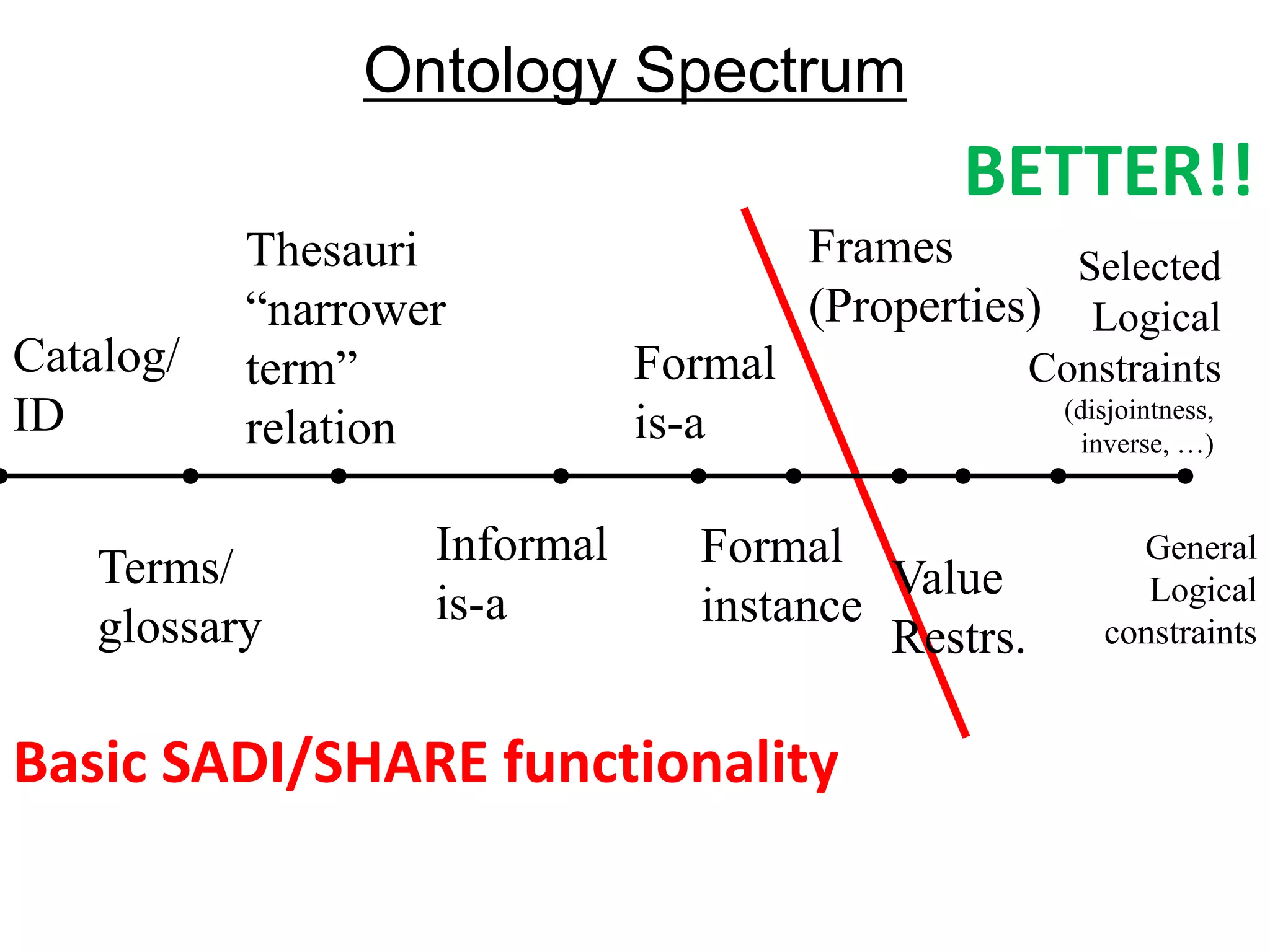
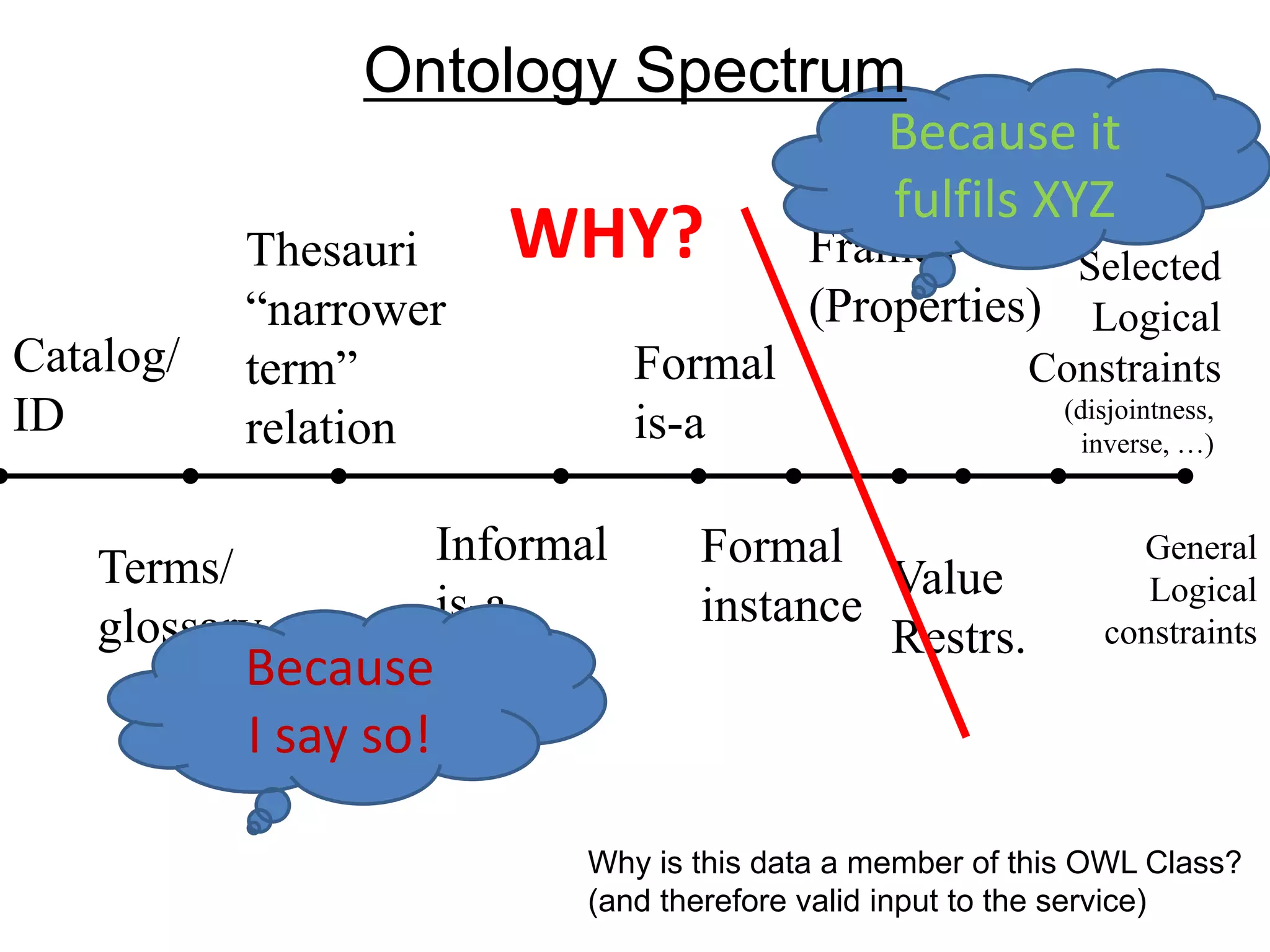
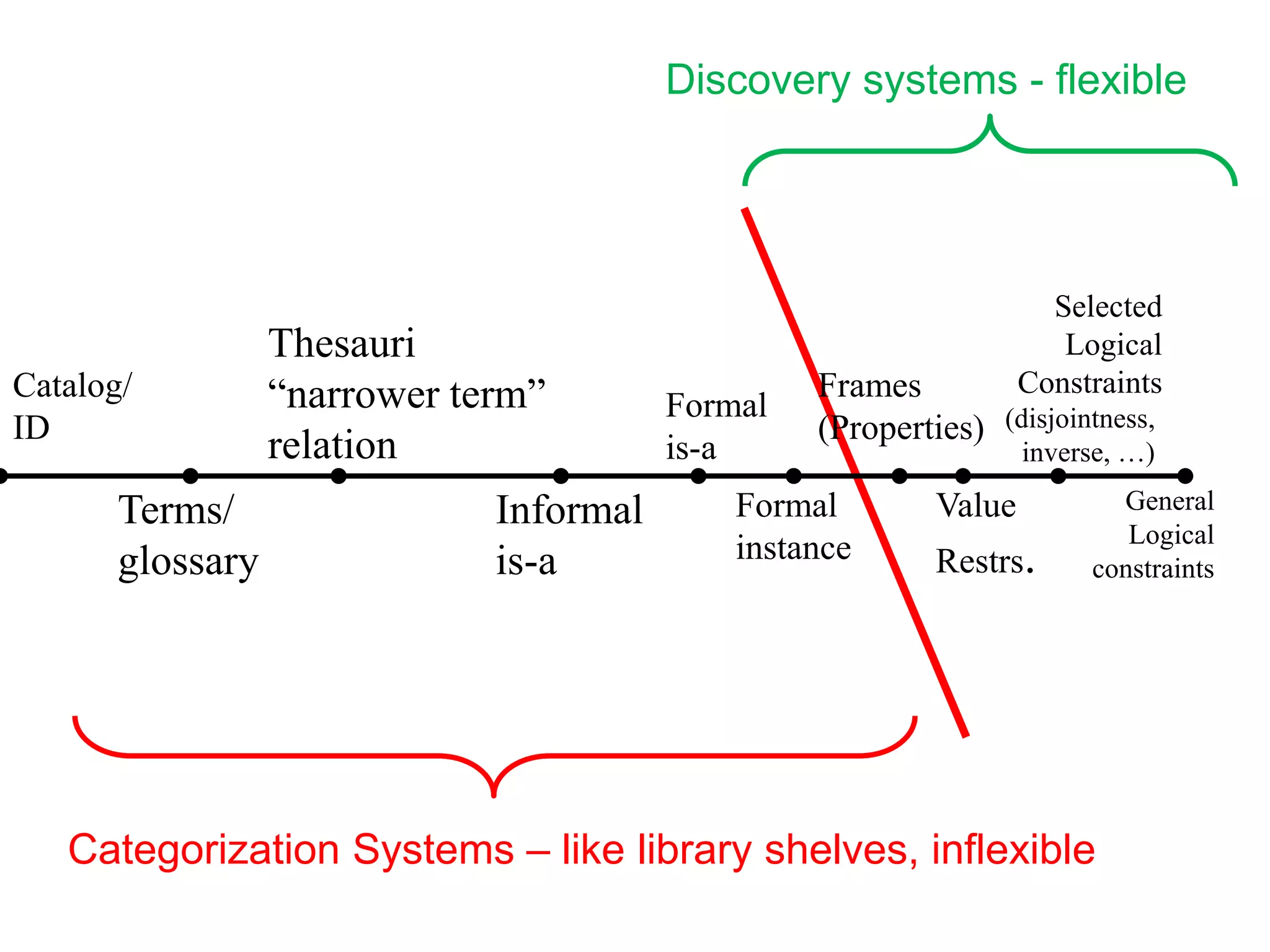



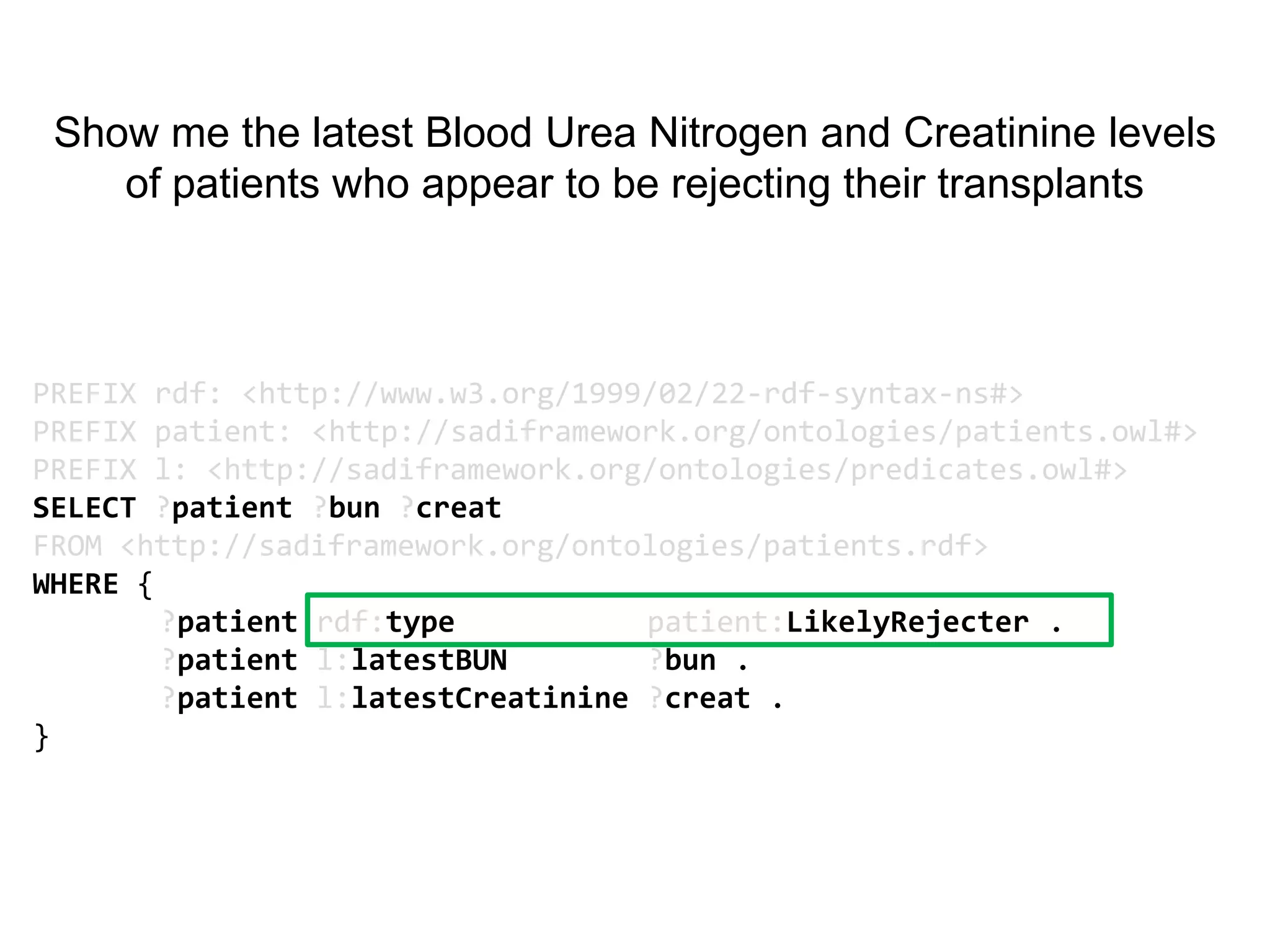


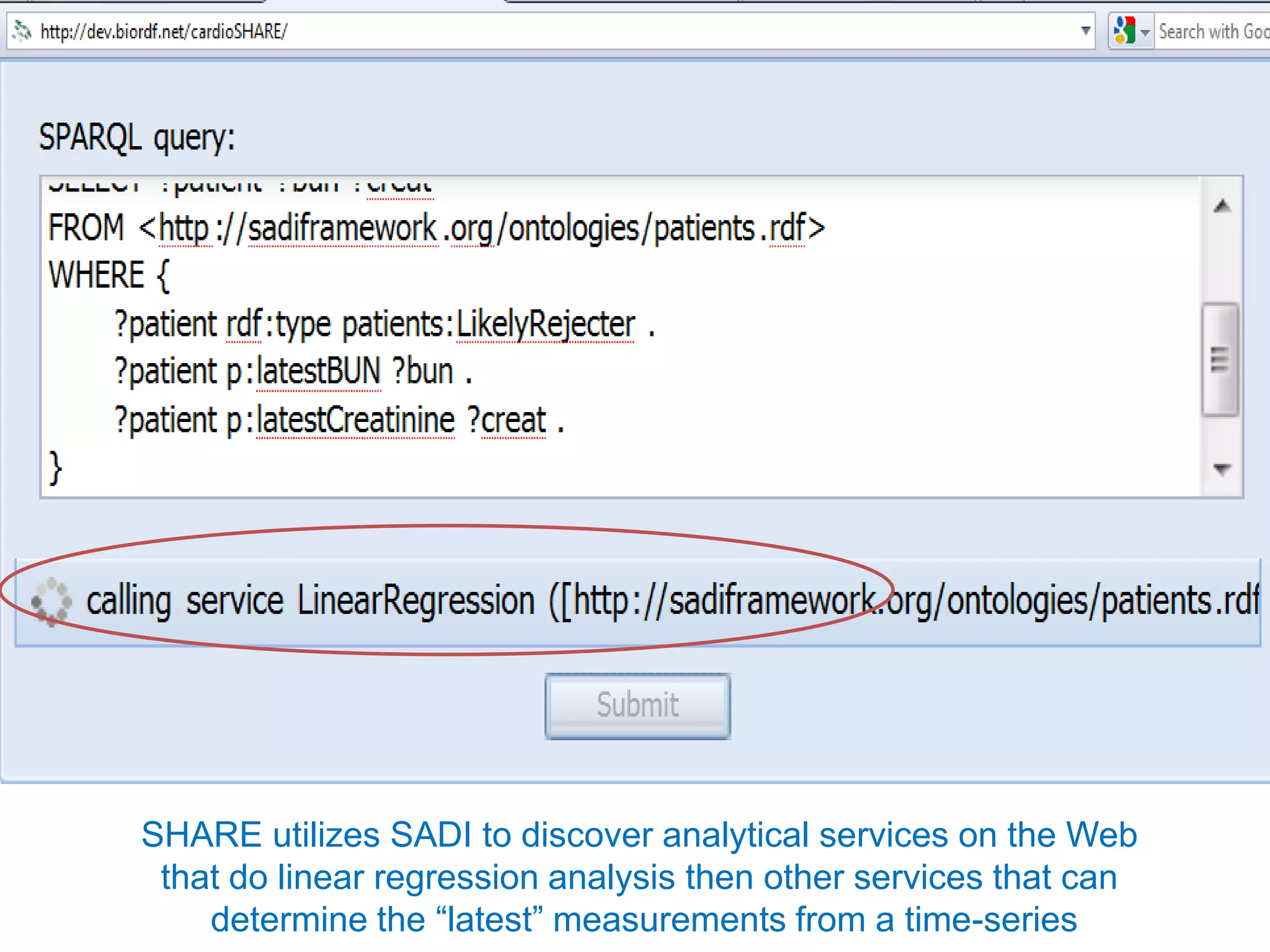



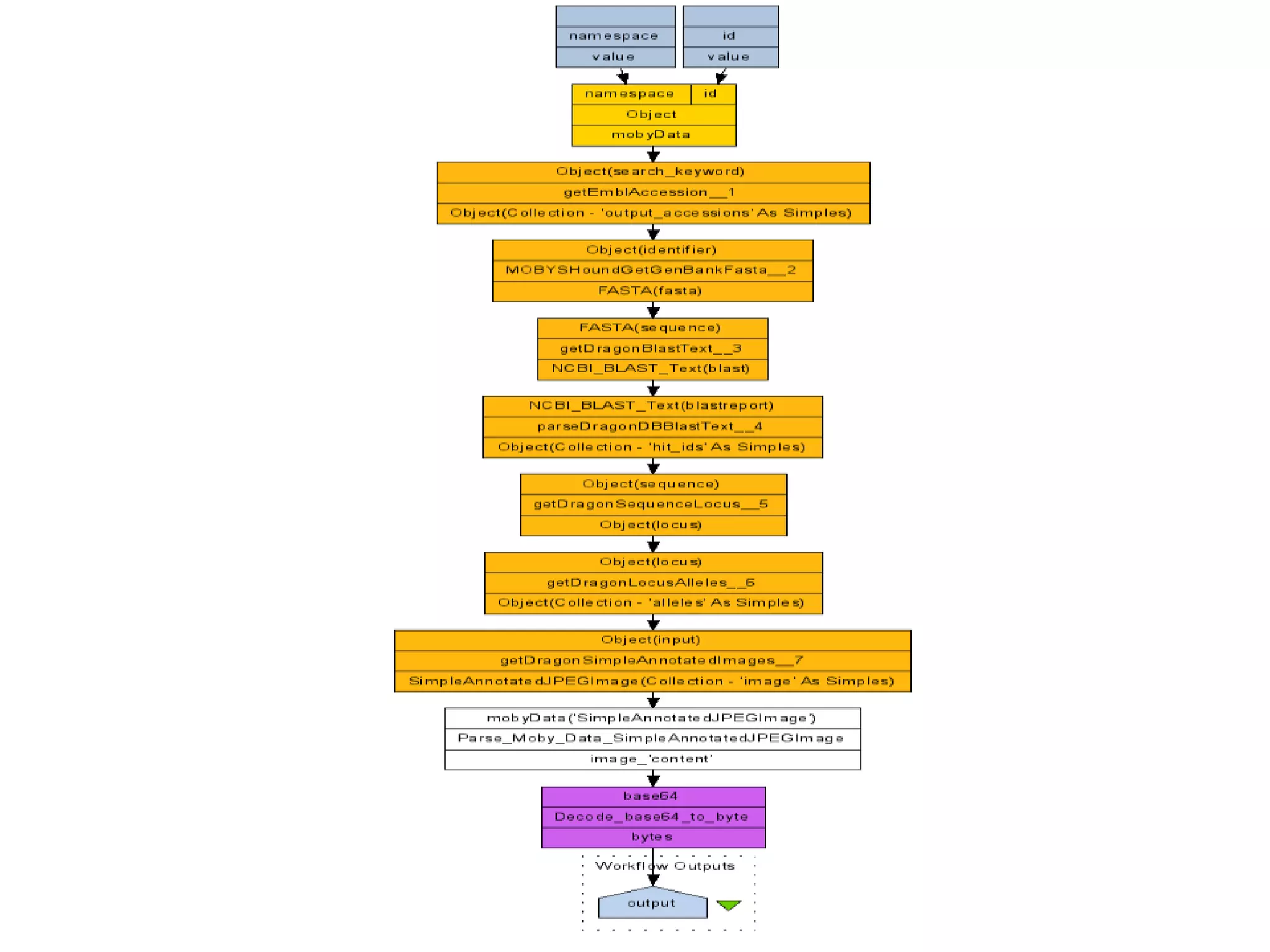
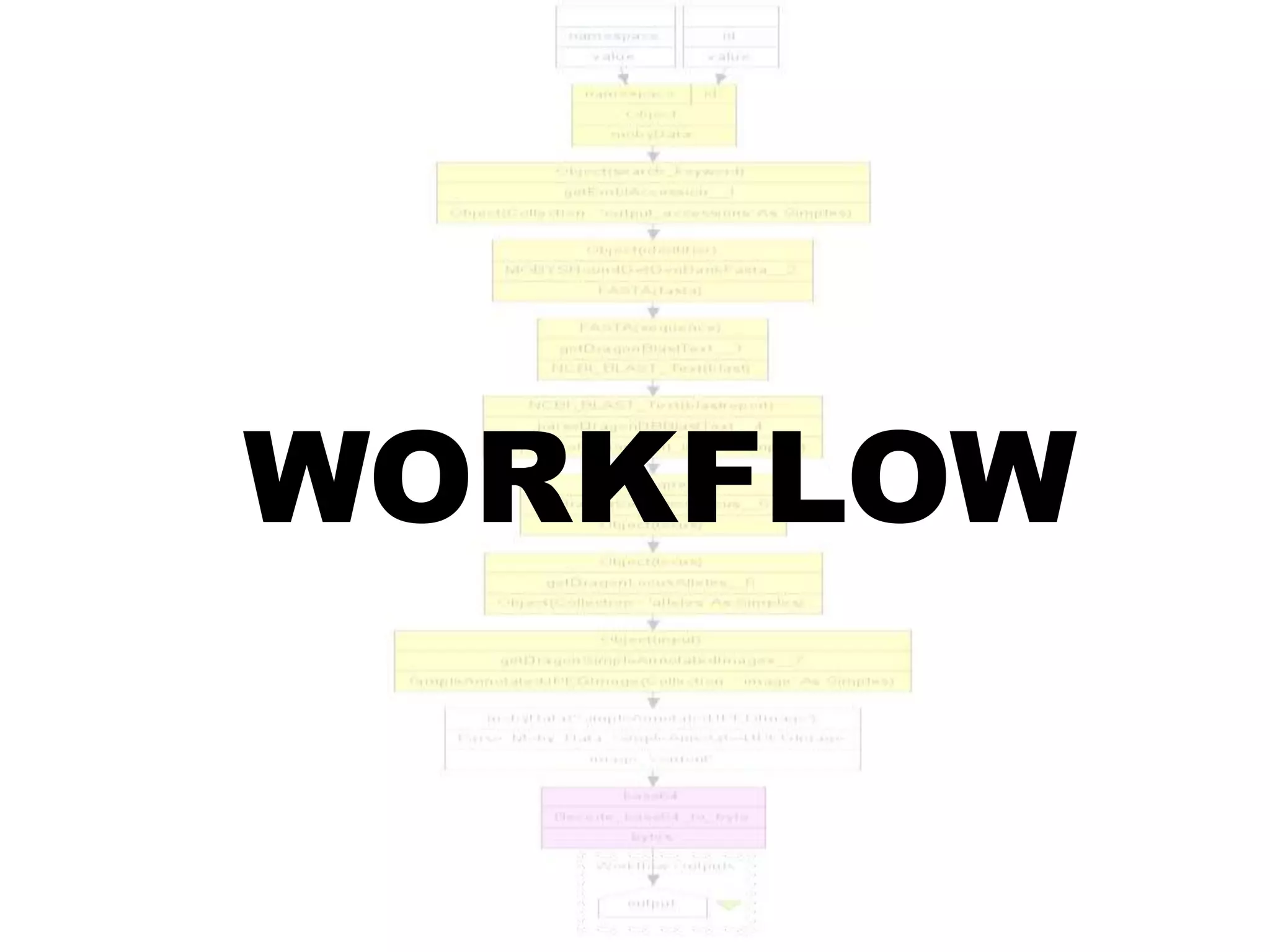

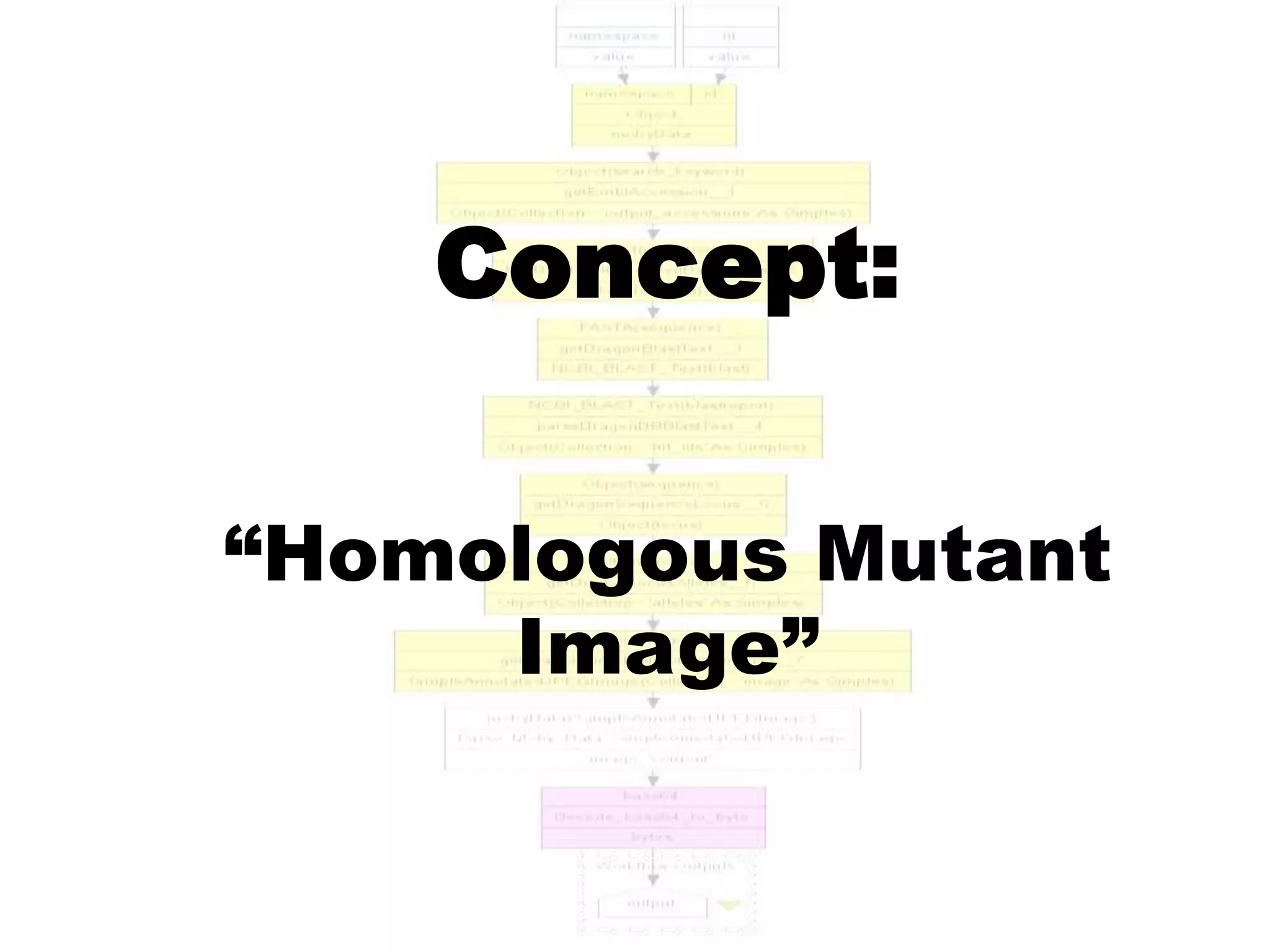




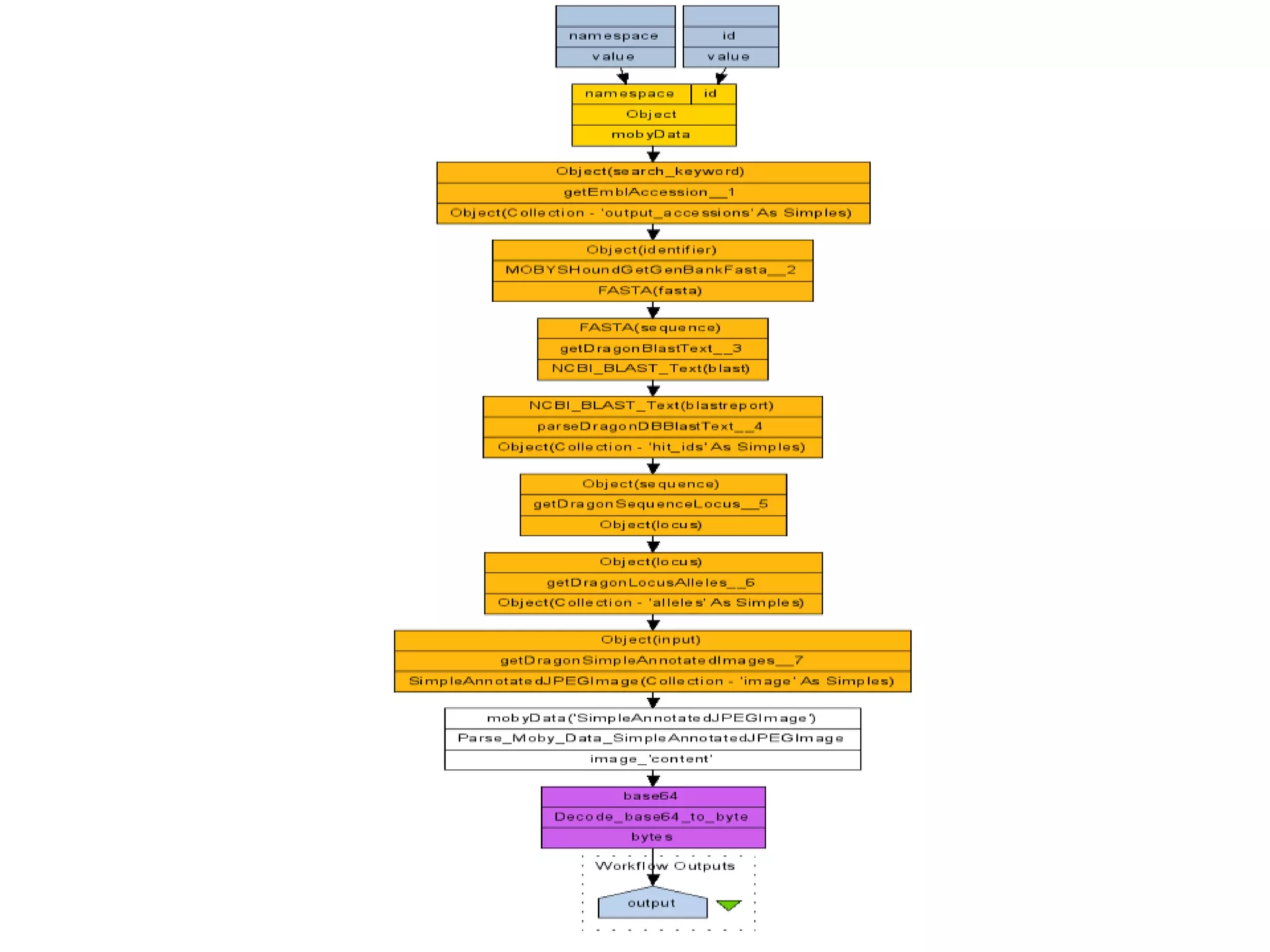


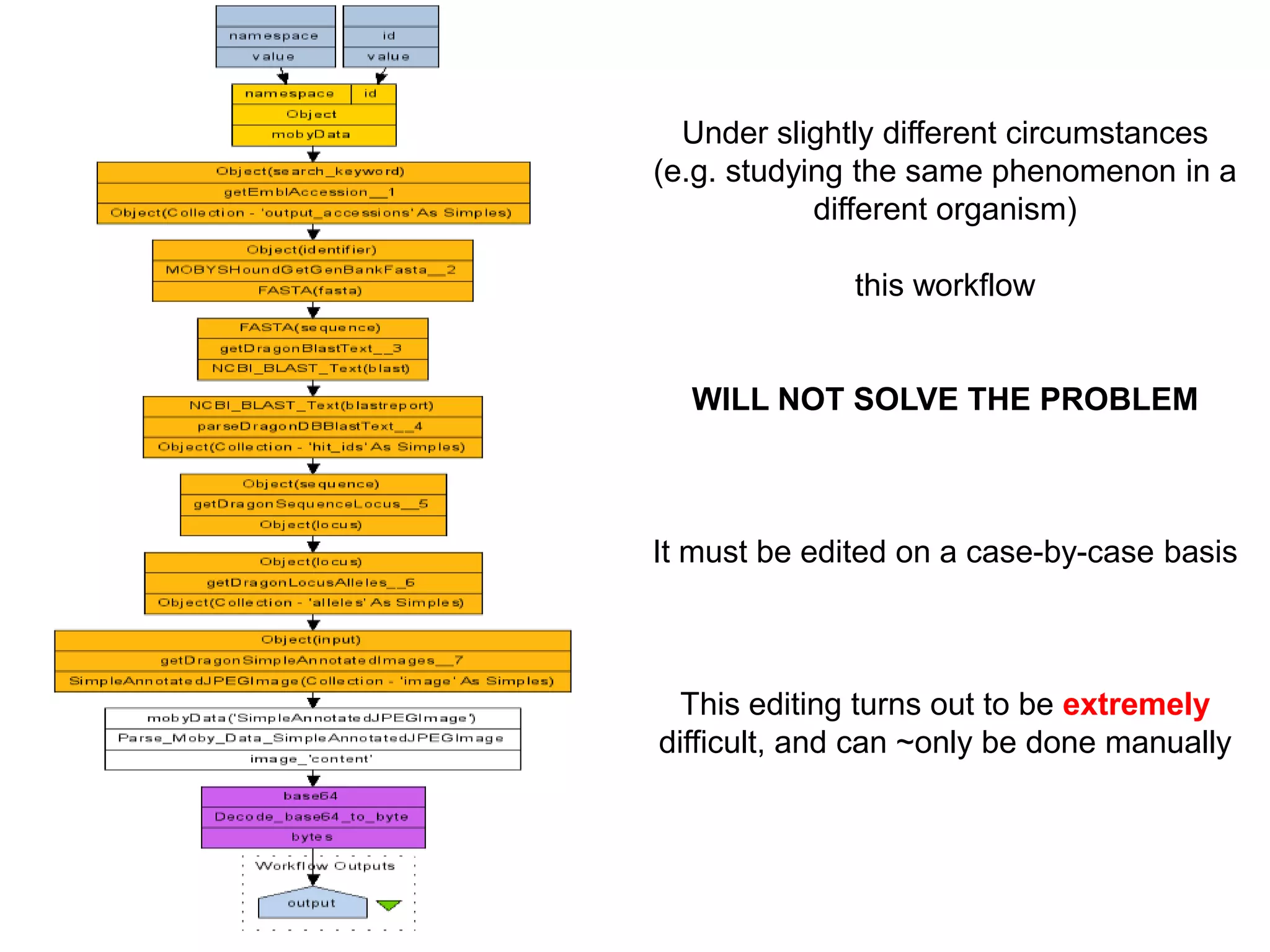

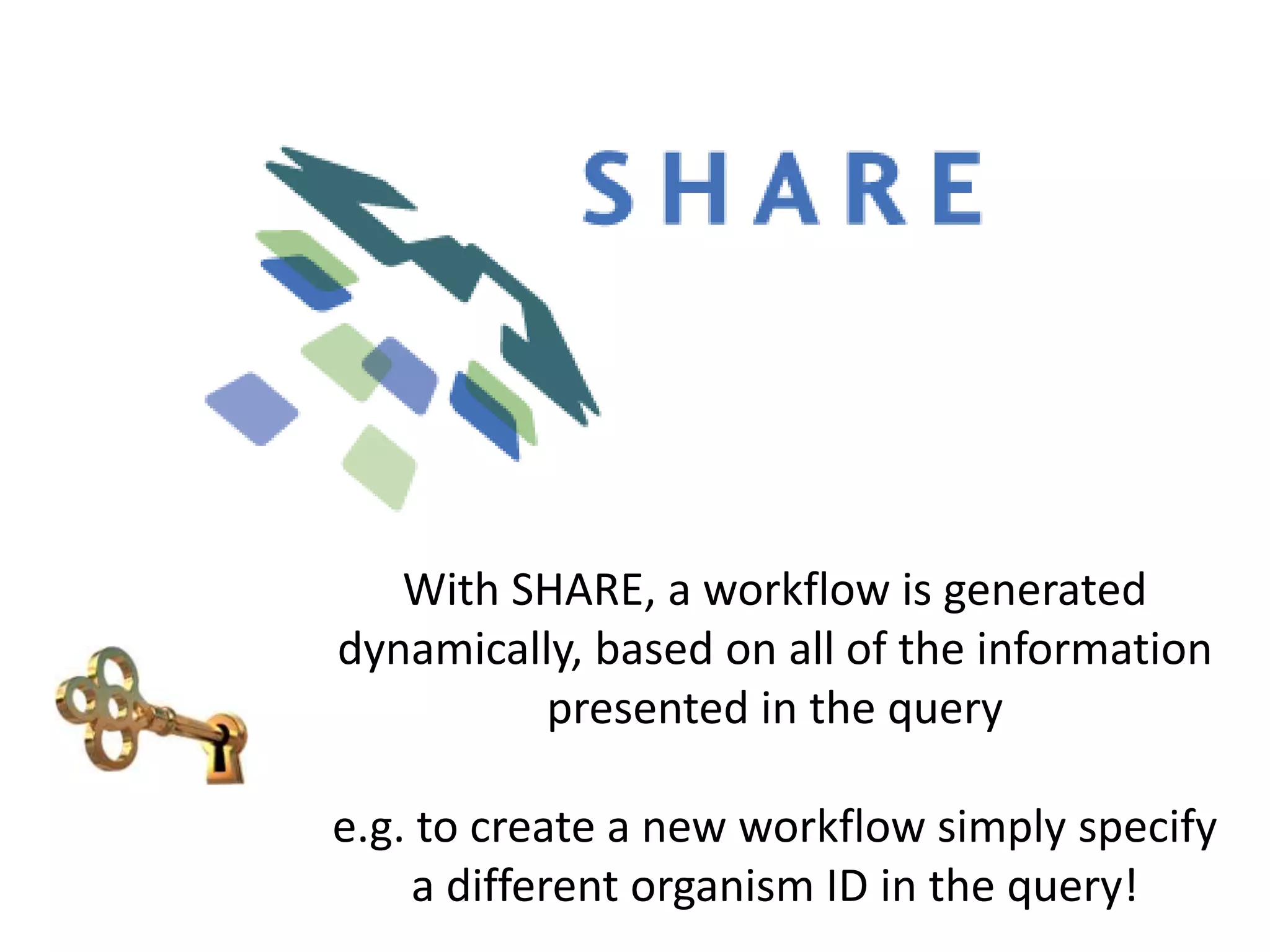
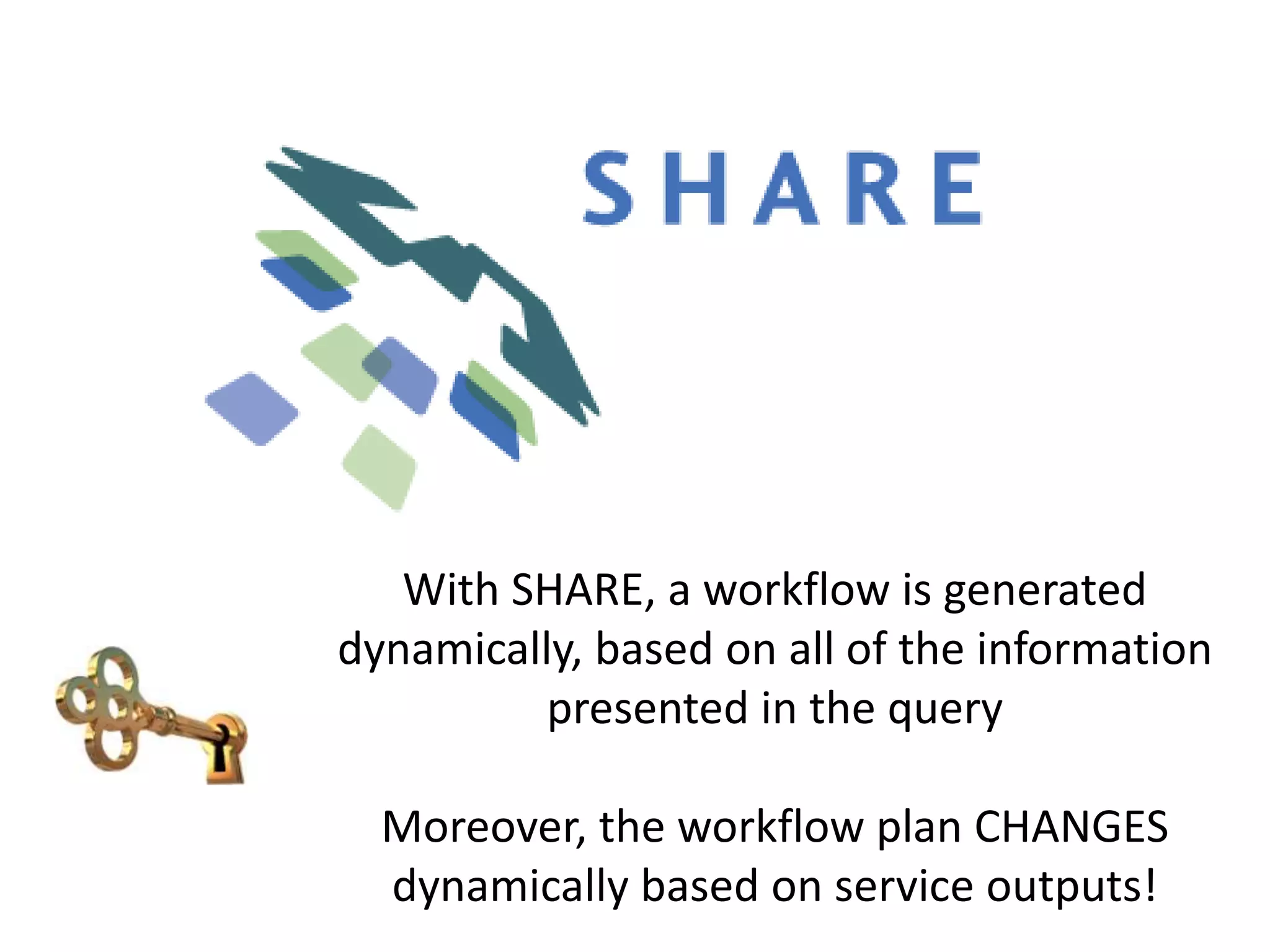

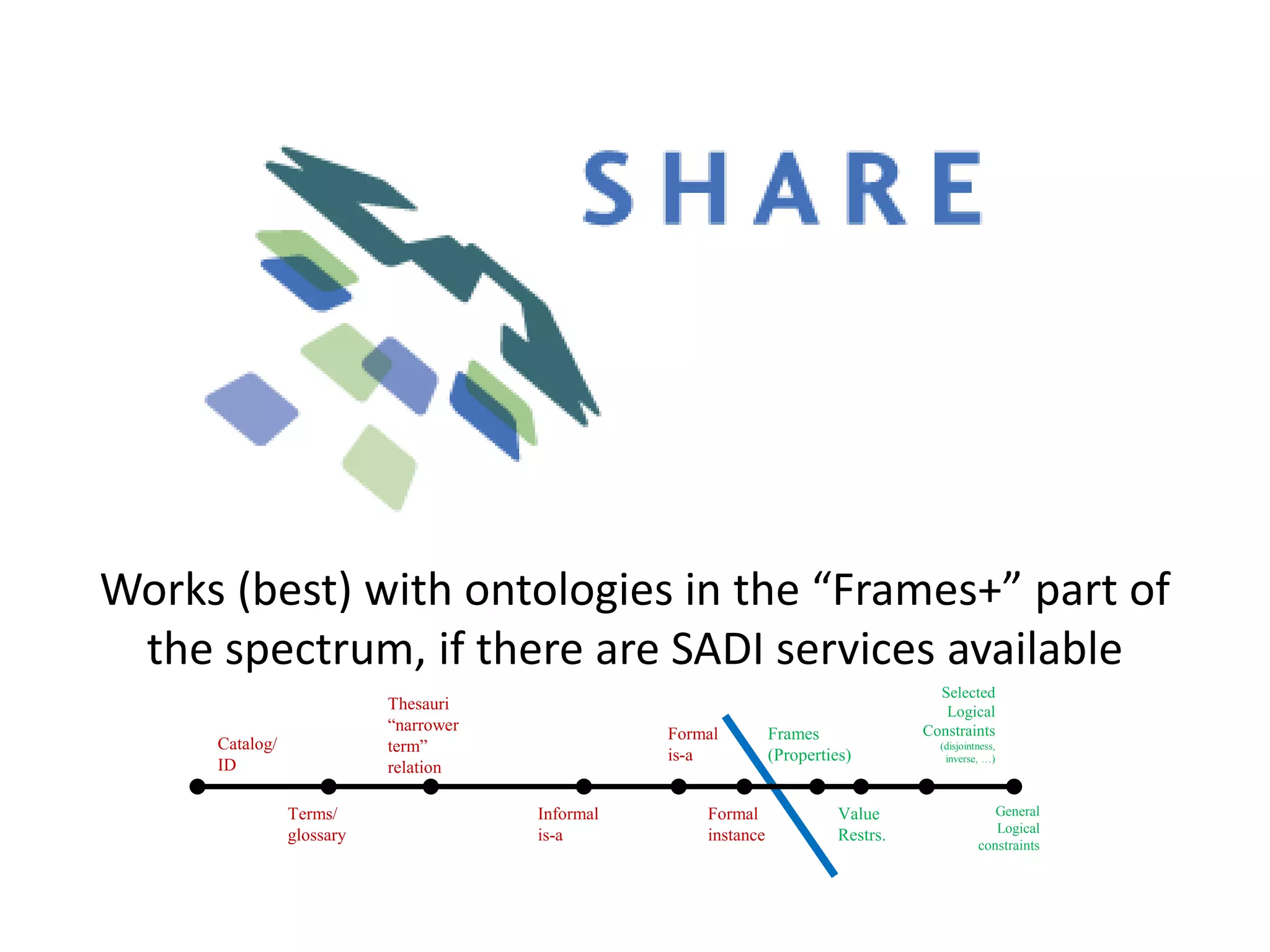

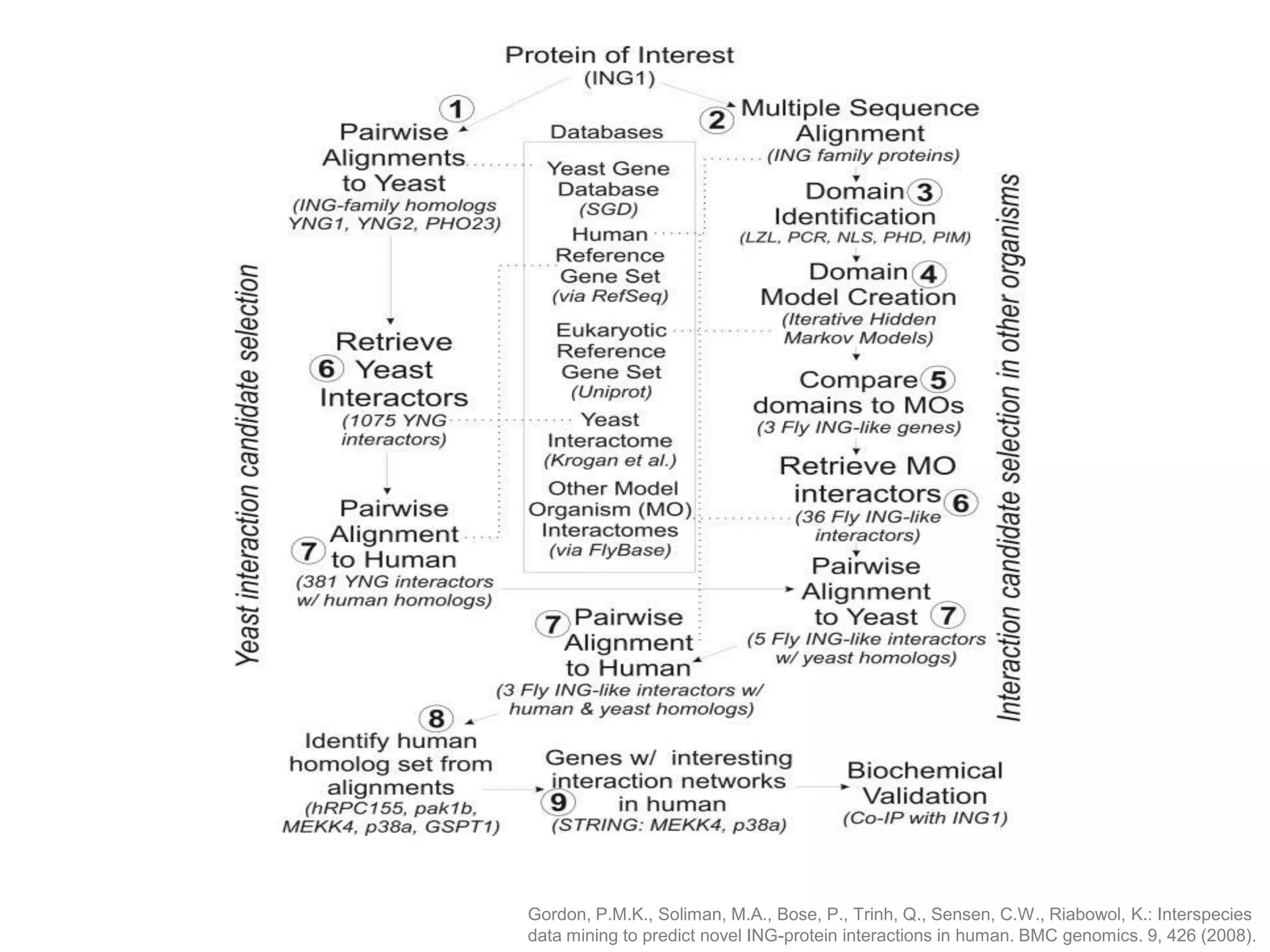










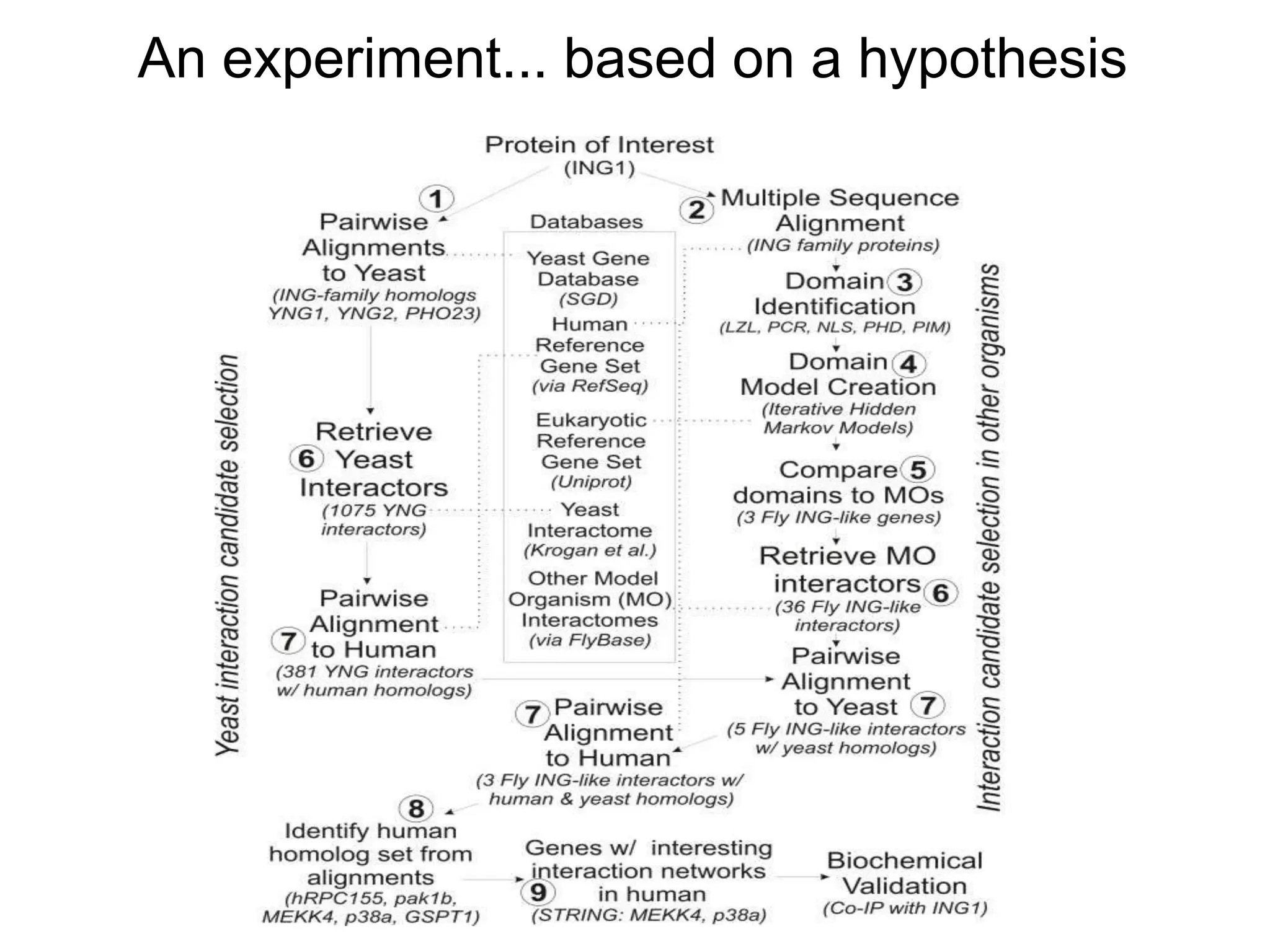
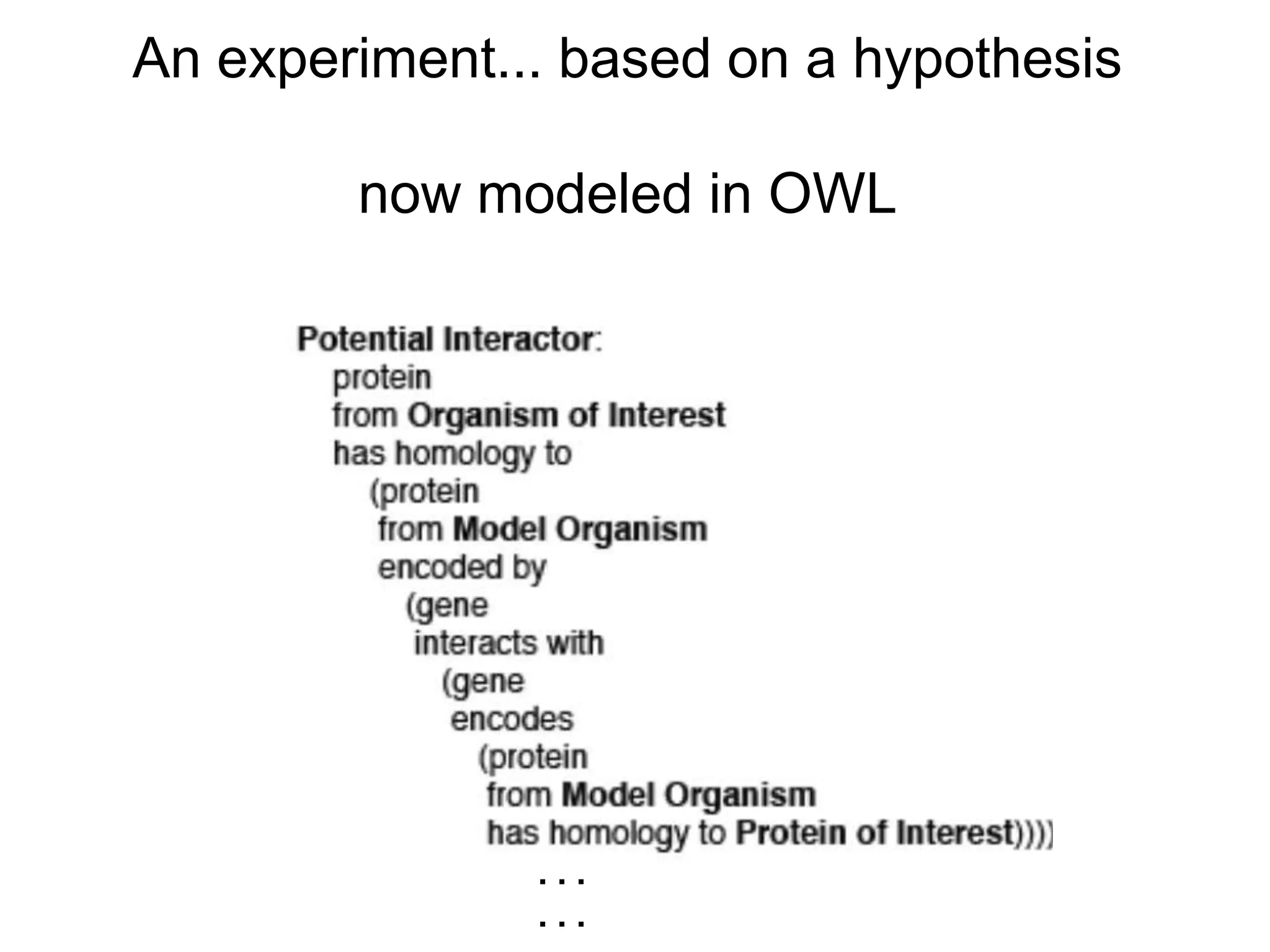


![Ontology = Hypothesis = Query = Workflow [= Materials and Methods ]
Most of your publication is done!
All you need to do now is interpret the results!
These can be automatically derived through
provenance information during workflow execution](https://image.slidesharecdn.com/flexibleworkflows-120516052642-phpapp01/75/Evaluating-Hypotheses-using-SPARQL-DL-as-an-abstract-workflow-language-to-choreograph-SADI-Semantic-Web-Services-137-2048.jpg)




