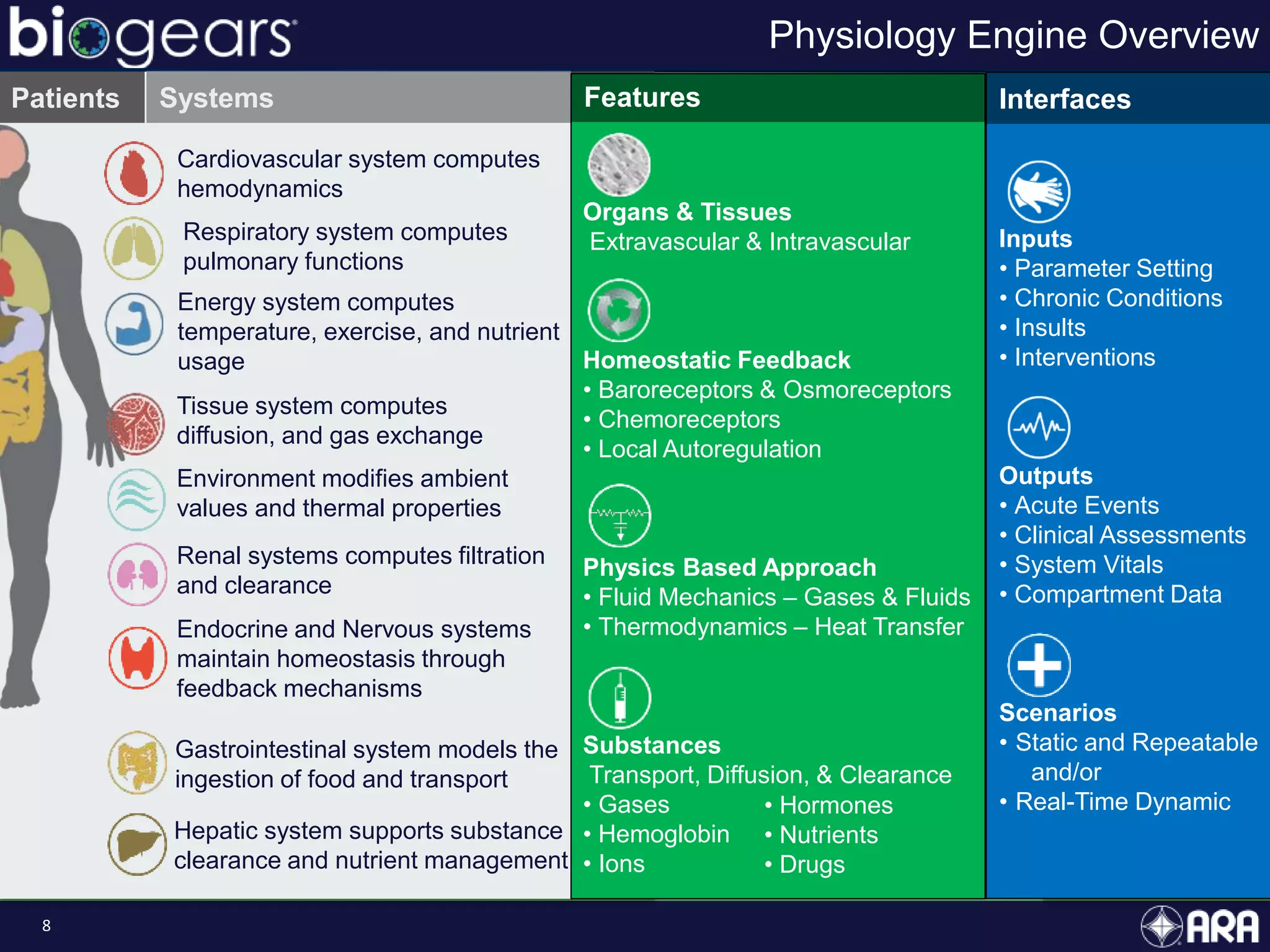
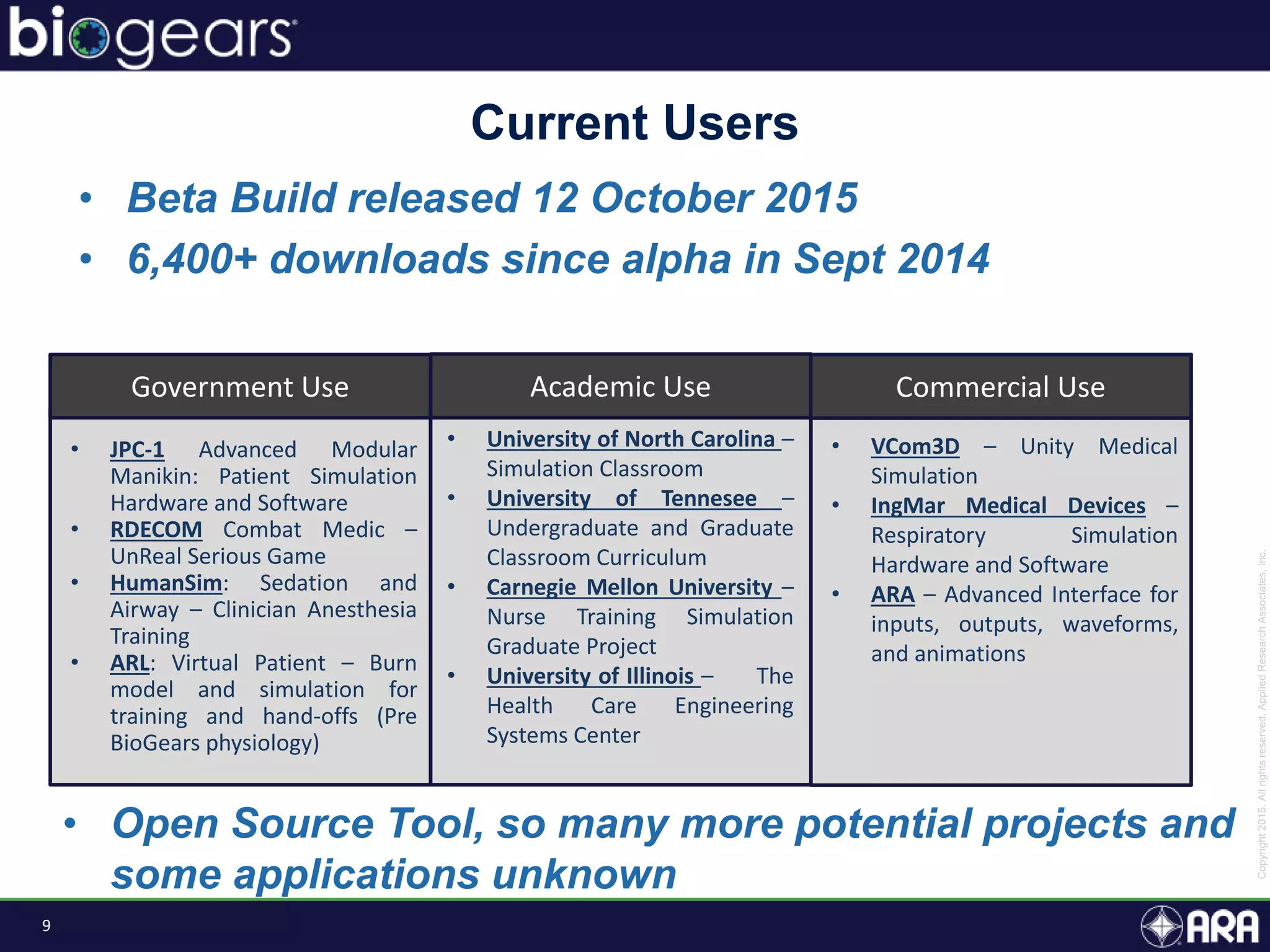
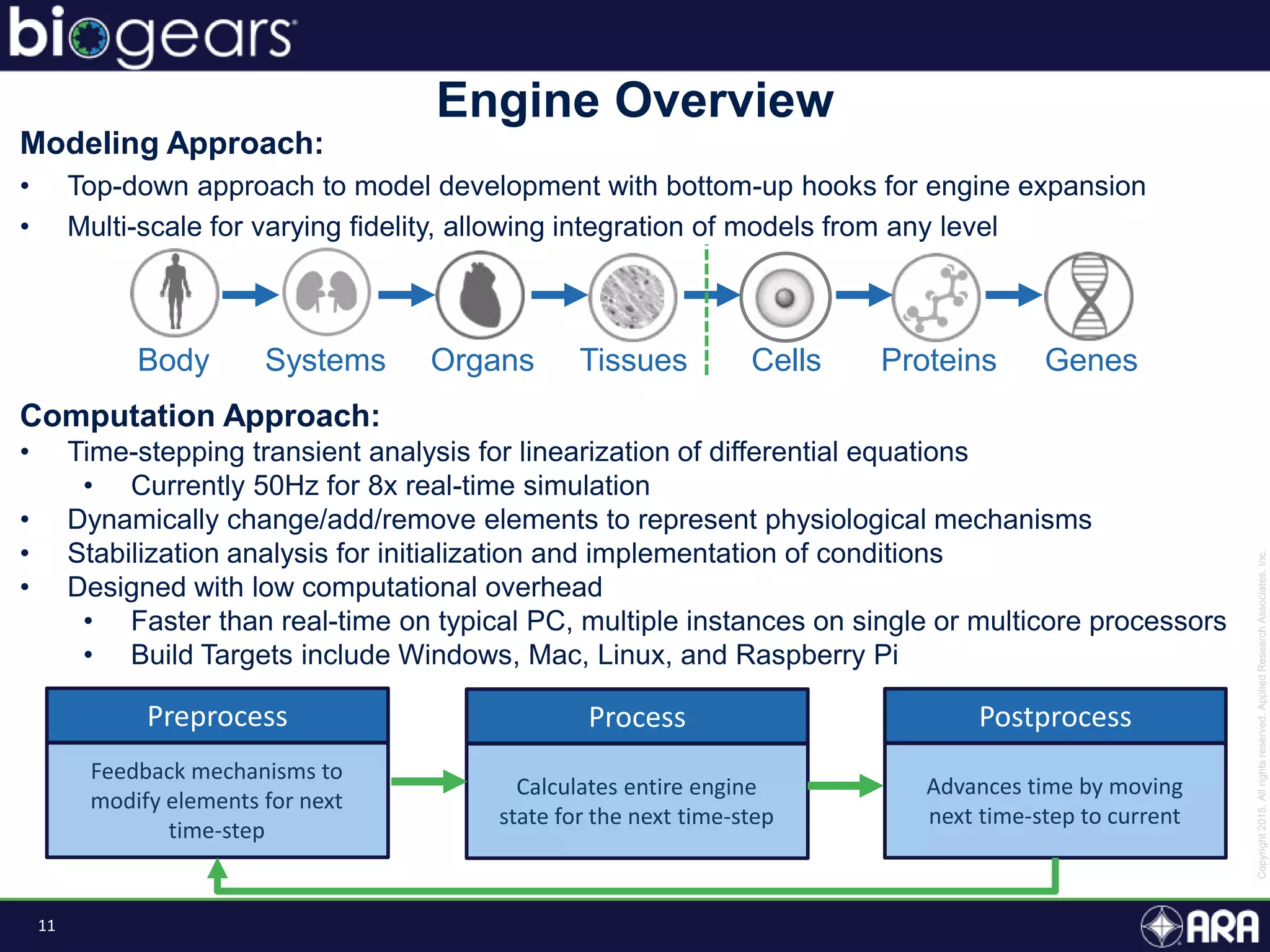
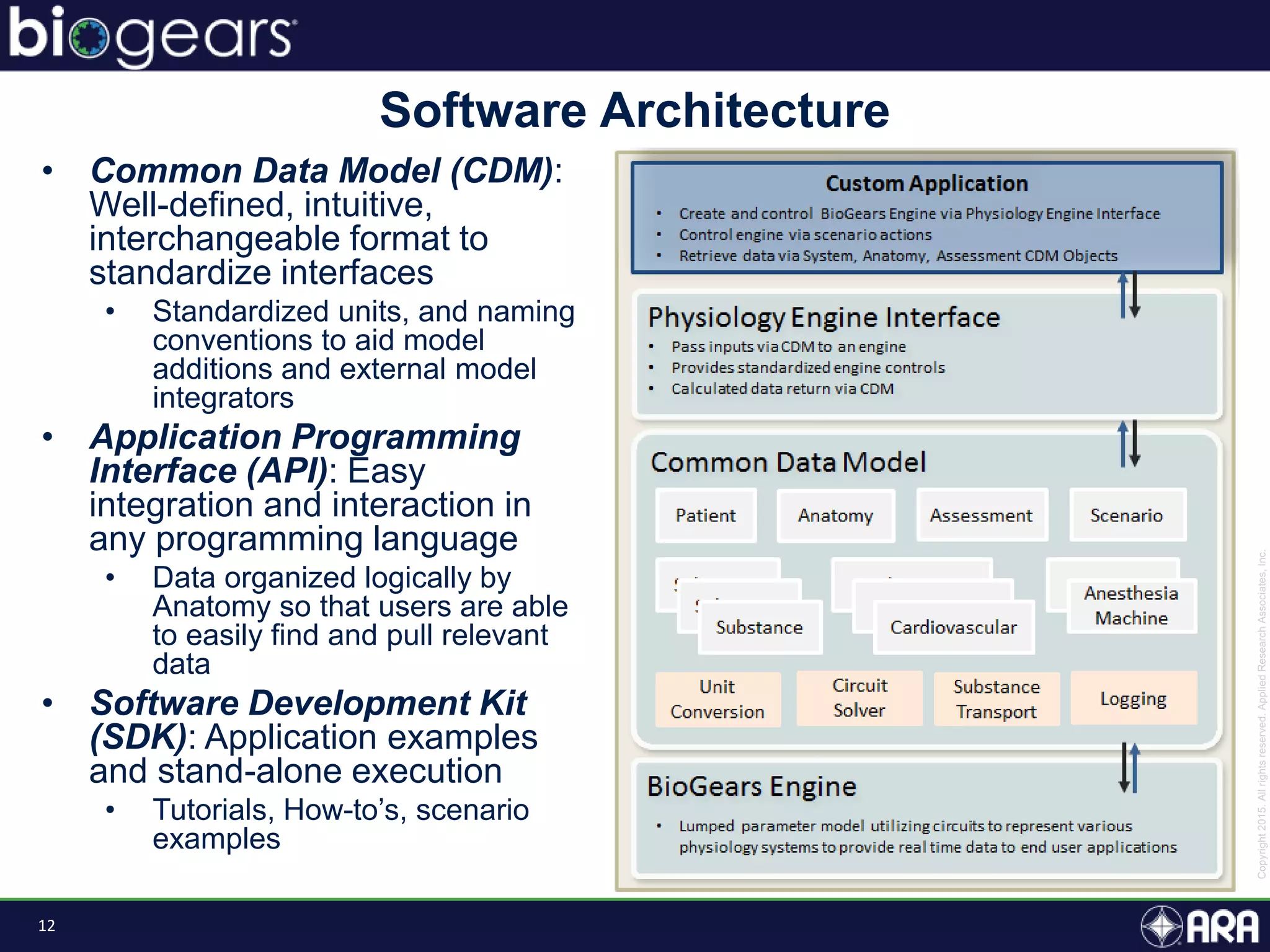
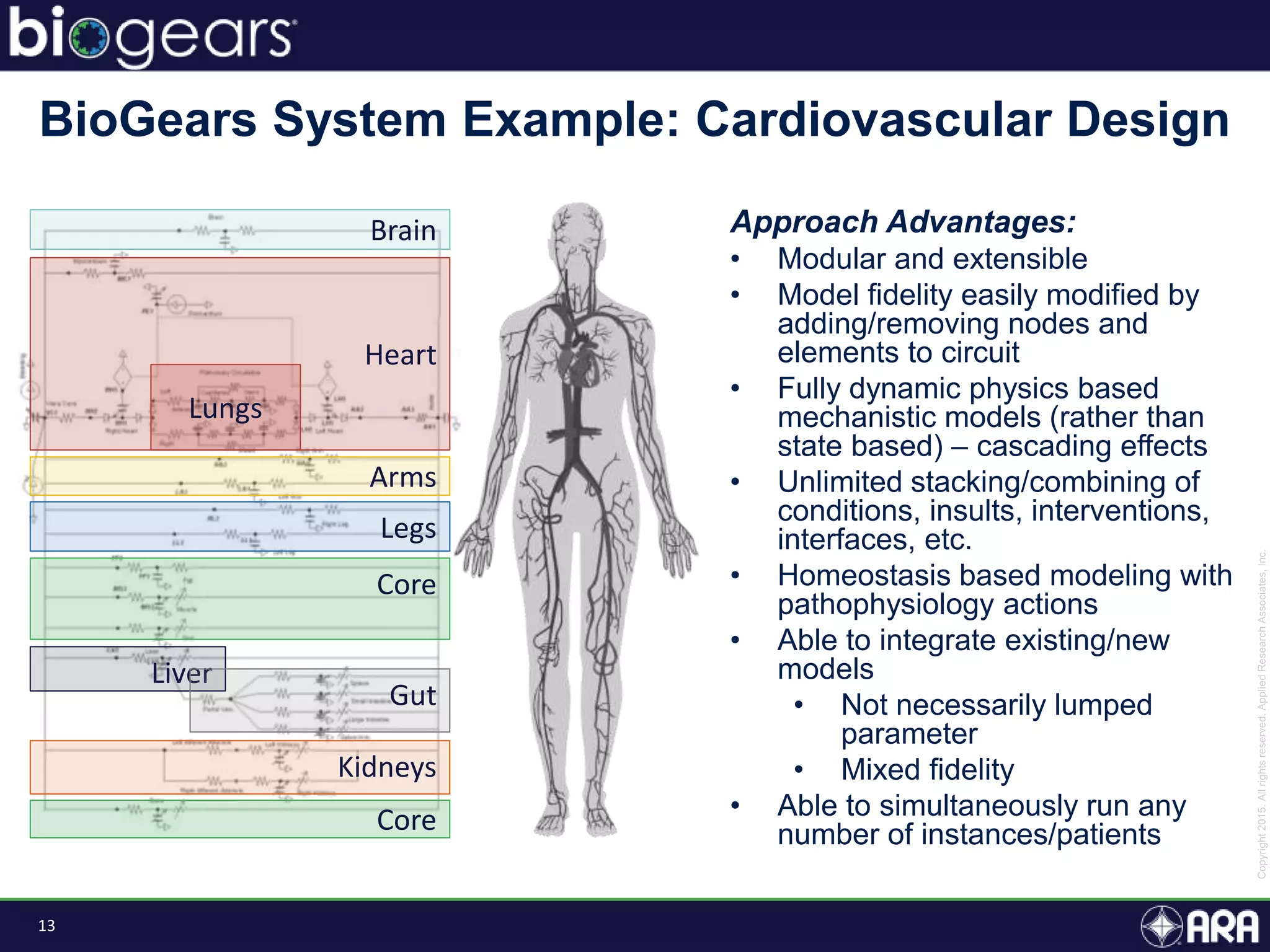
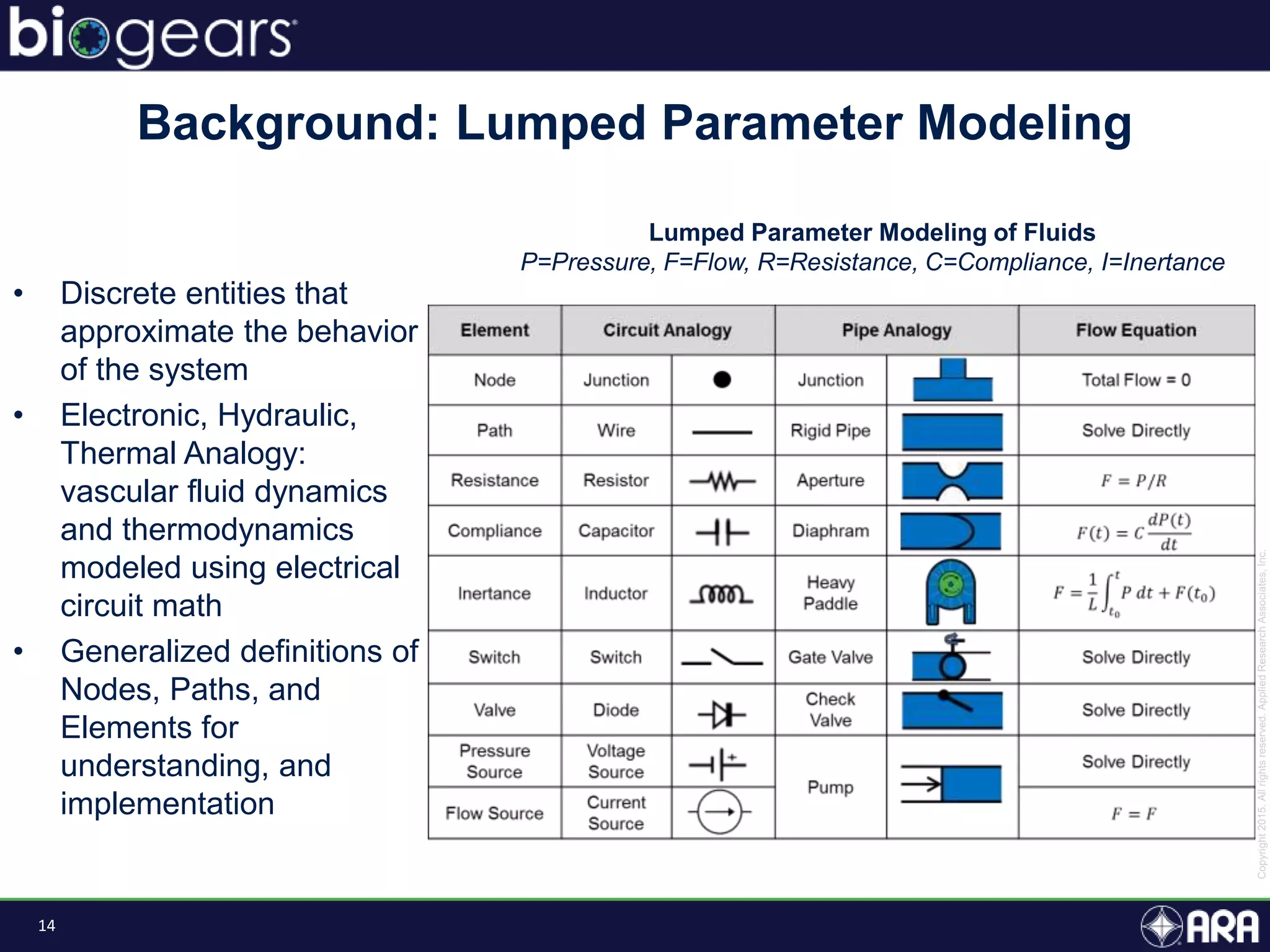
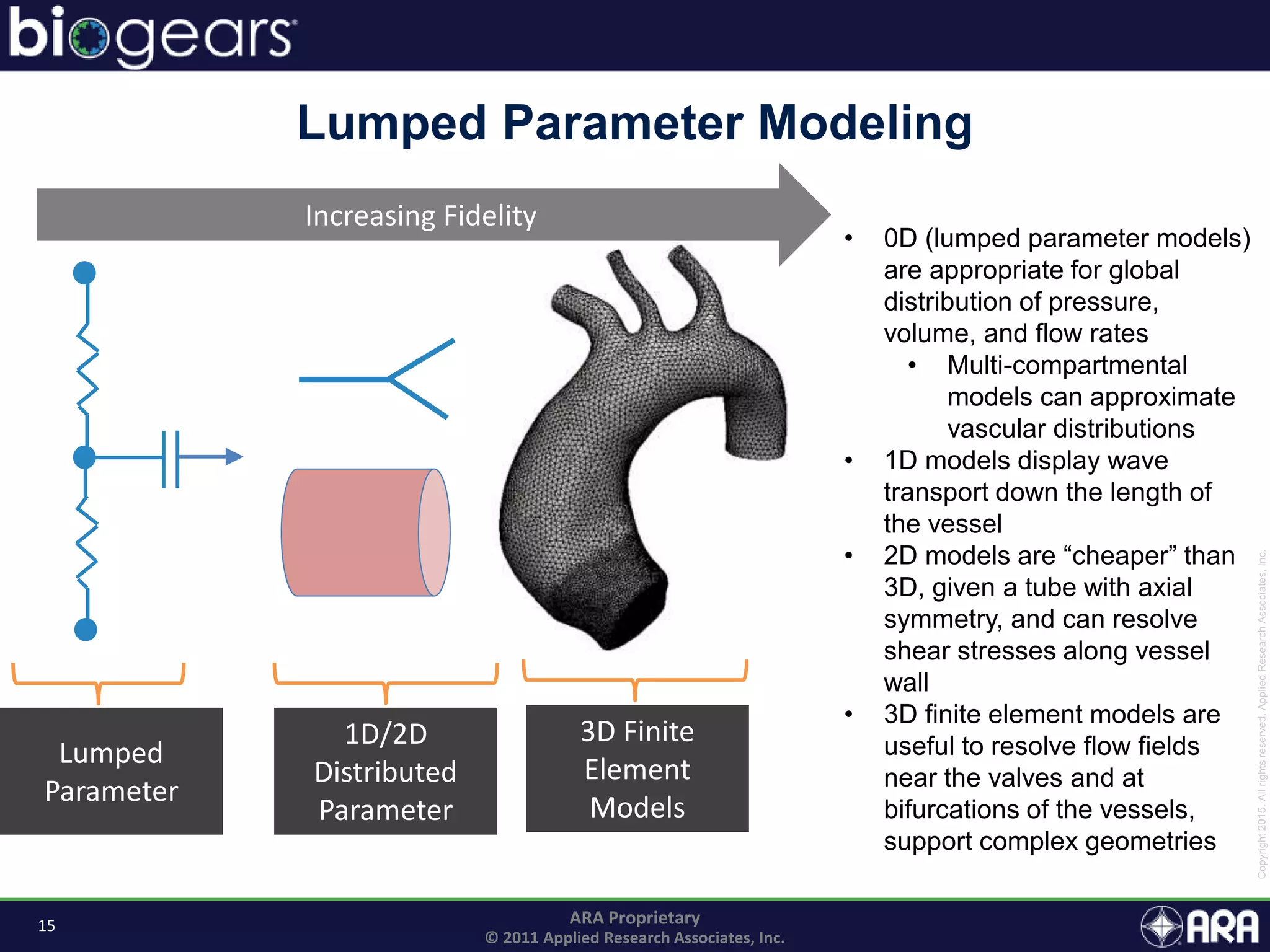
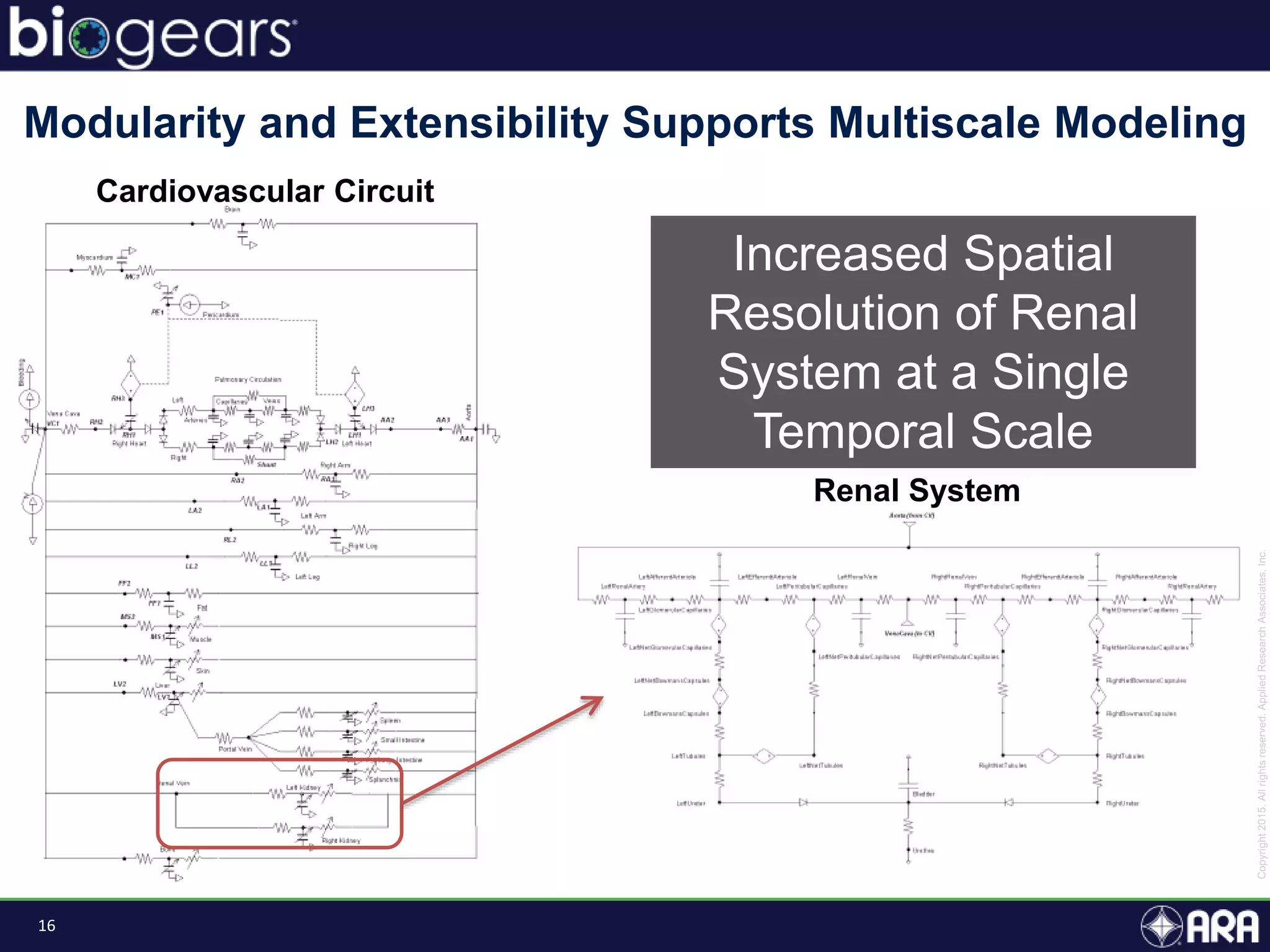
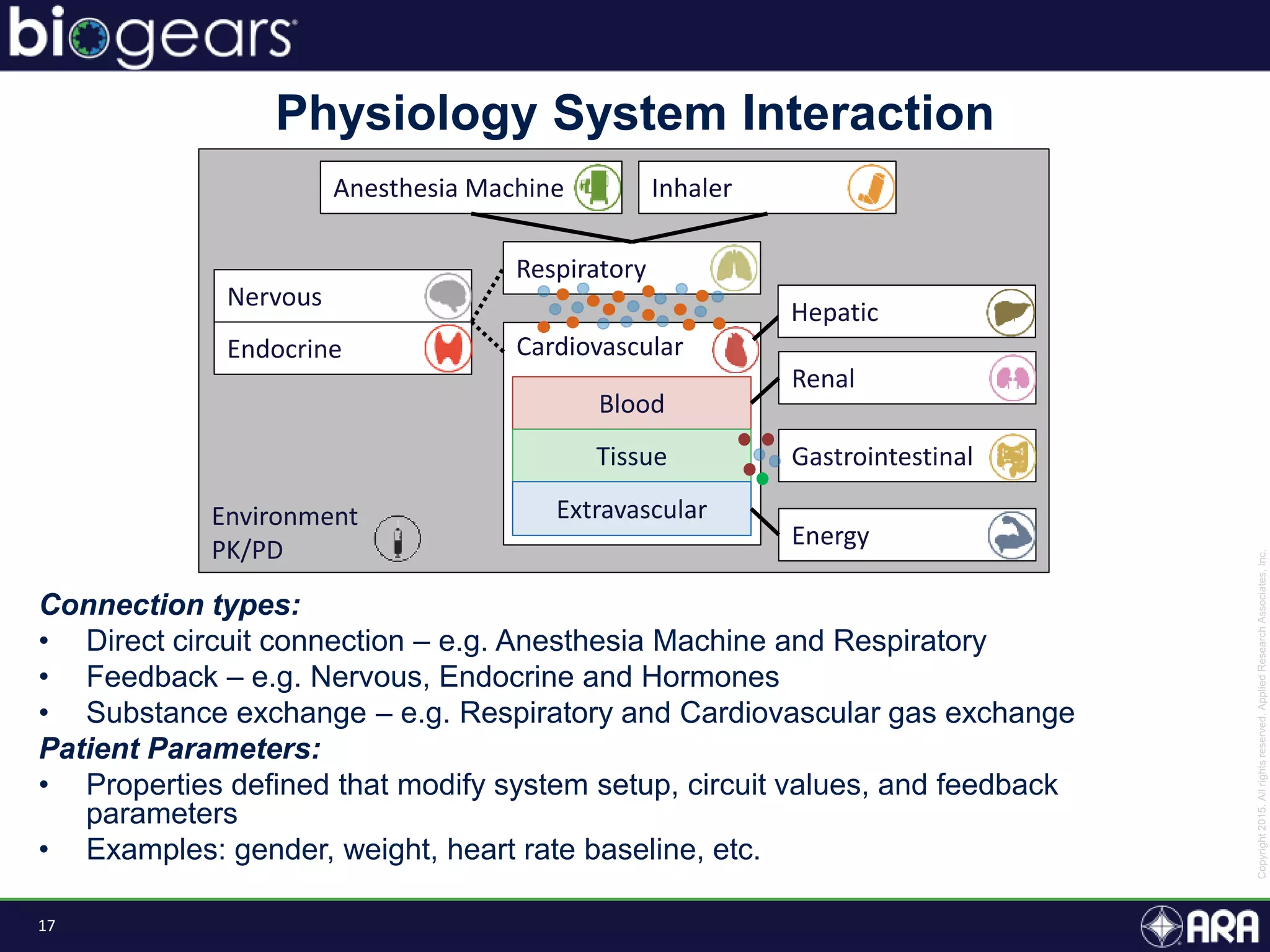
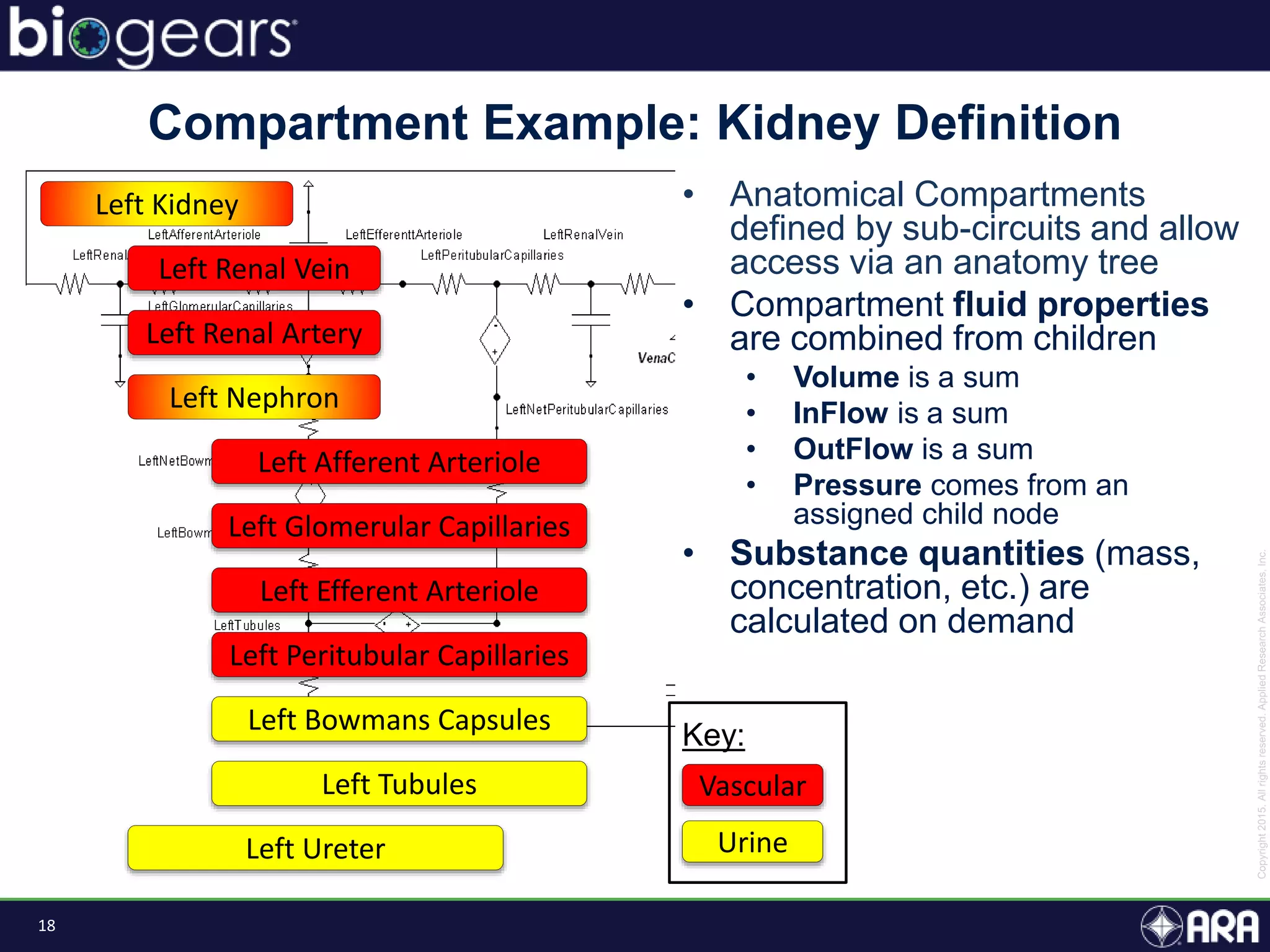
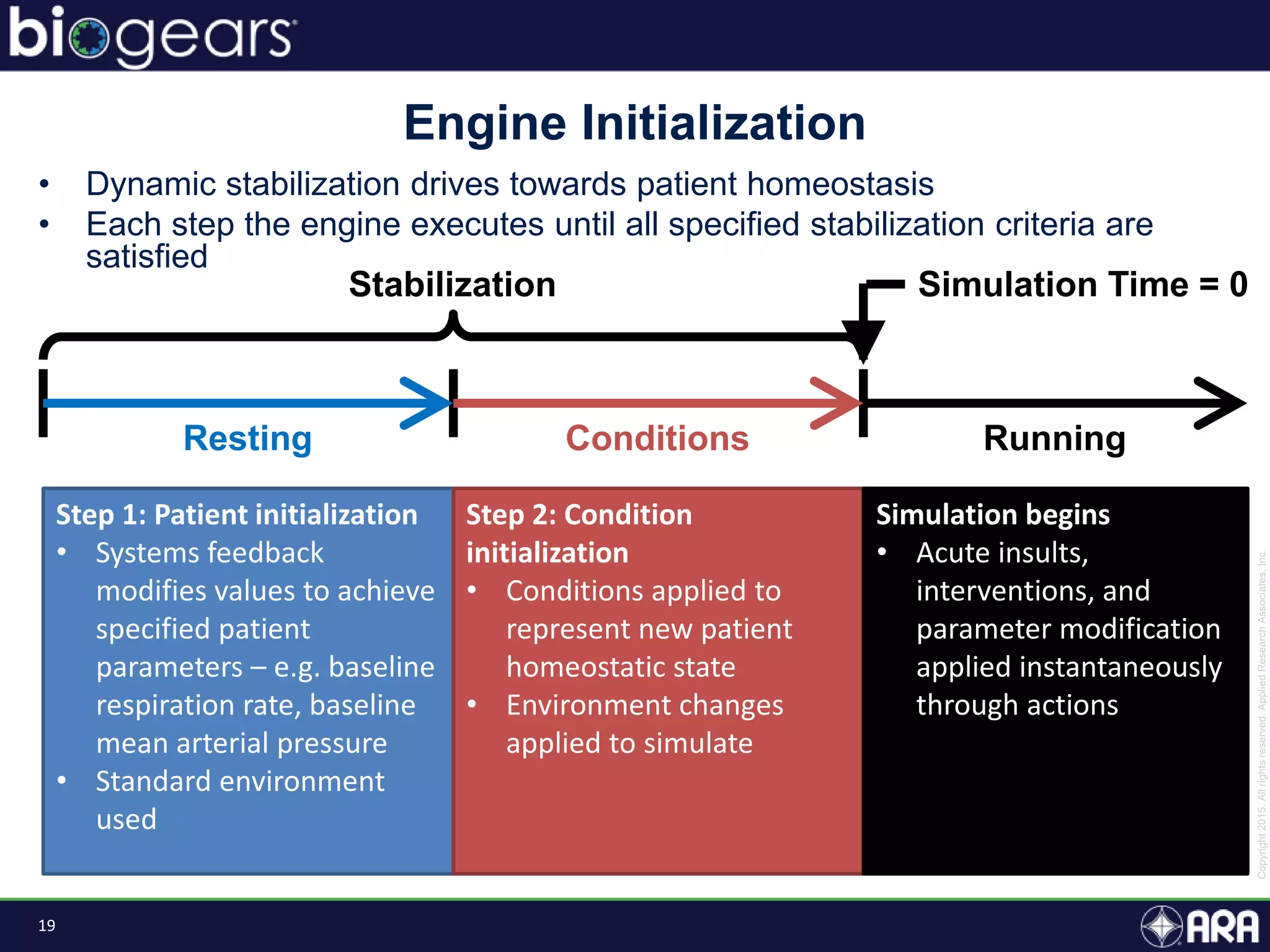
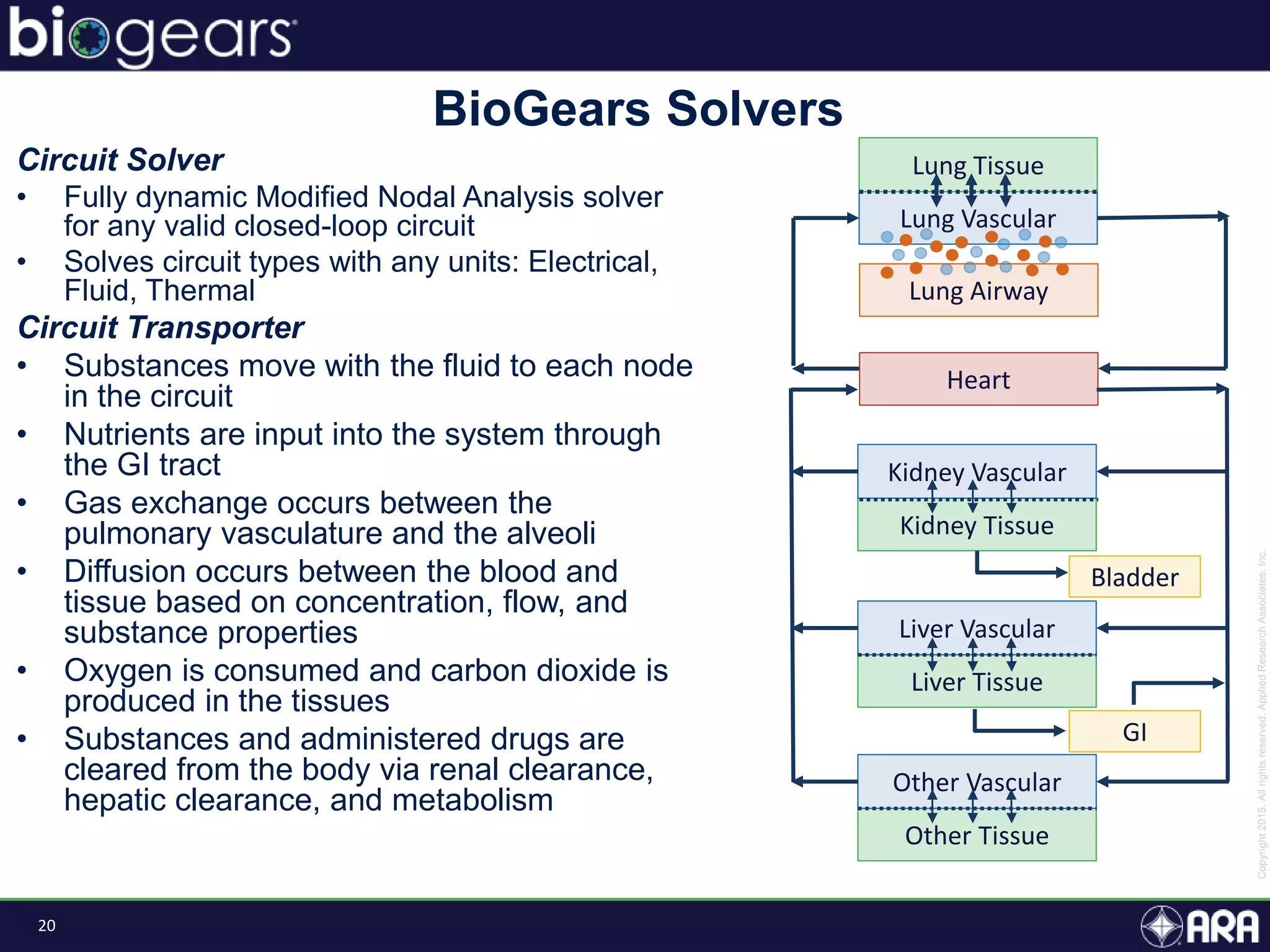
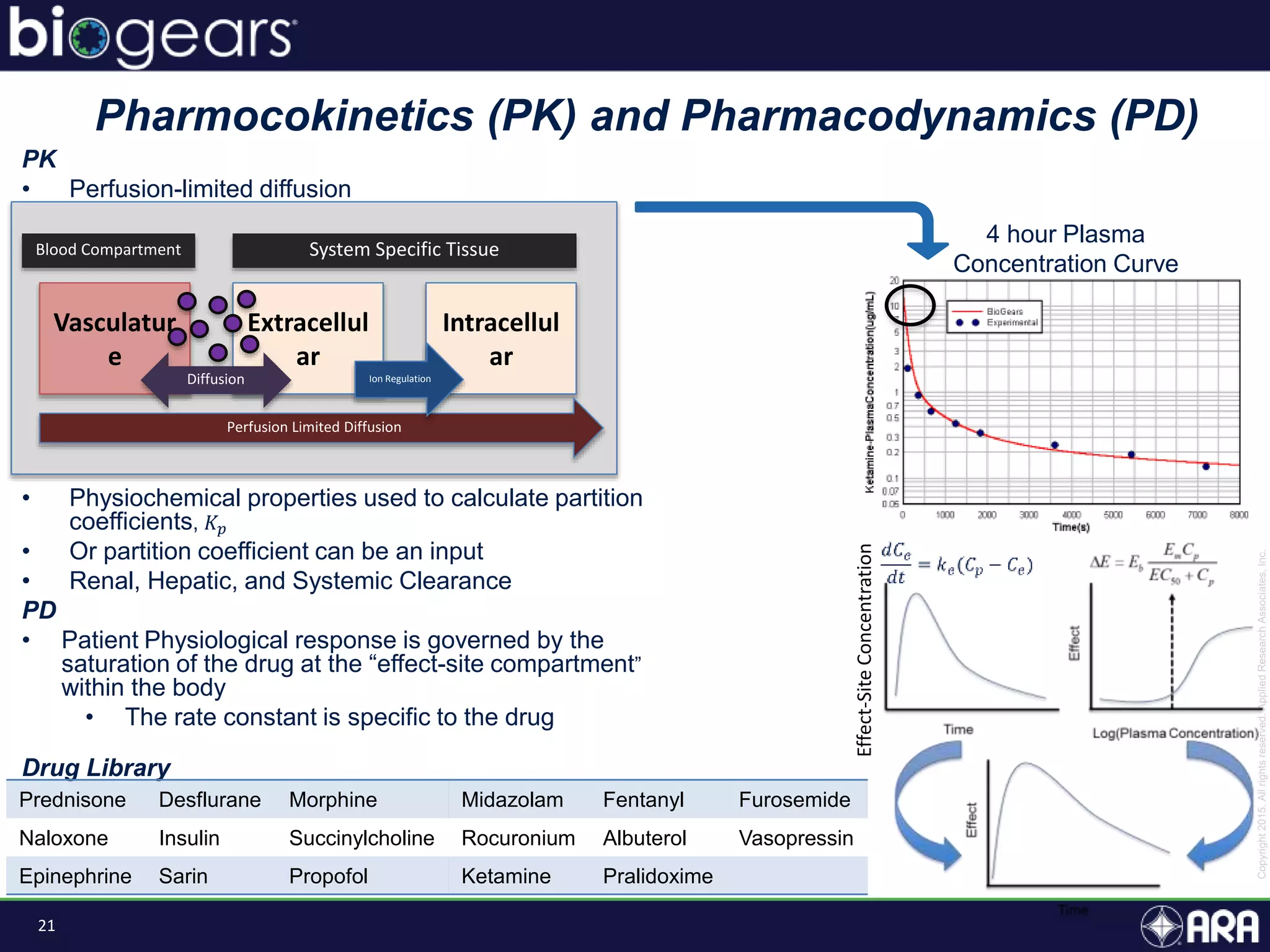
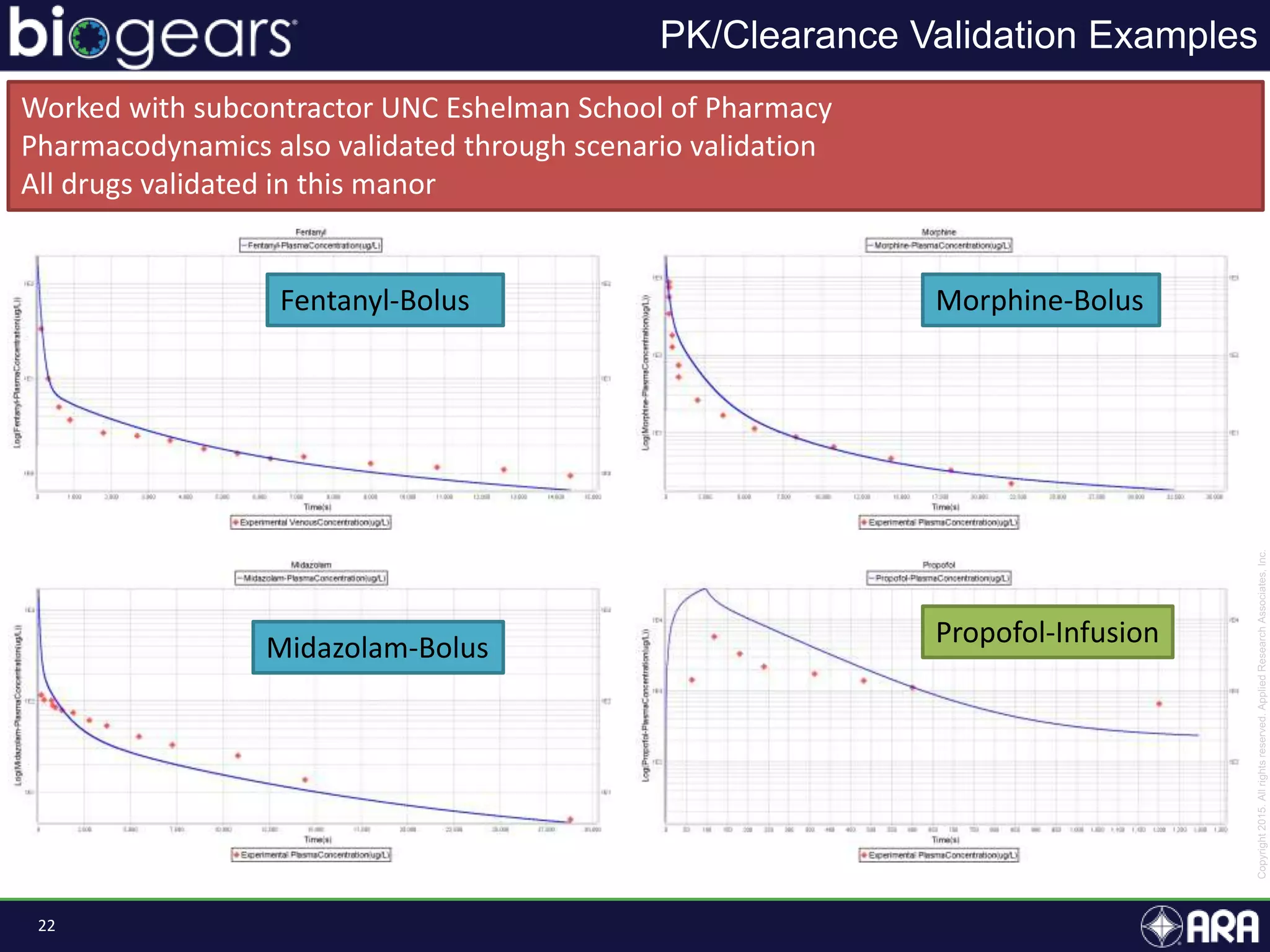
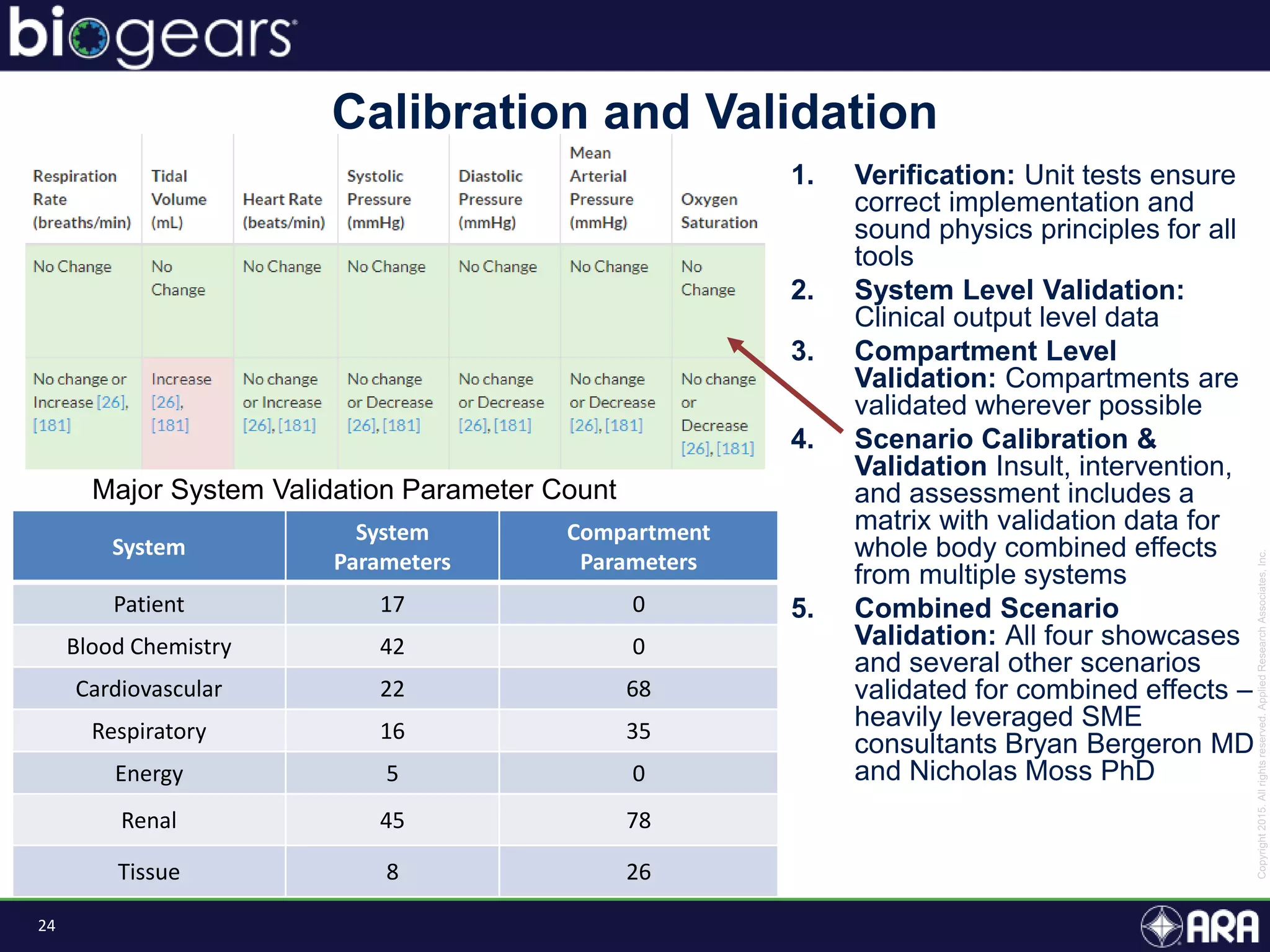
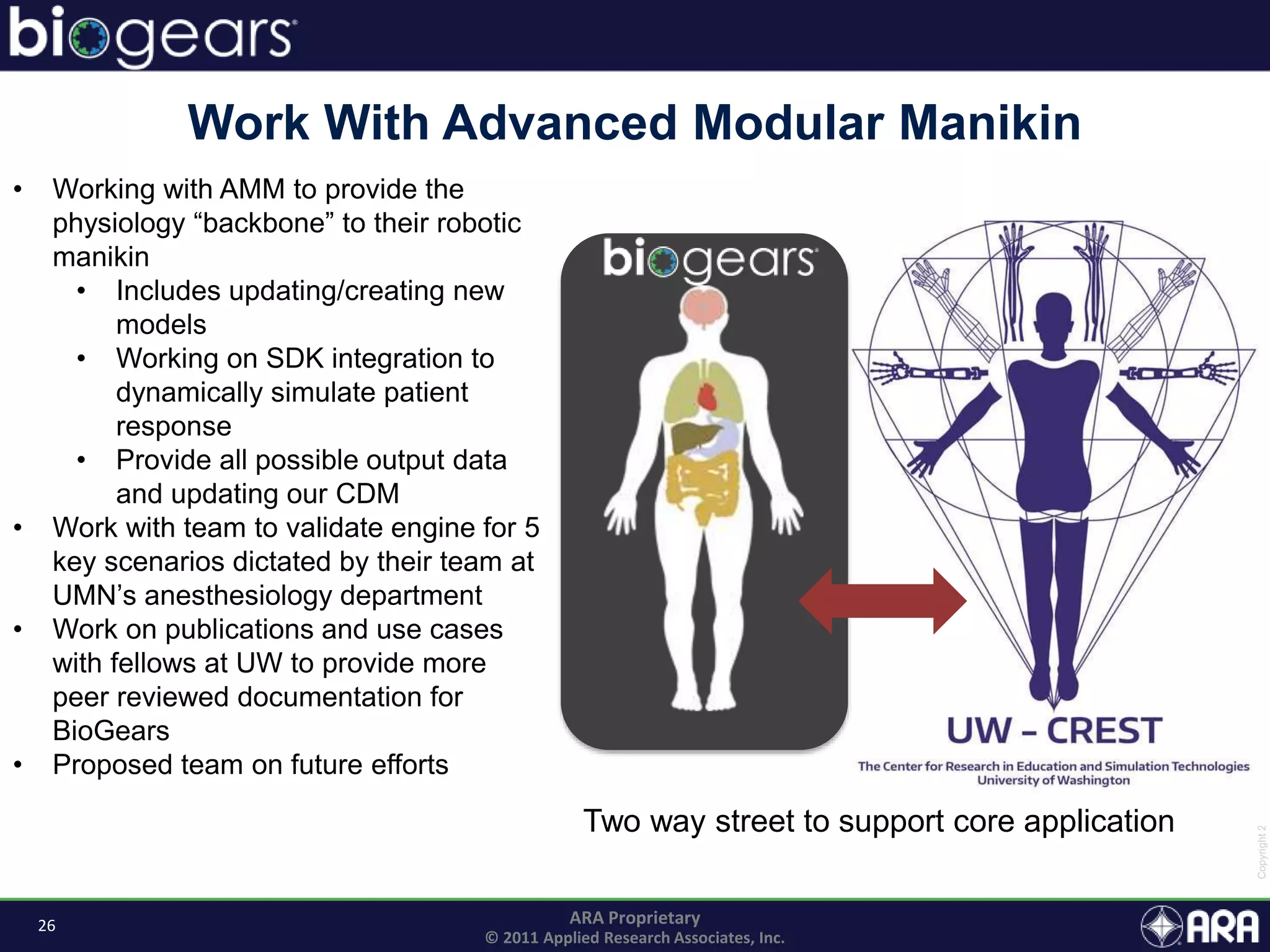
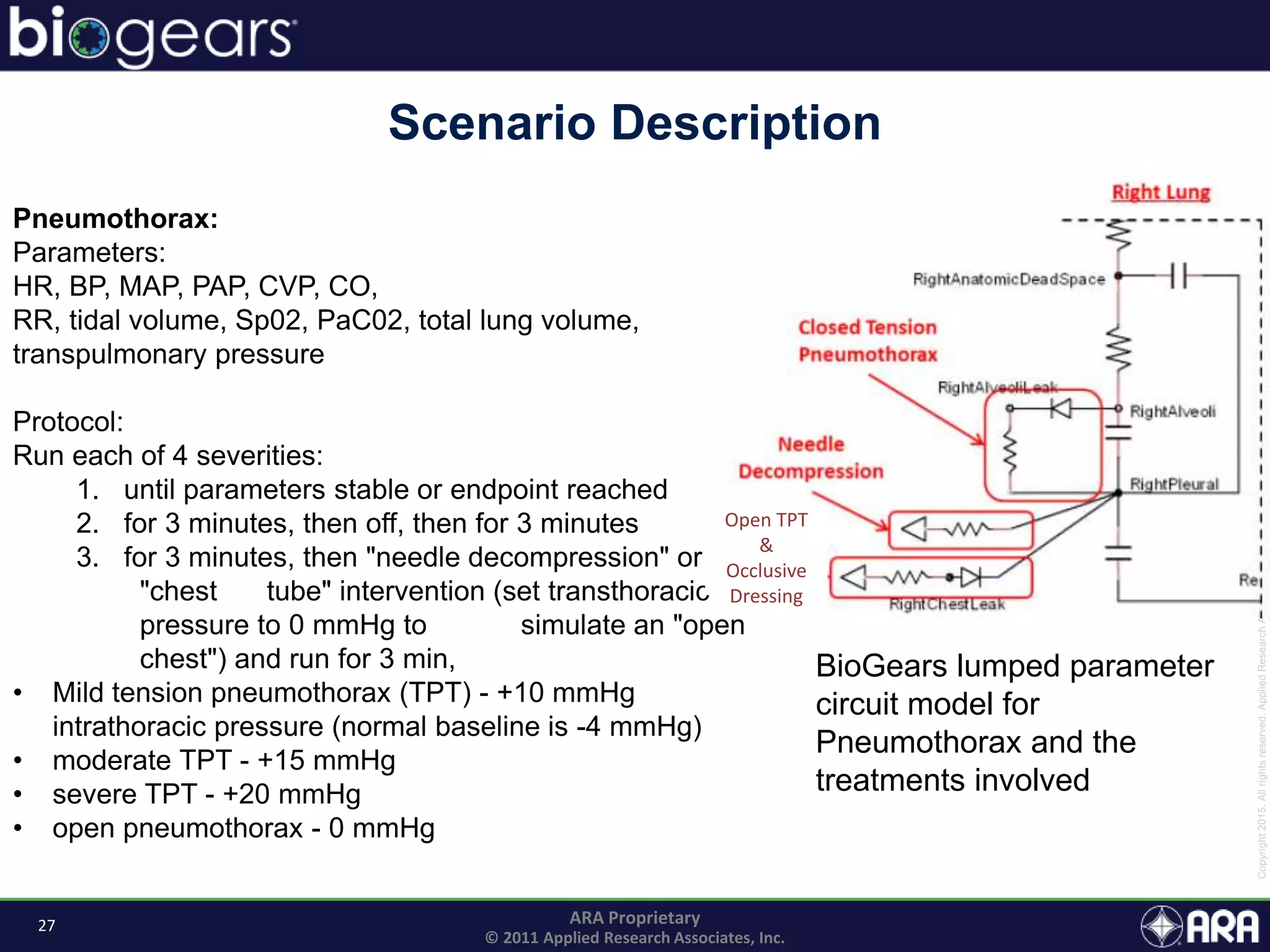
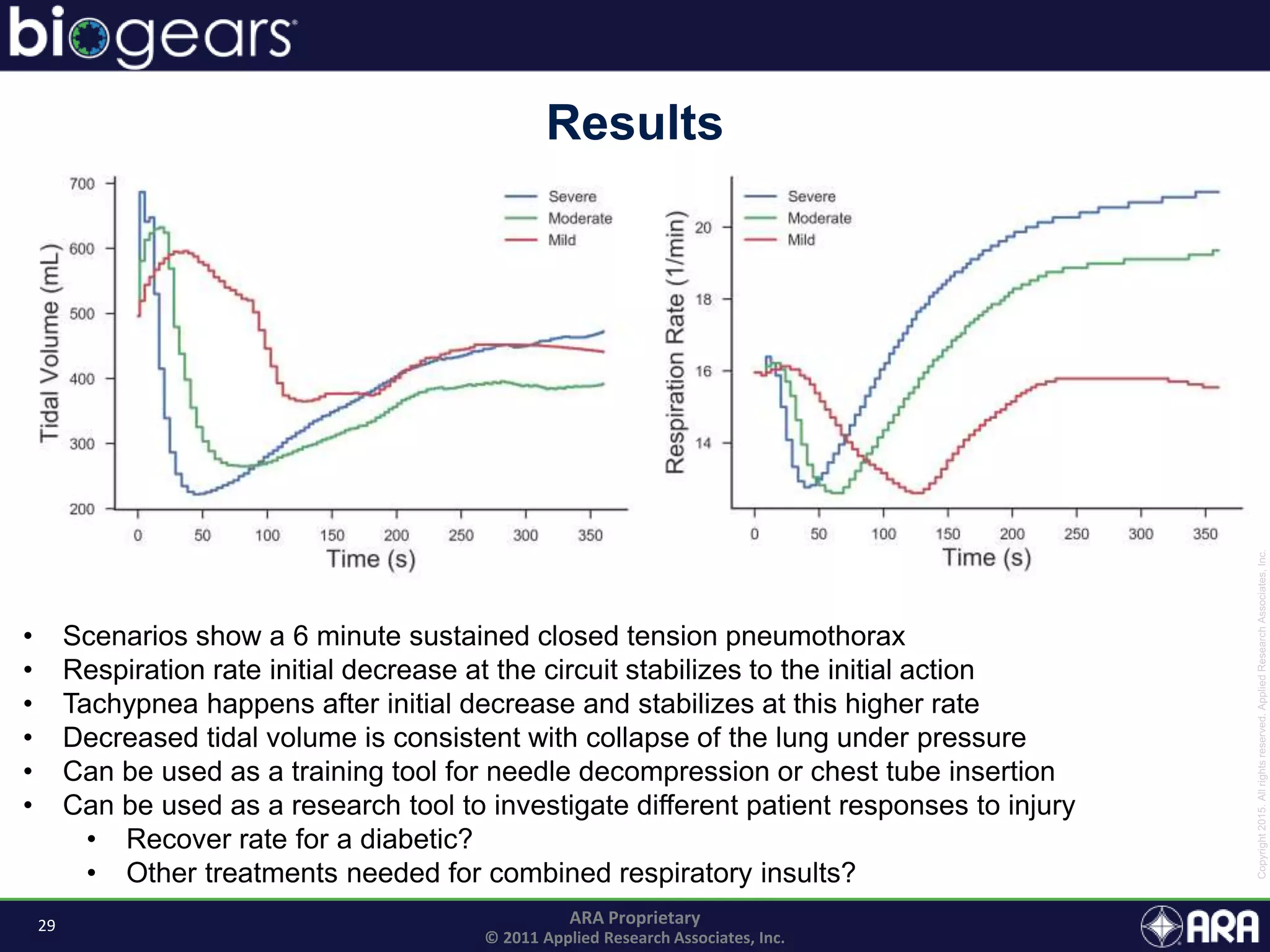
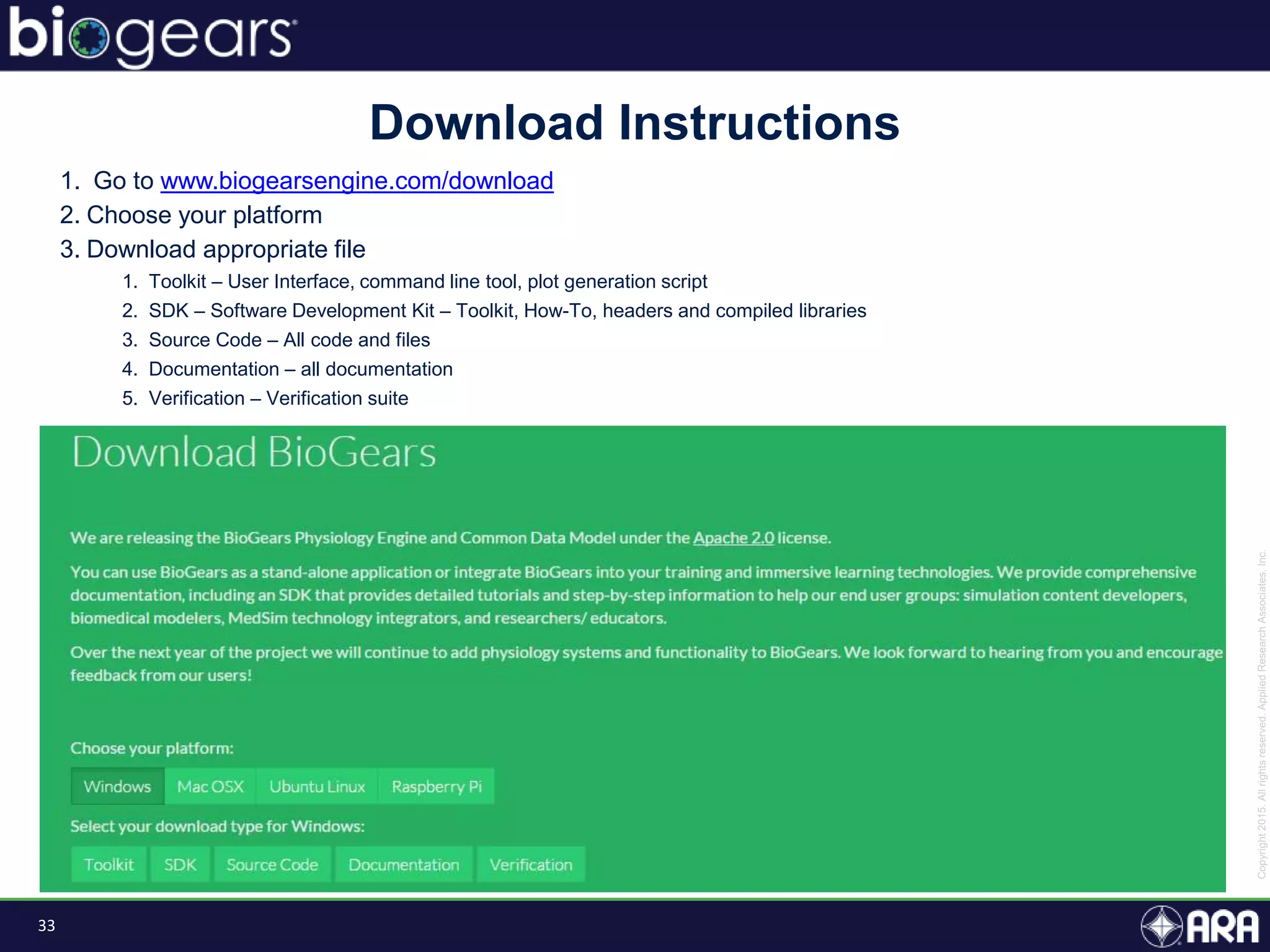
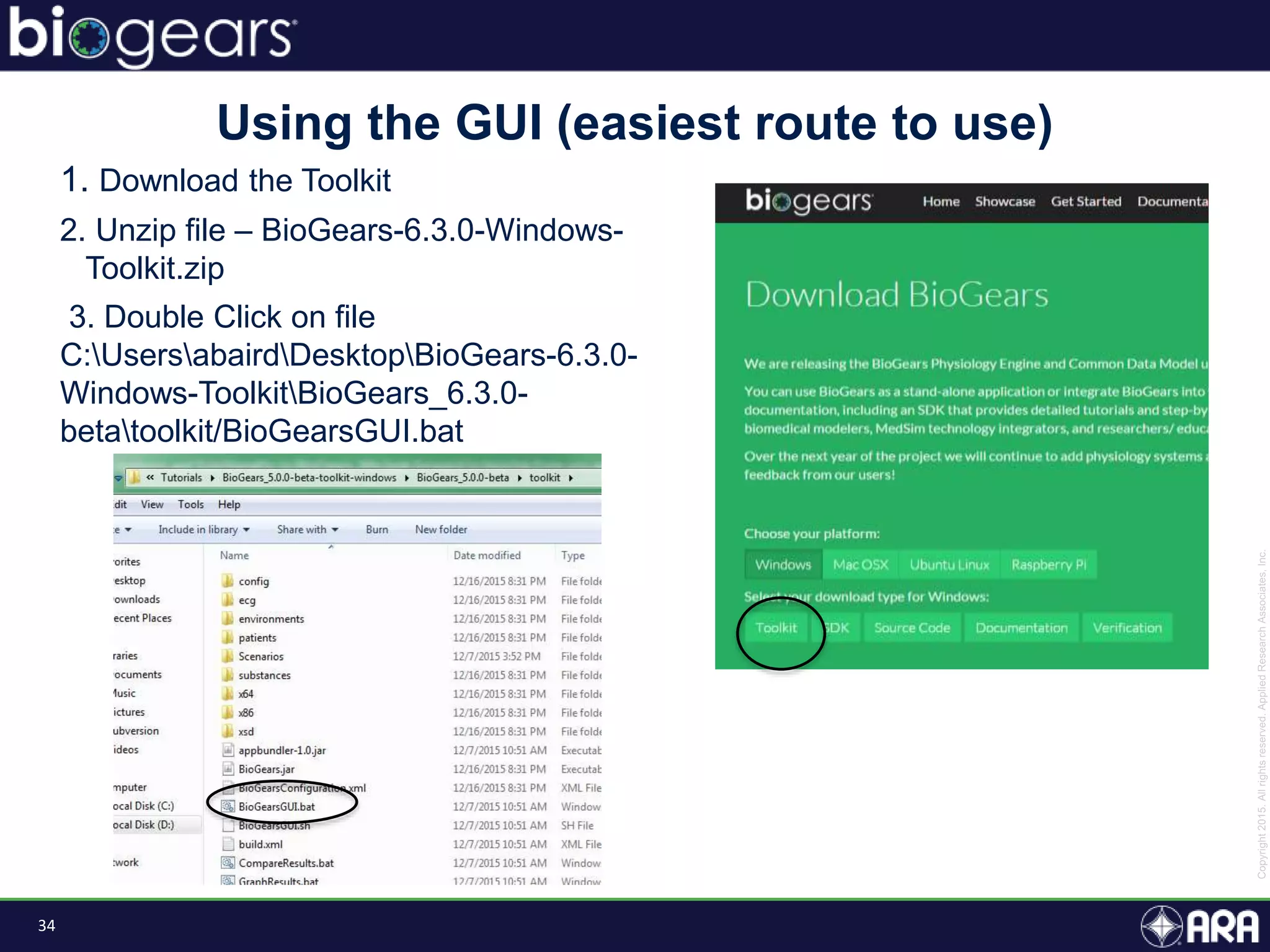
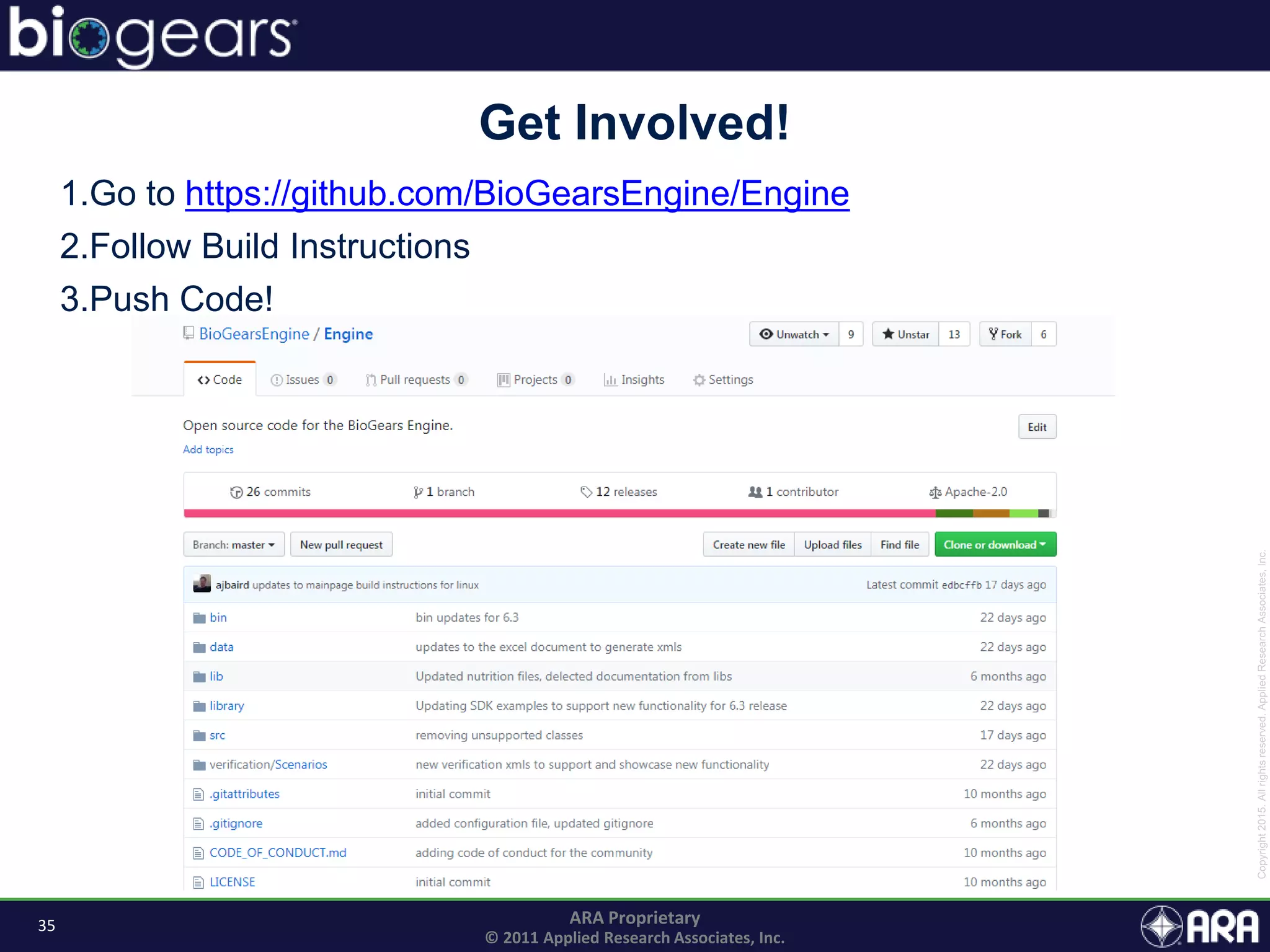
The document discusses Biogears, a multiscale physiology modeling framework developed by Applied Research Associates, Inc., to enhance medical training and simulation. With a budget of $6,959,593, the project aims to create an accessible physiology research platform and improve simulated physiological accuracy across various applications. Key features include a modular modeling approach, pharmacokinetics and dynamics integration, and open source accessibility for users, fostering collaboration in medical education.