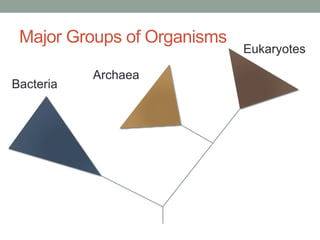
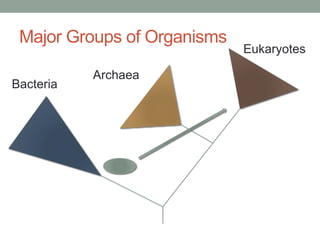
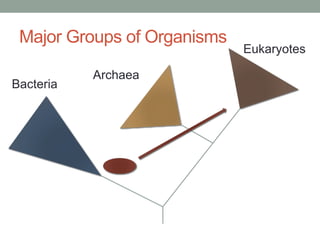
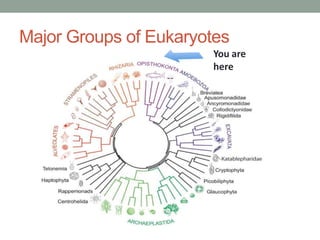
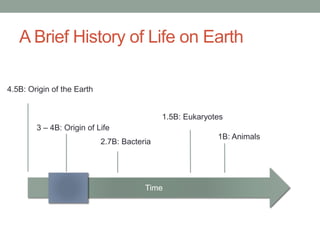
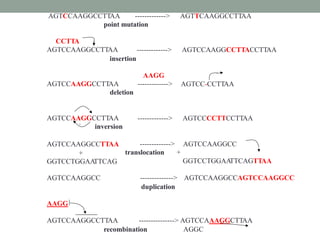
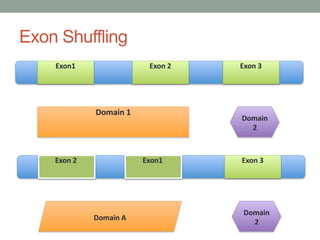
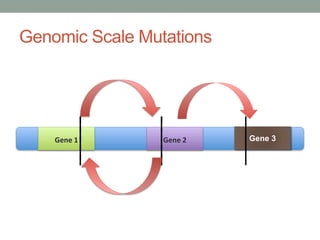
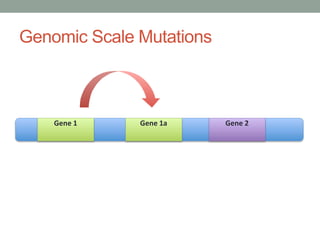
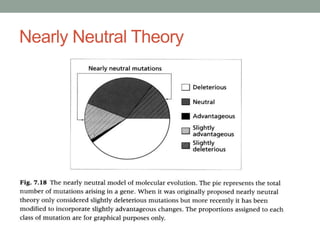
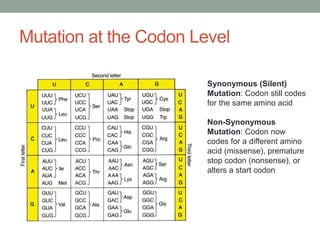
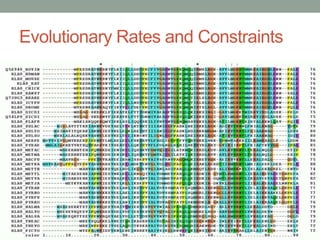
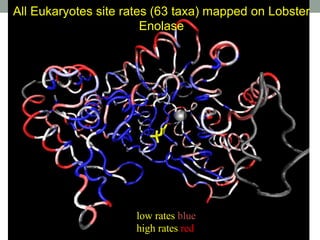
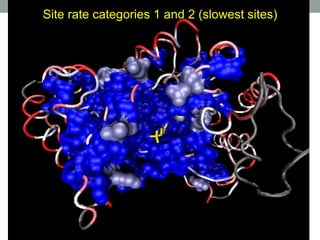
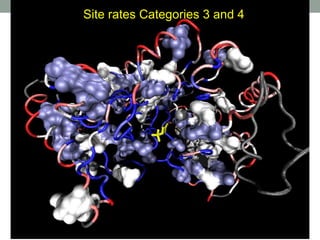
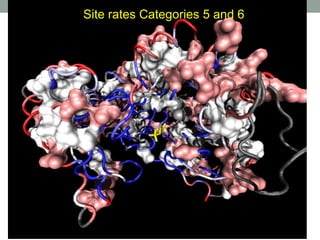
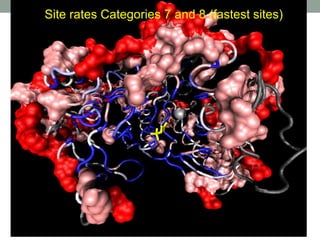
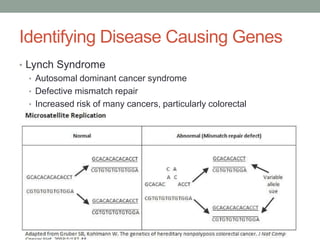
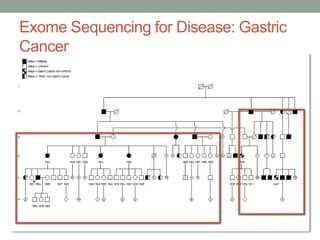
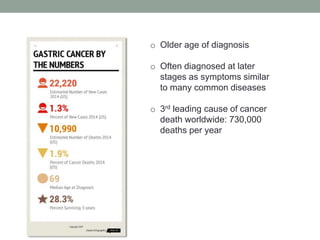
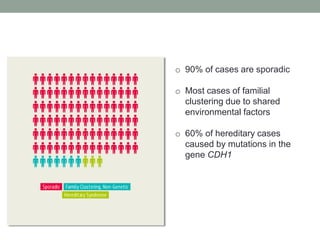
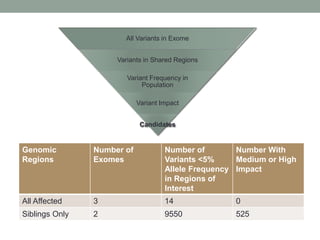
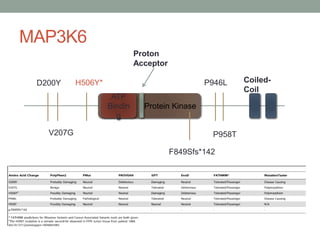
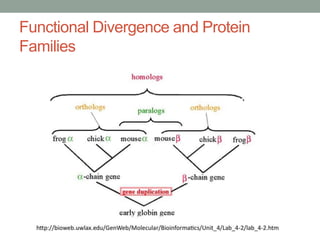
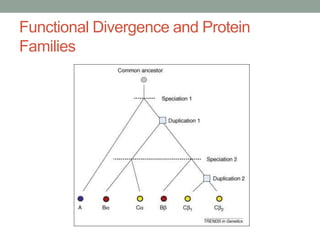
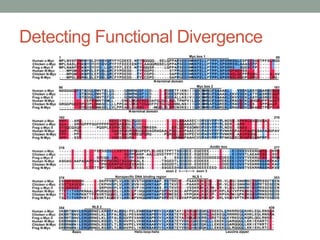
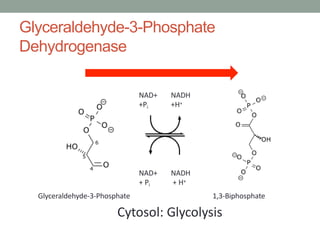
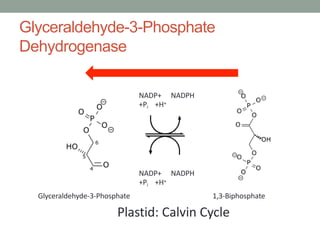
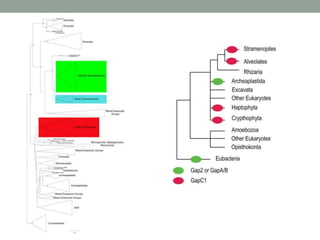
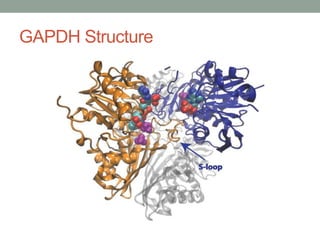
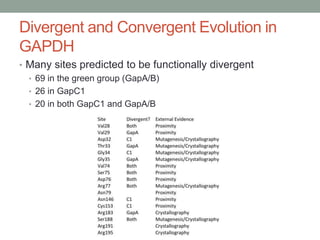
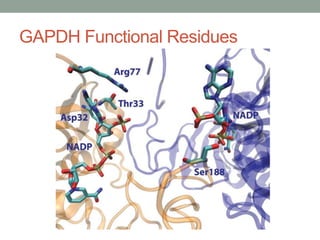
The document discusses protein evolution, function, and human health. It provides an overview of why protein evolution matters for scientific curiosity, understanding disease, and more. It then covers major topics in protein evolution including processes like mutation and selection, as well as how protein function and structure impact evolution. Finally, it discusses practical applications like identifying disease-causing genes and mutations through an evolutionary lens.