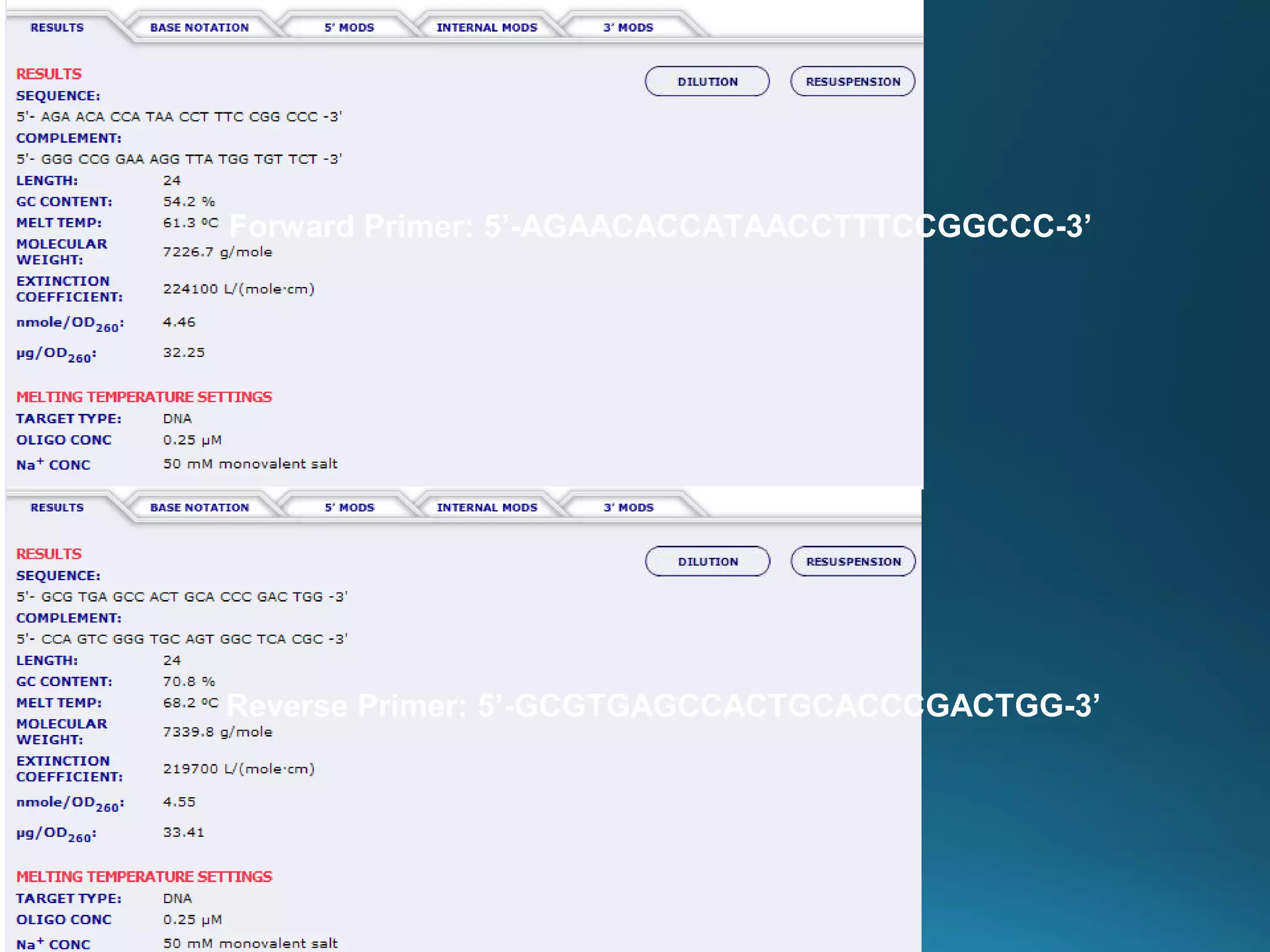
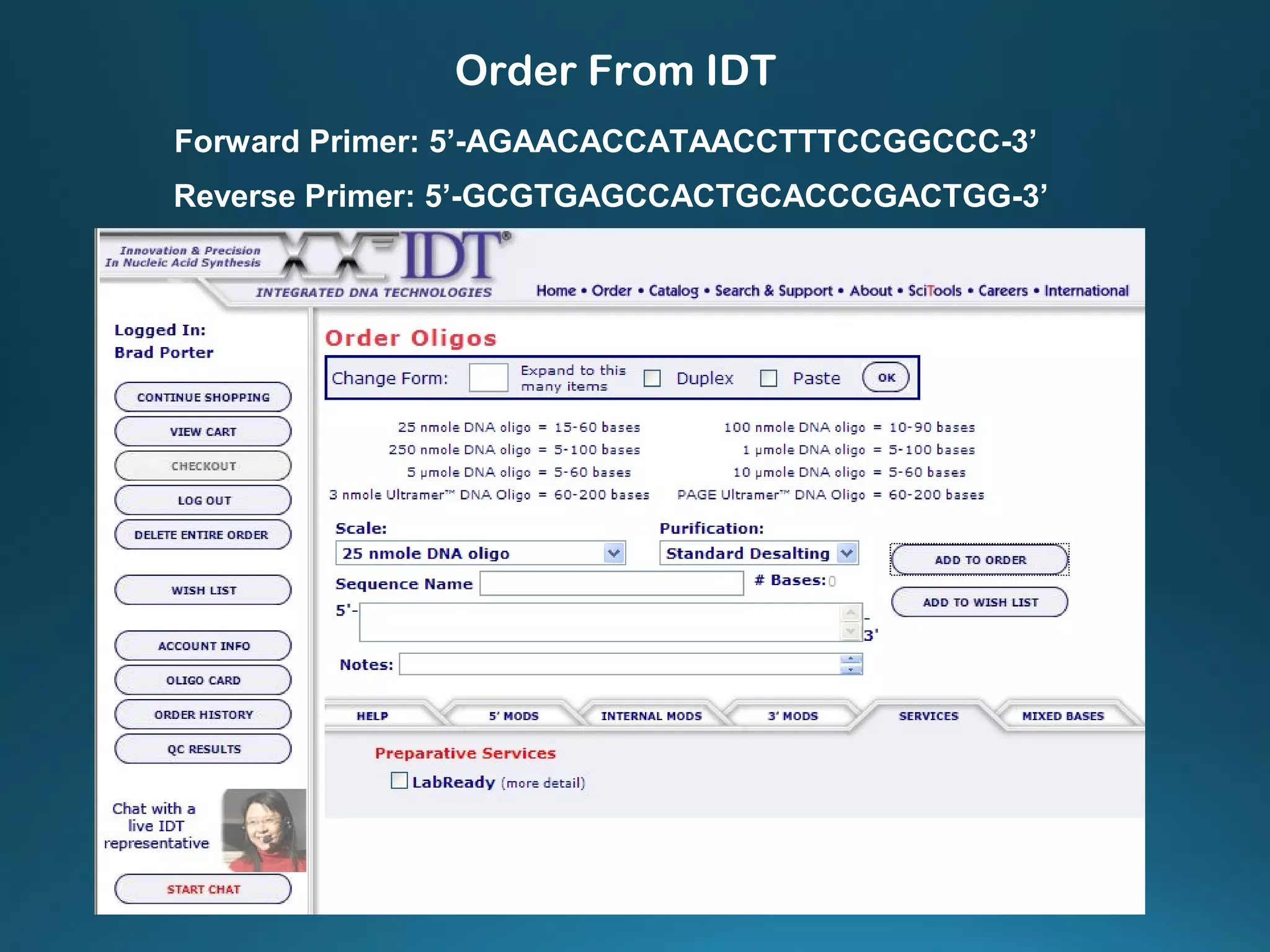
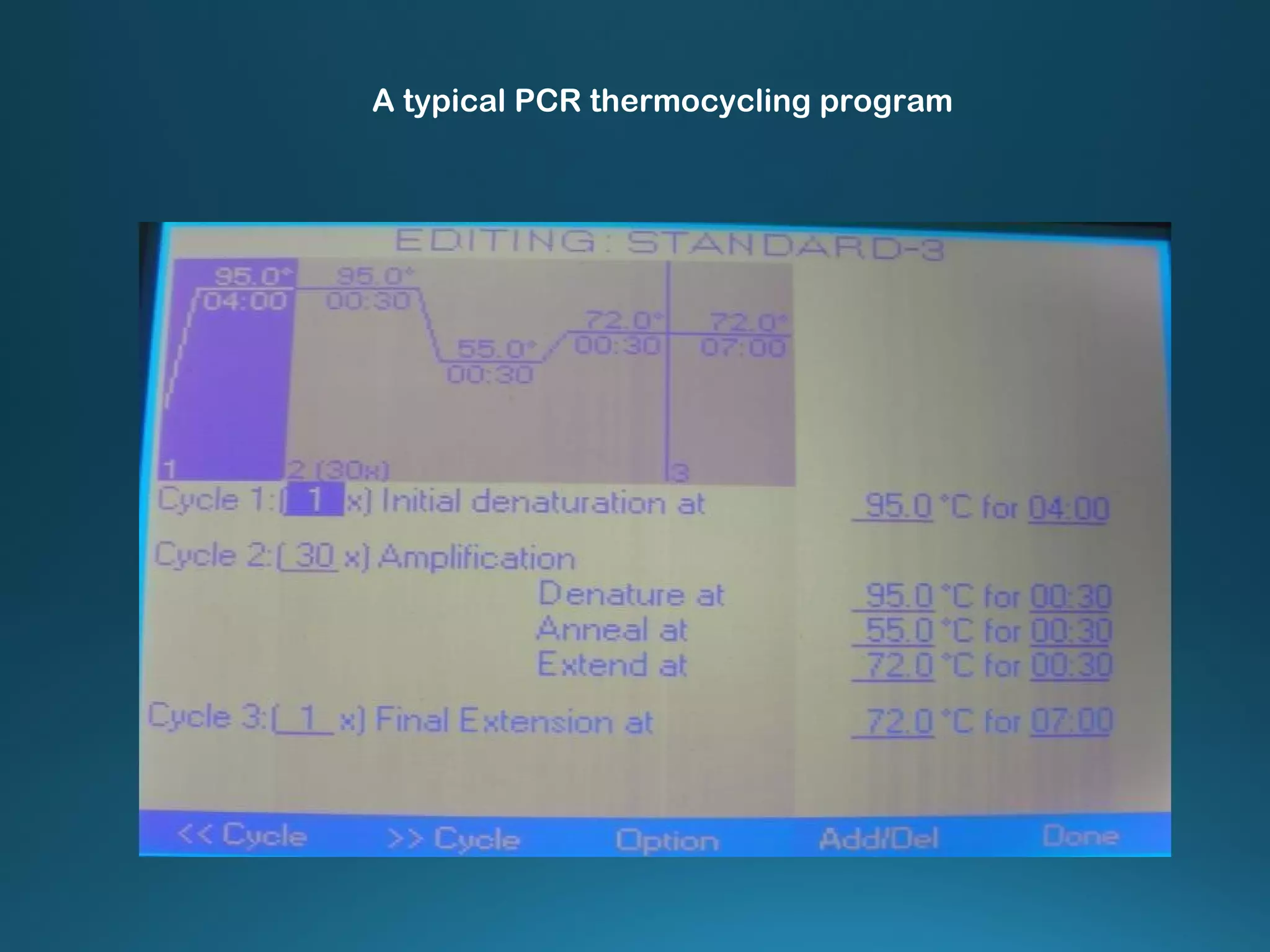
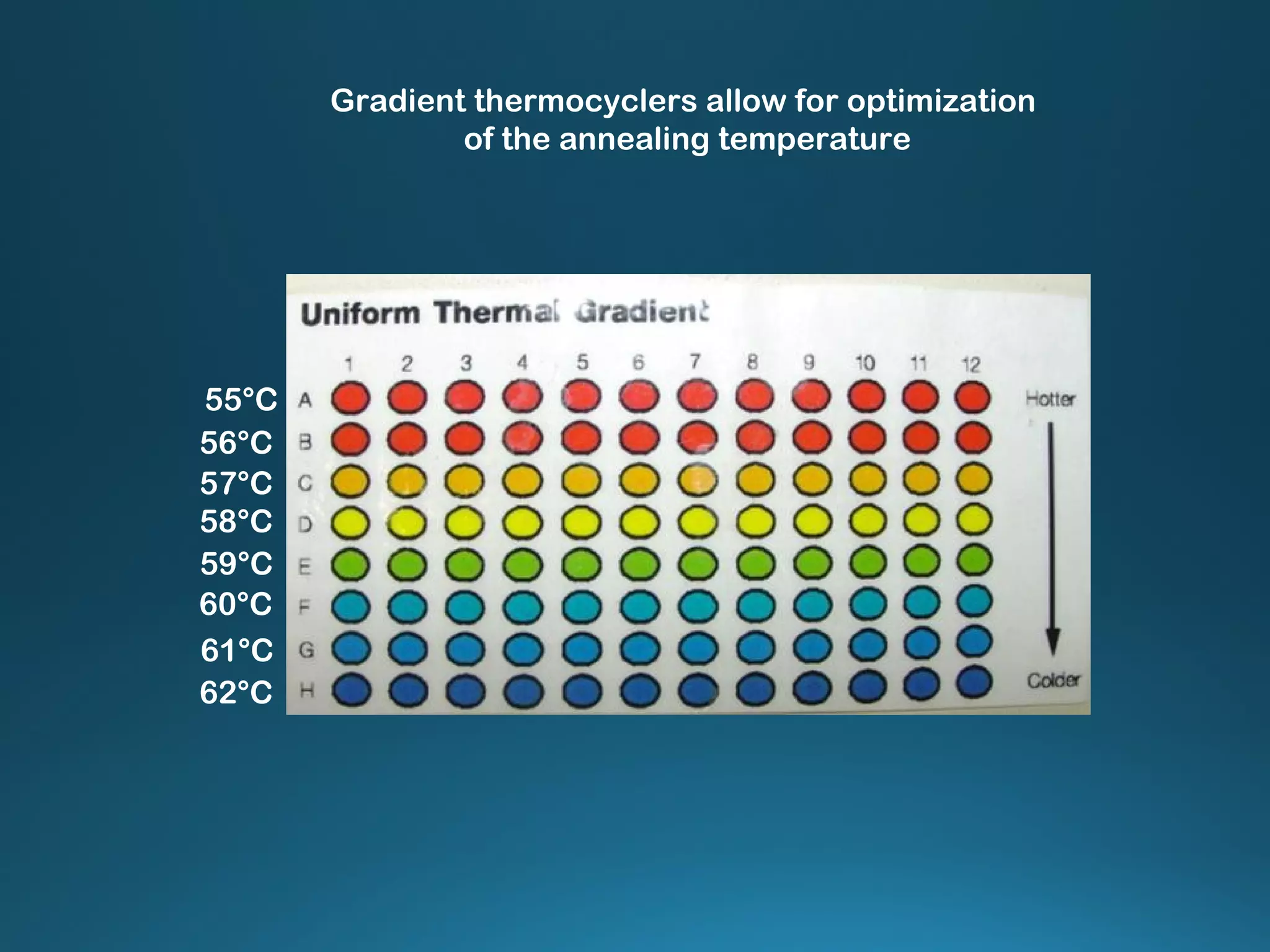
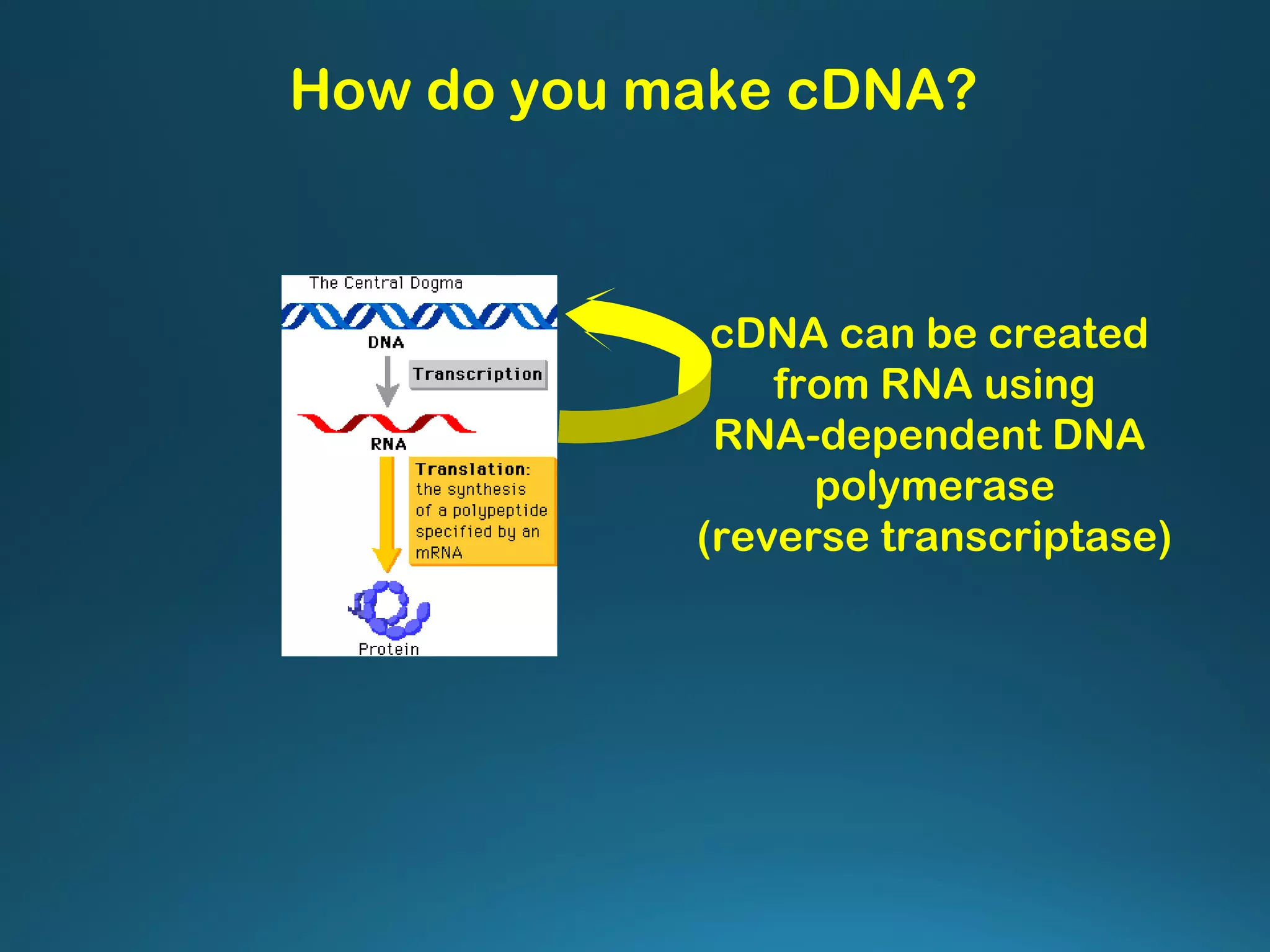
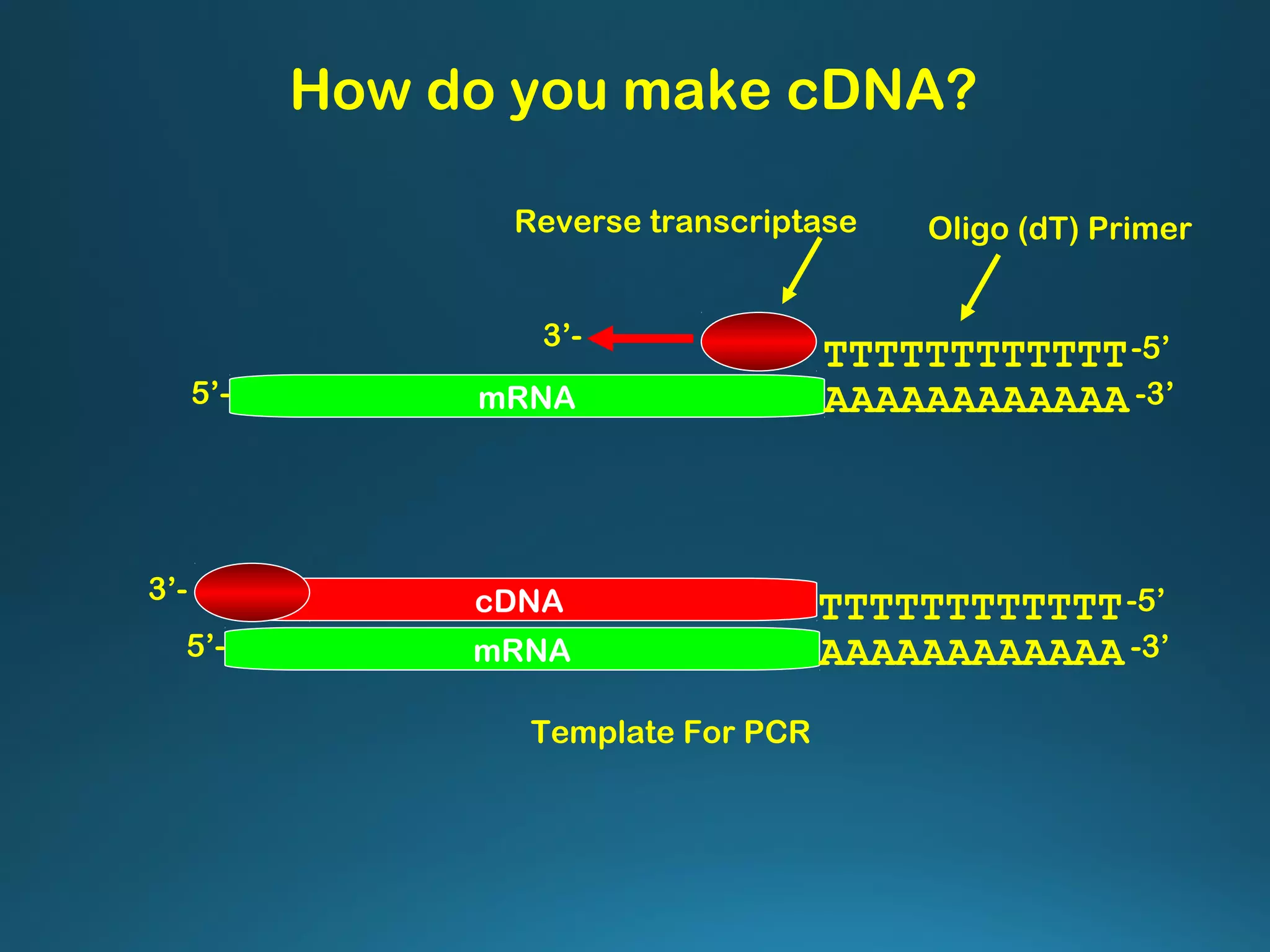
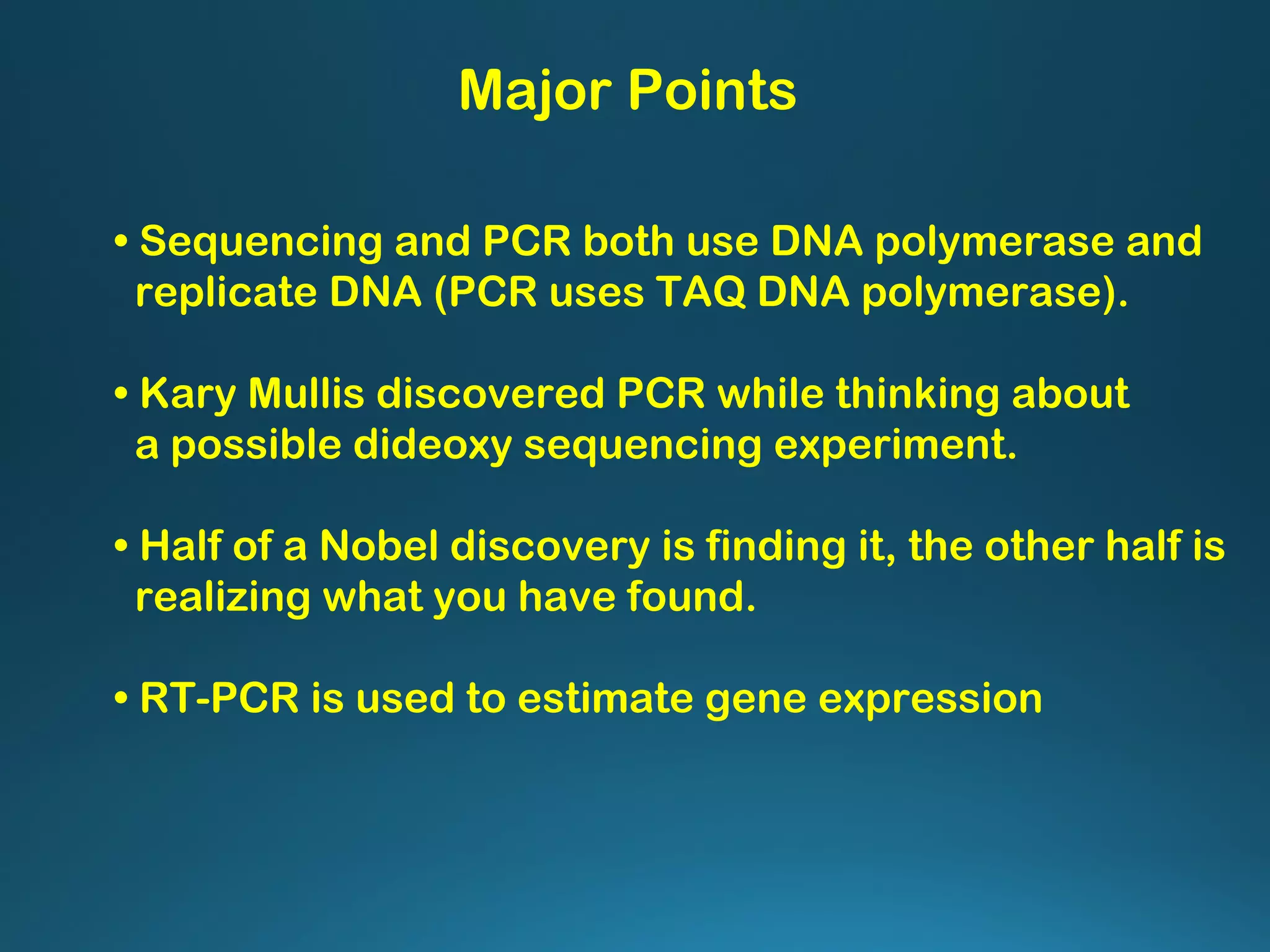
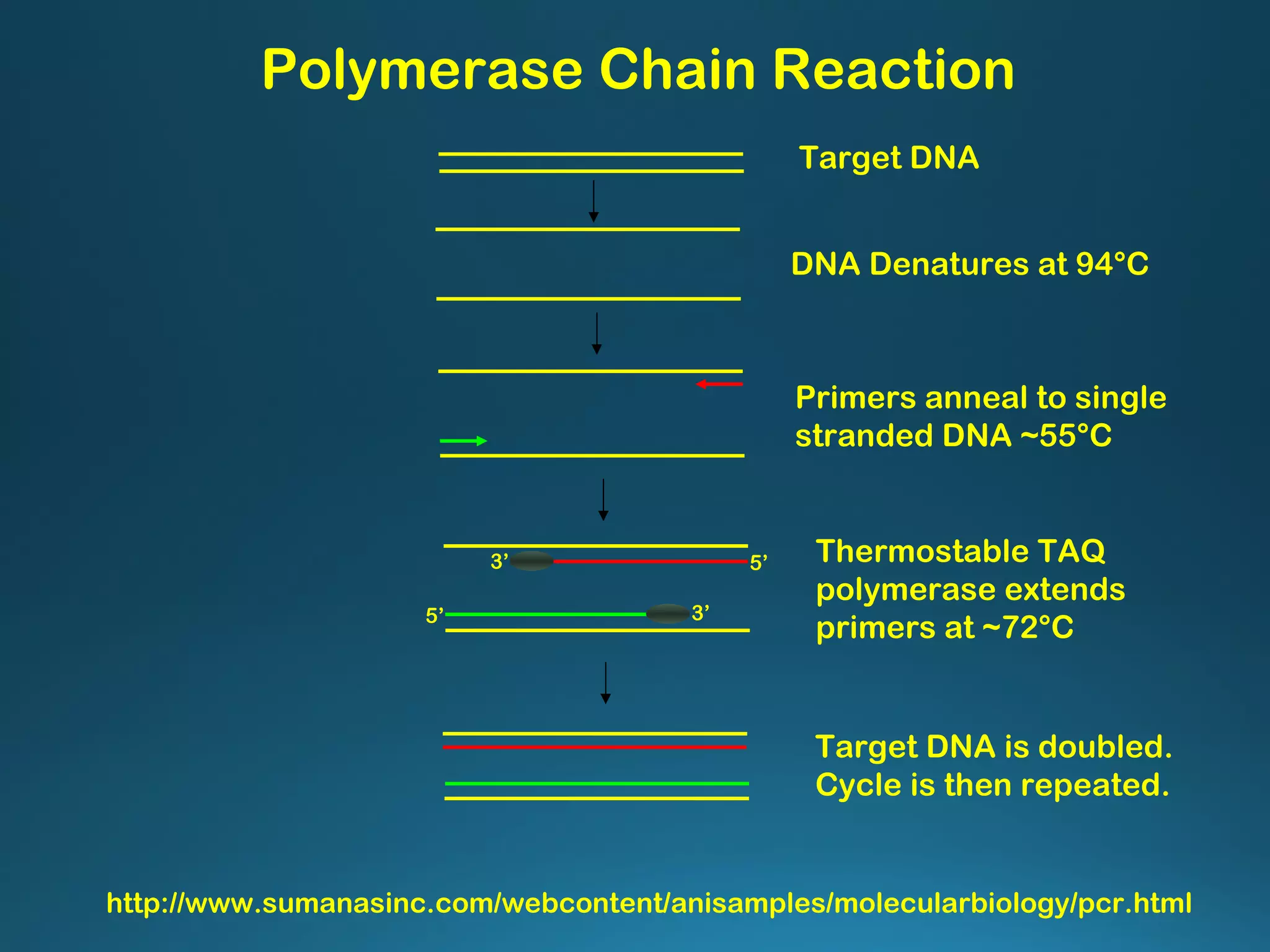
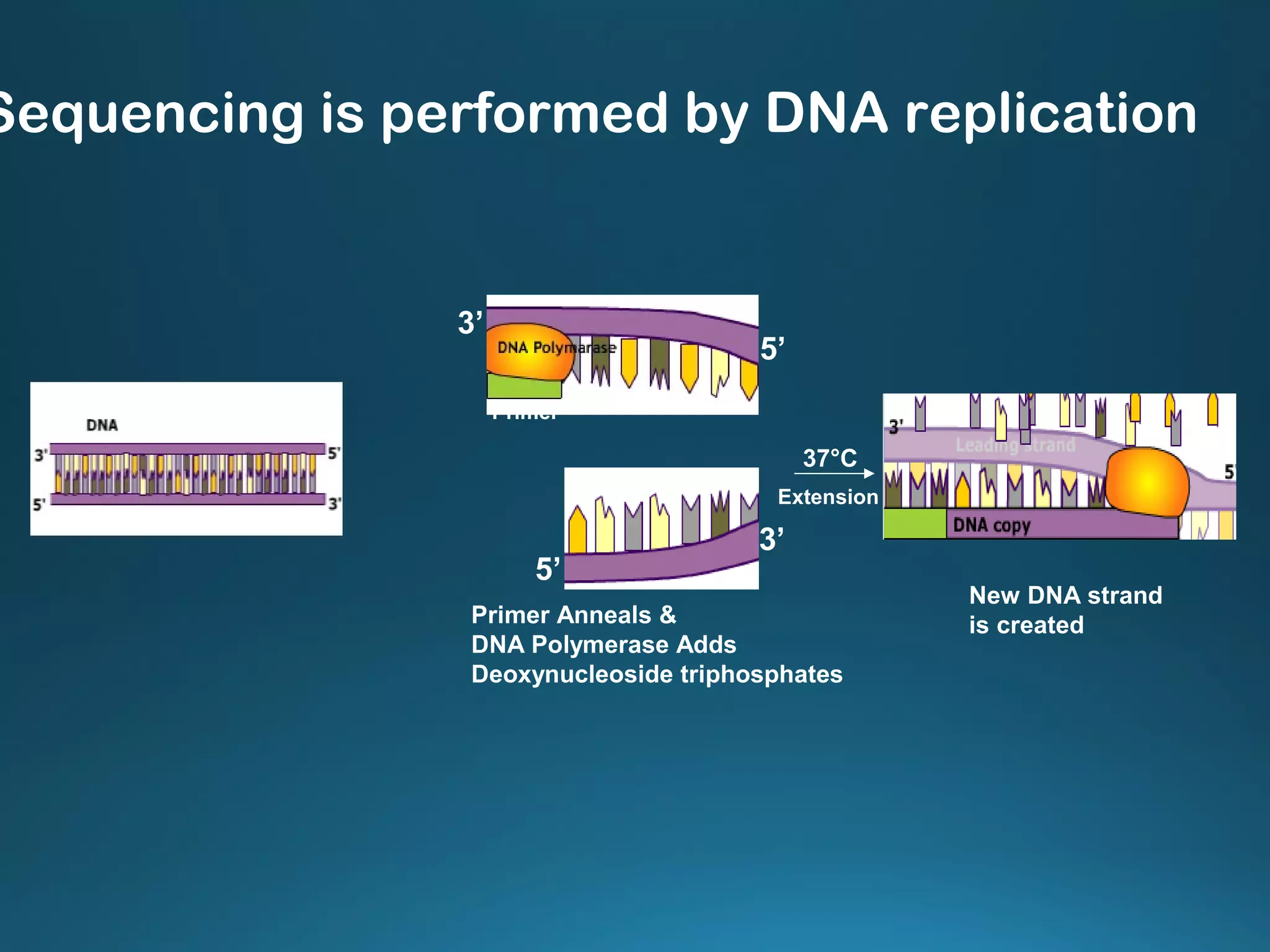
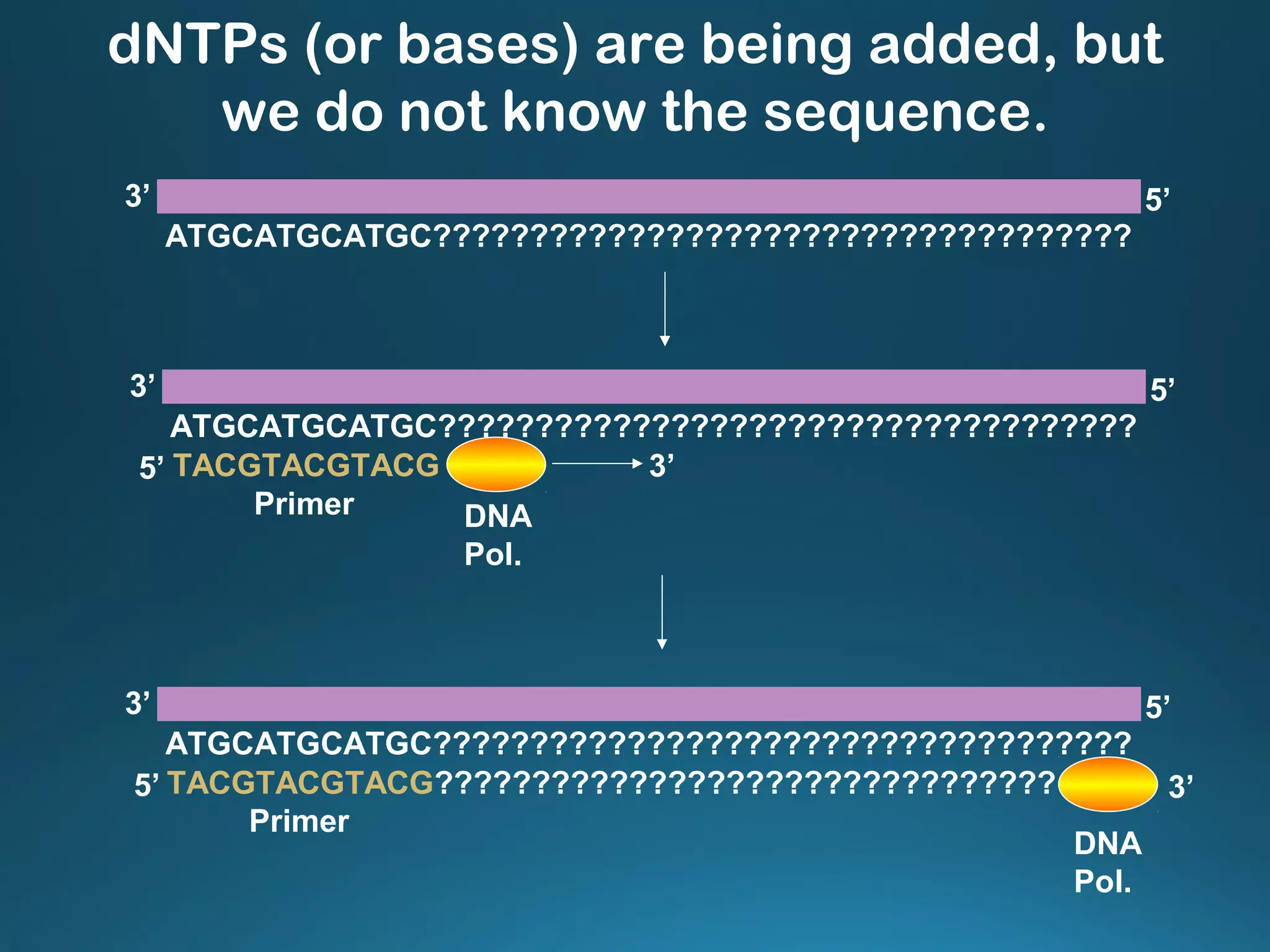
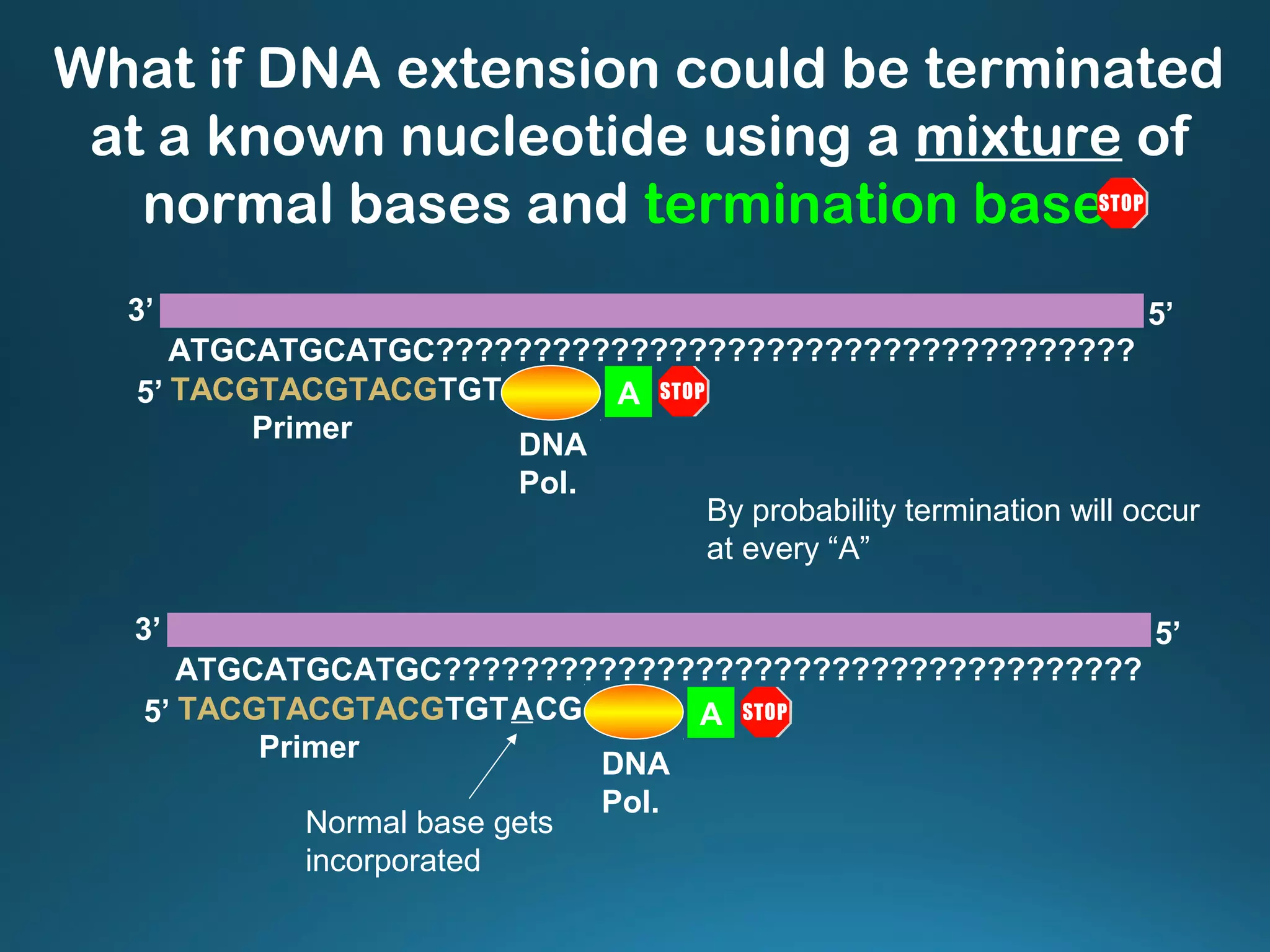
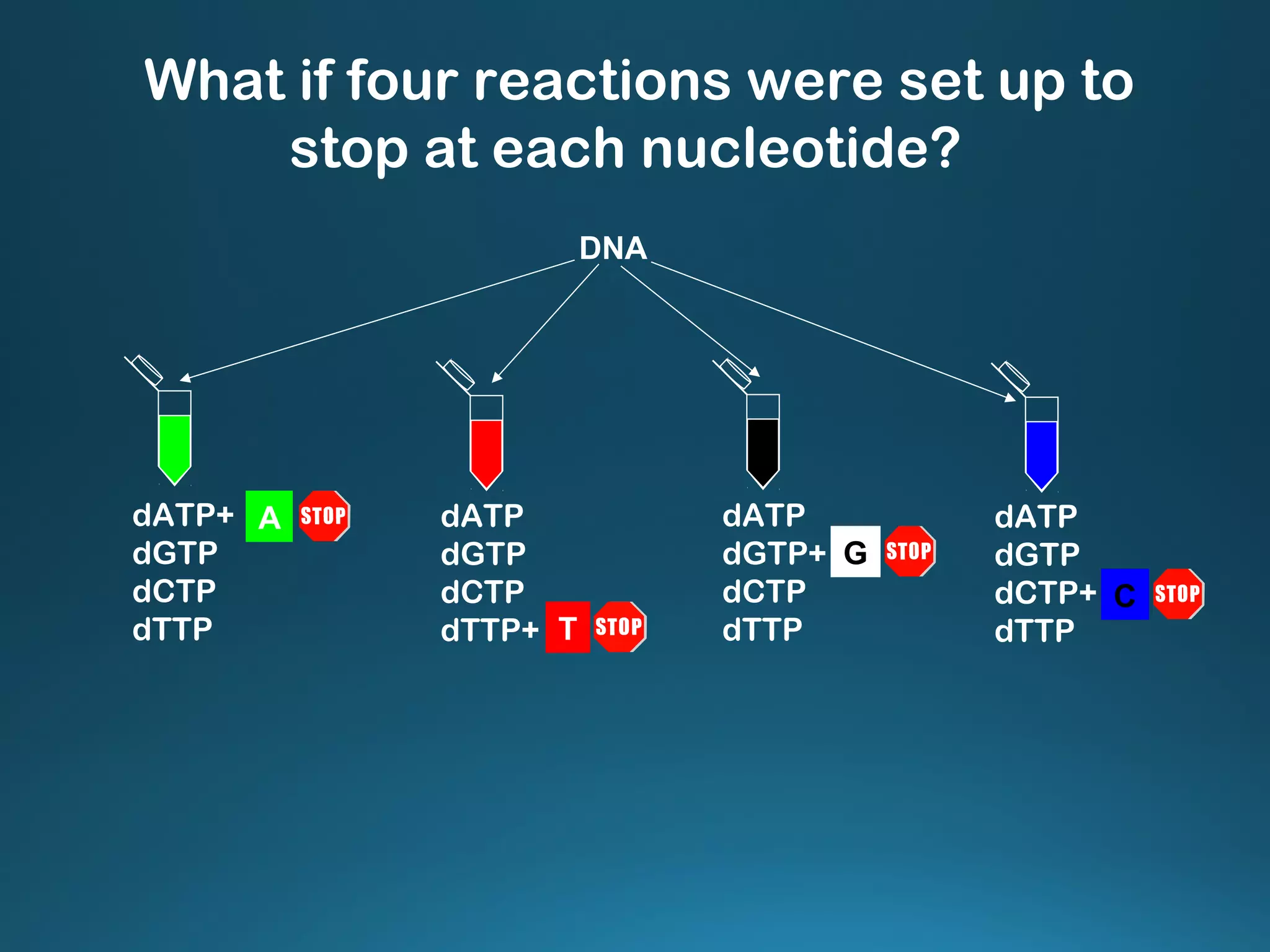
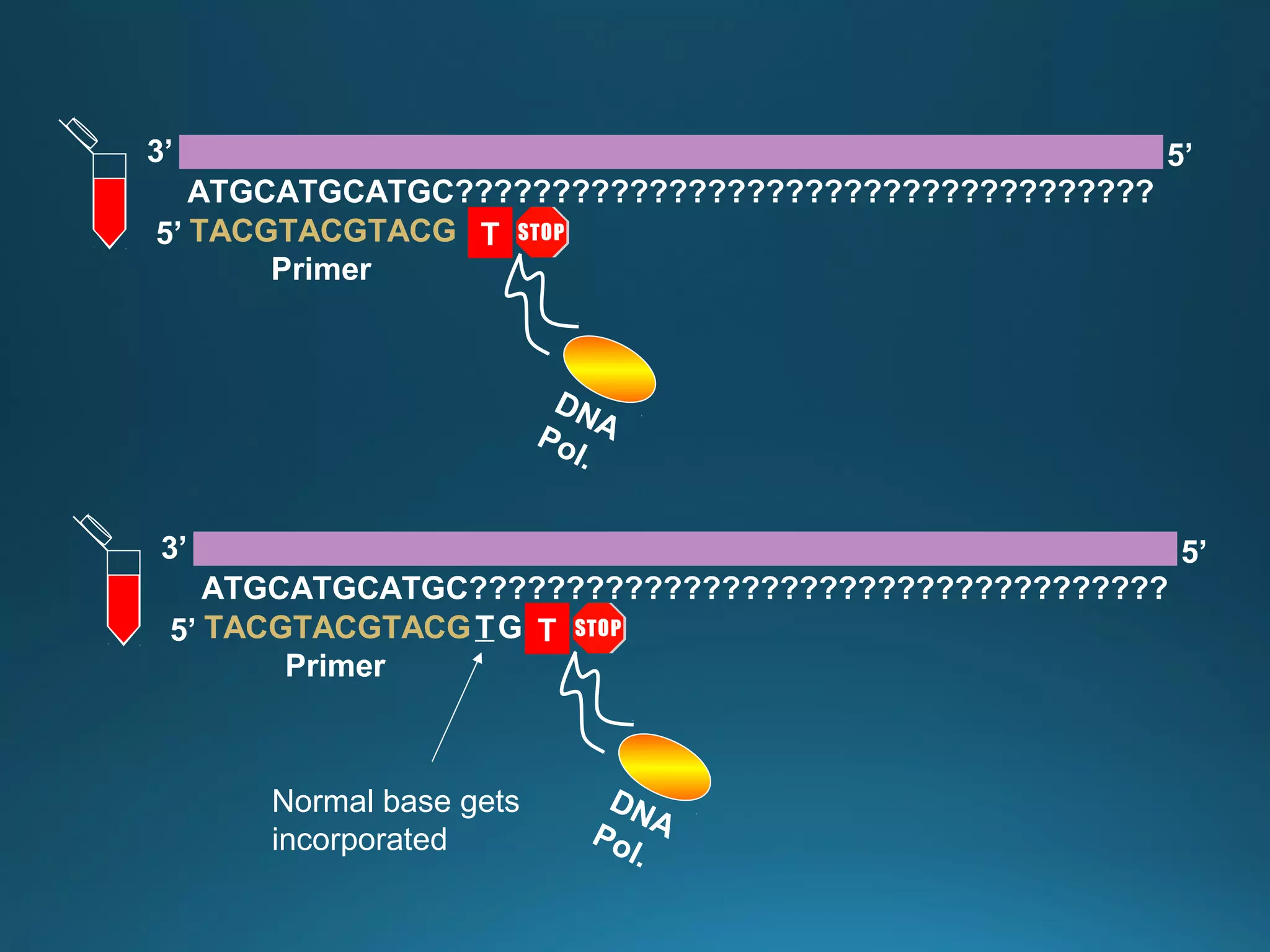
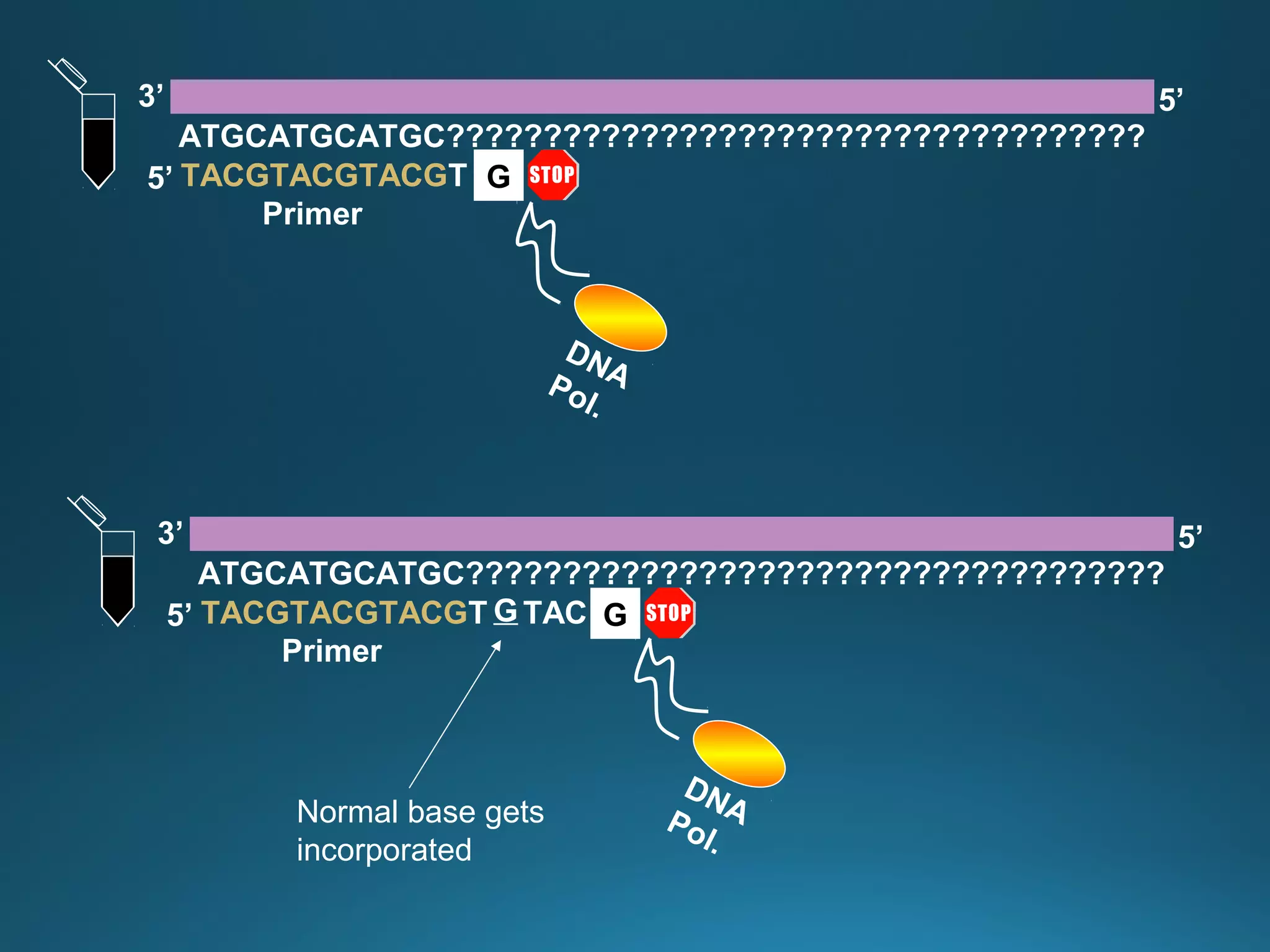
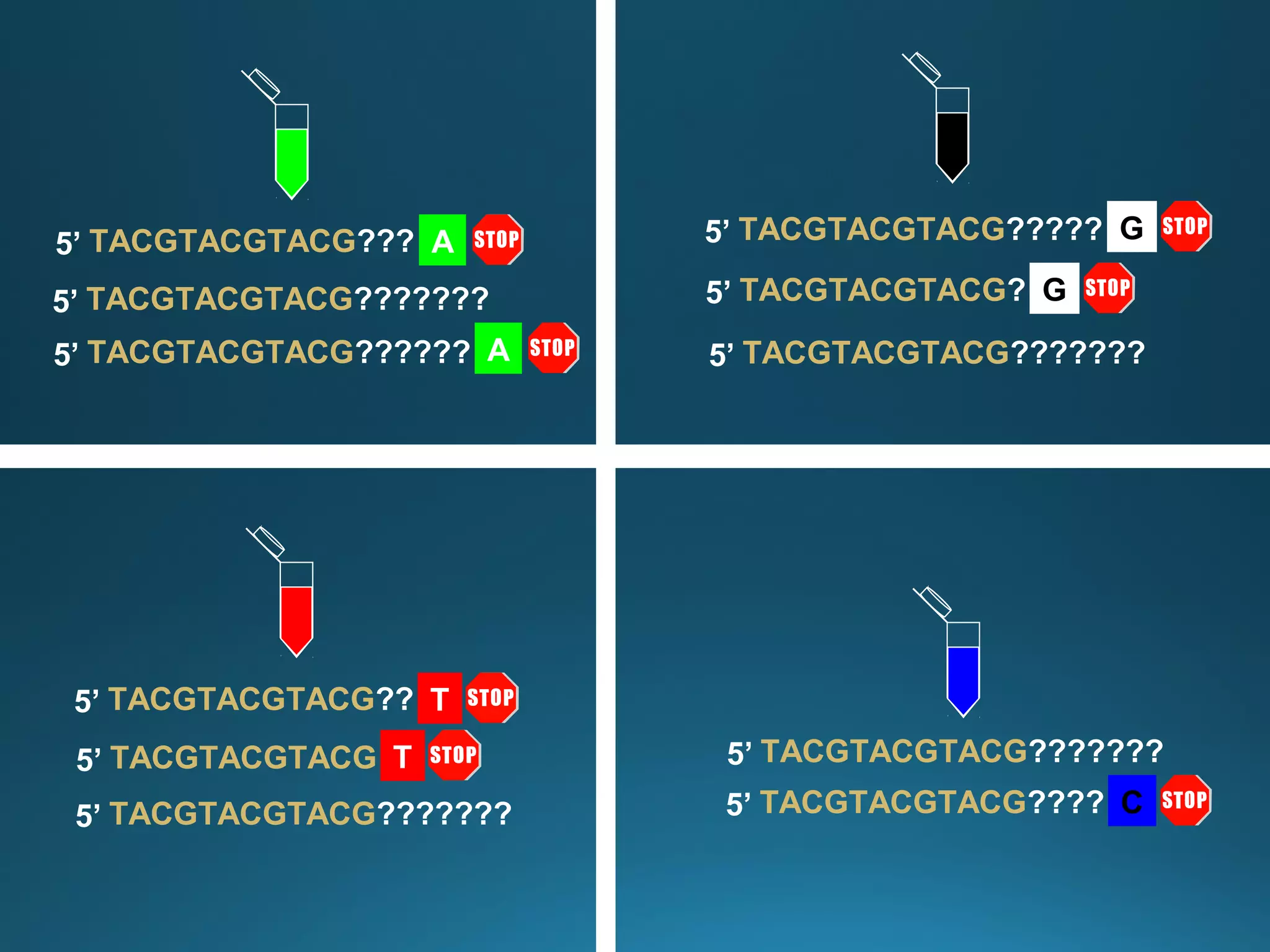
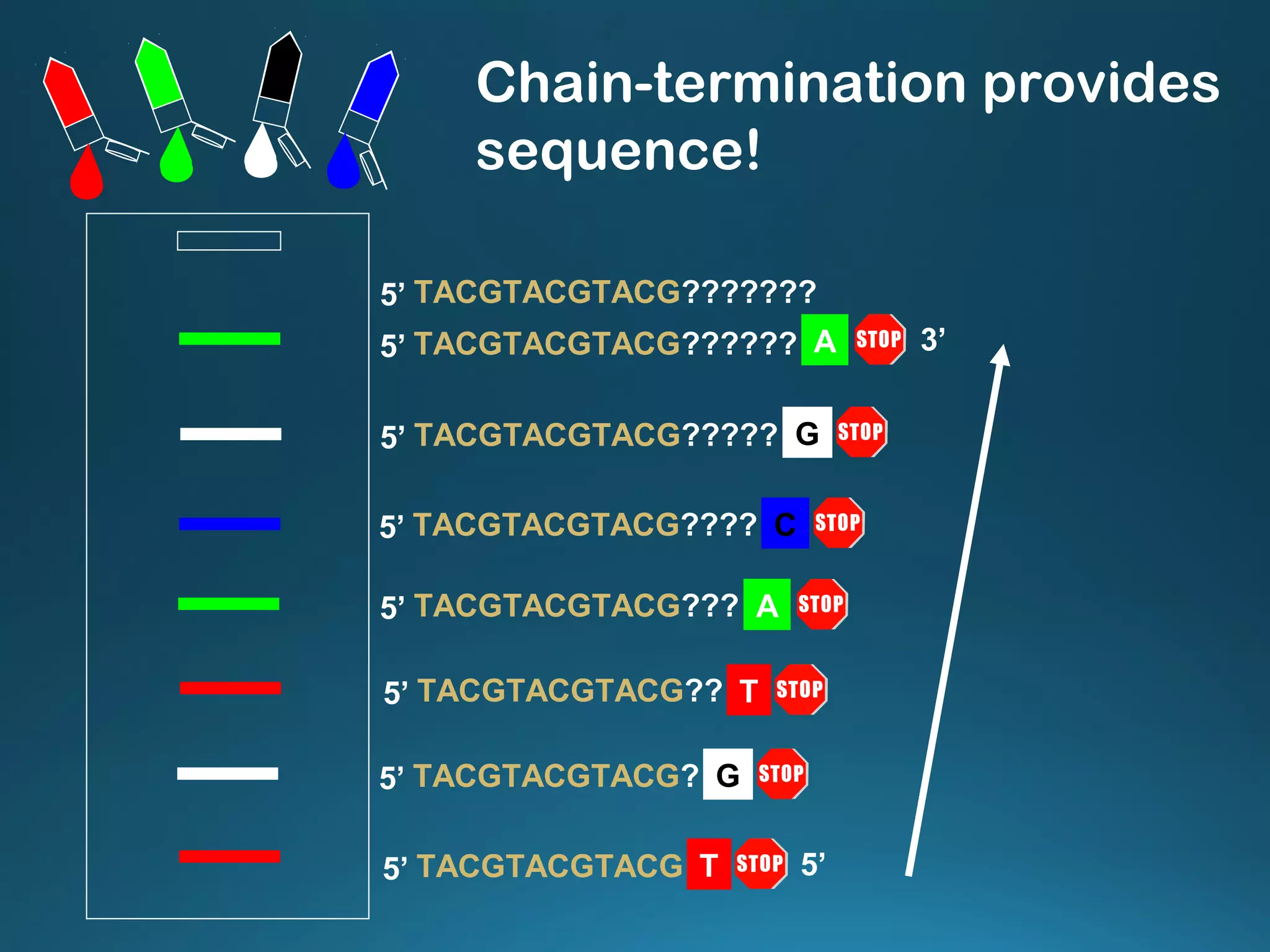
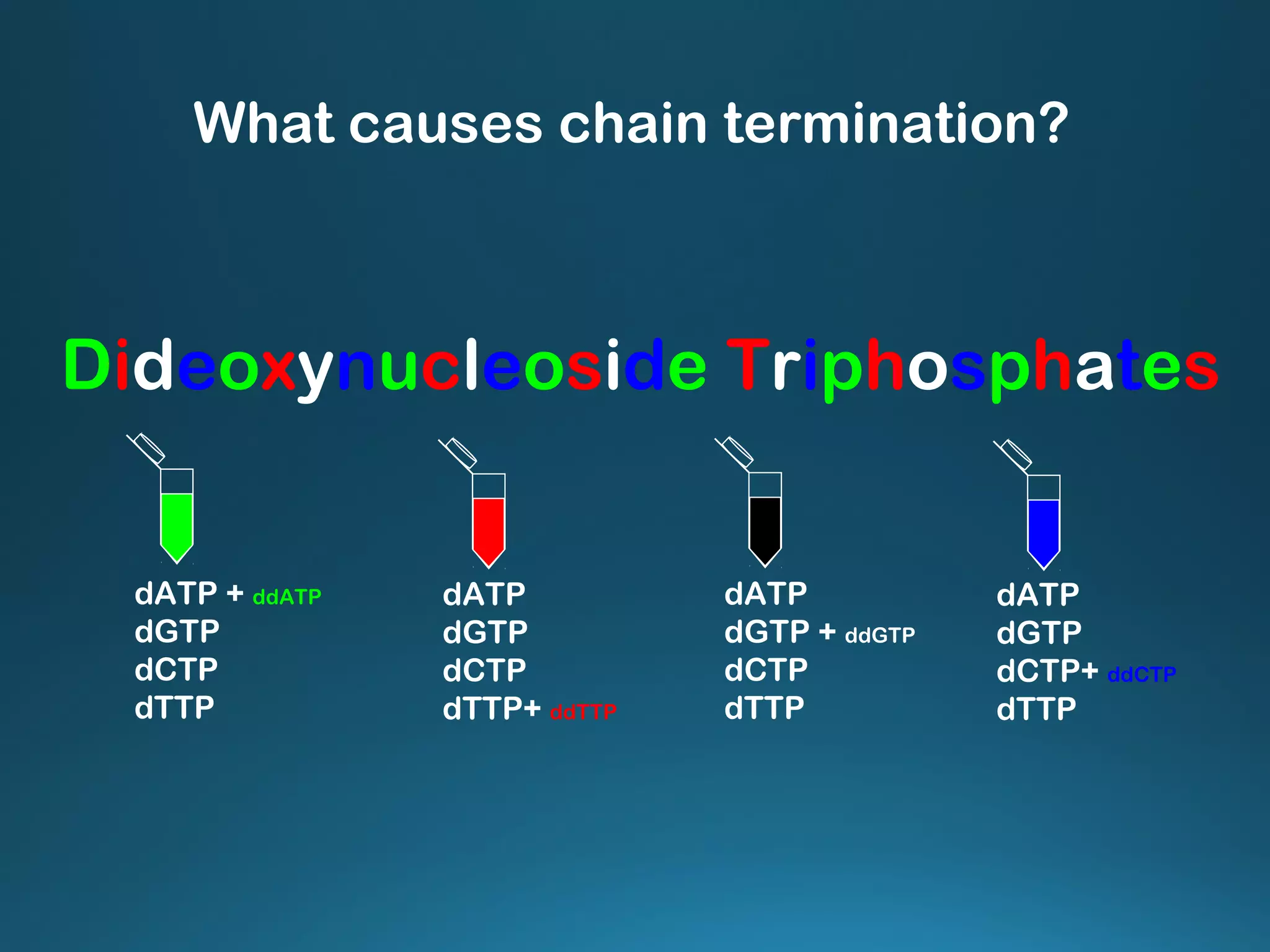
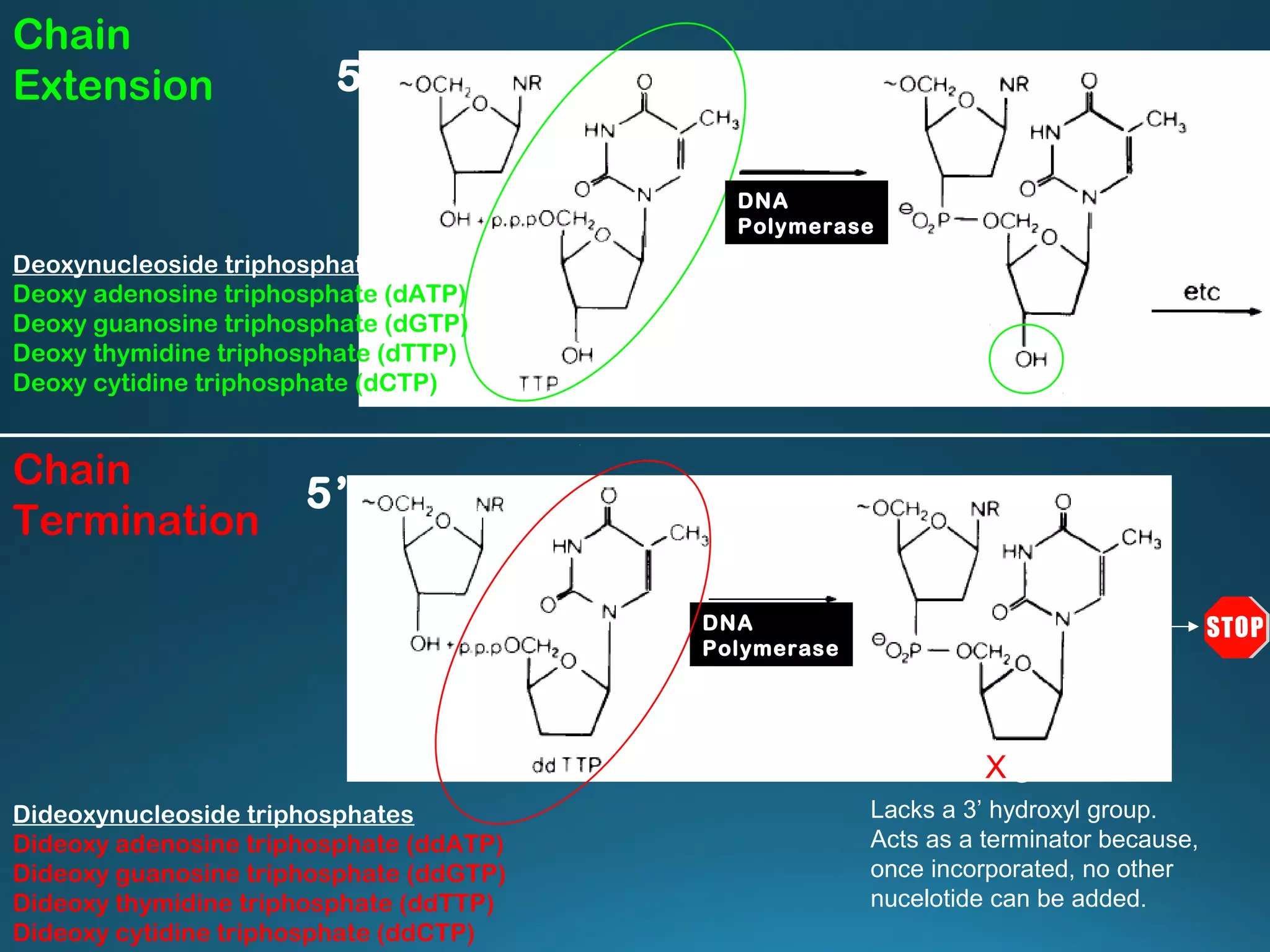
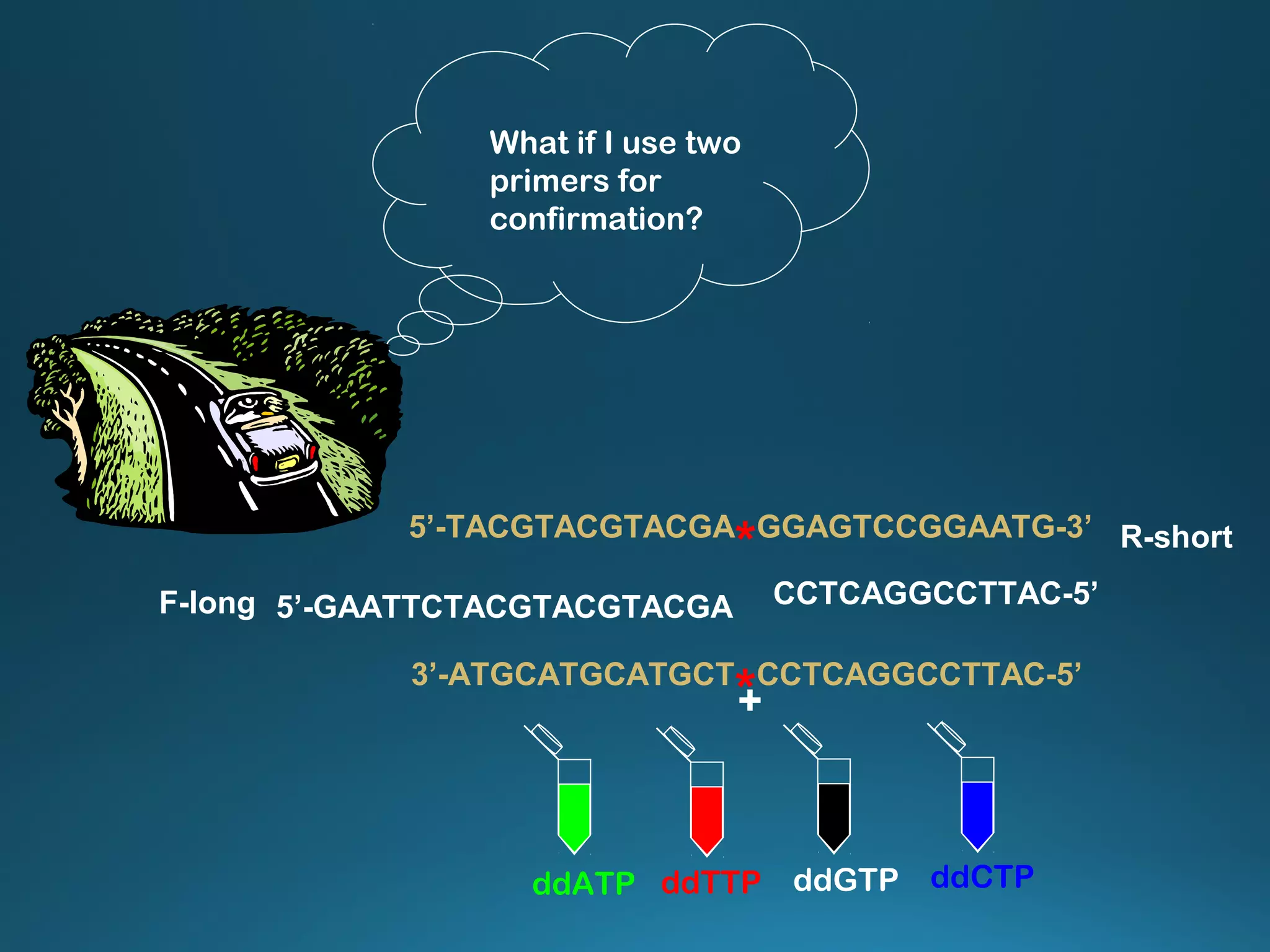
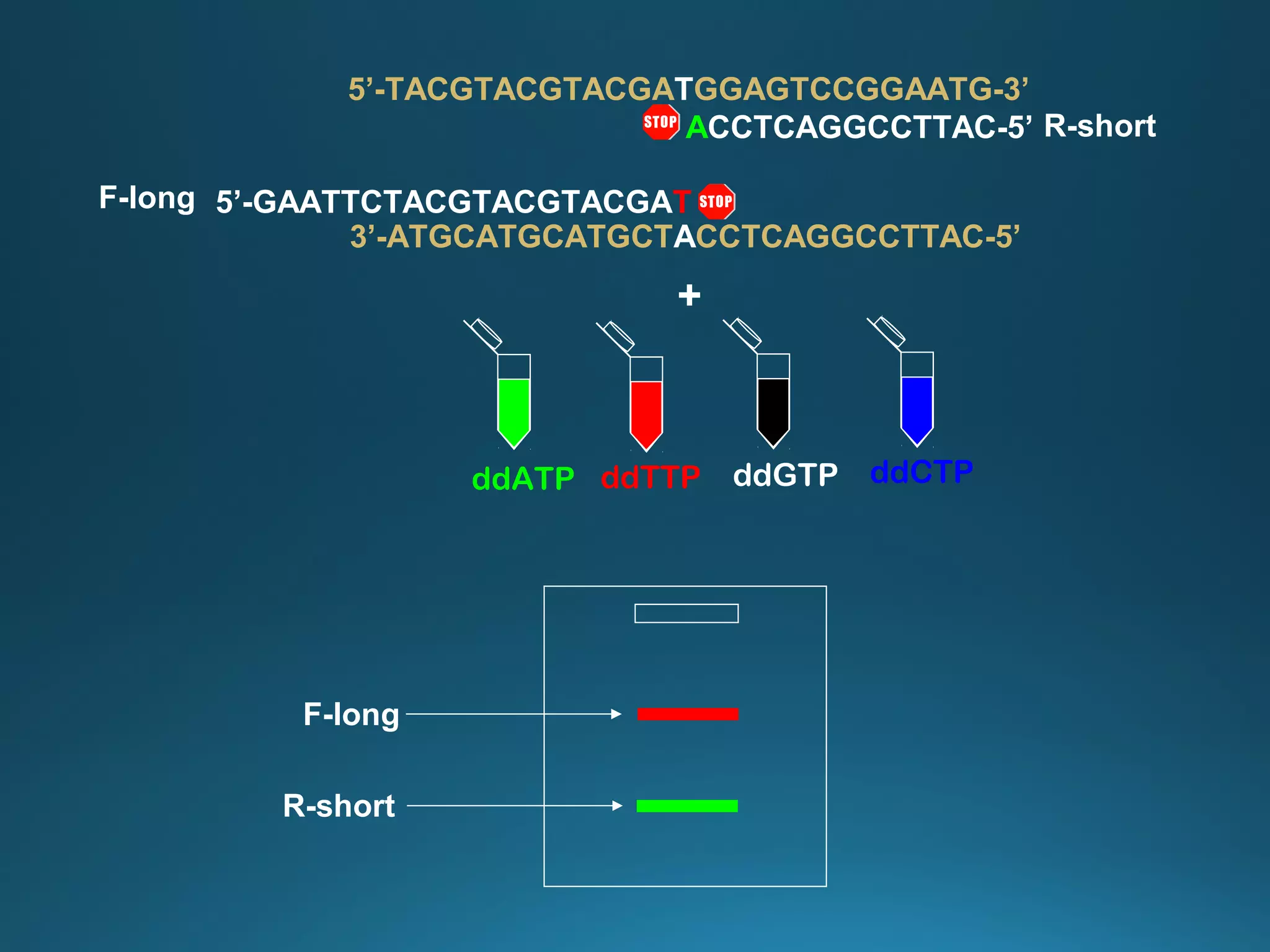
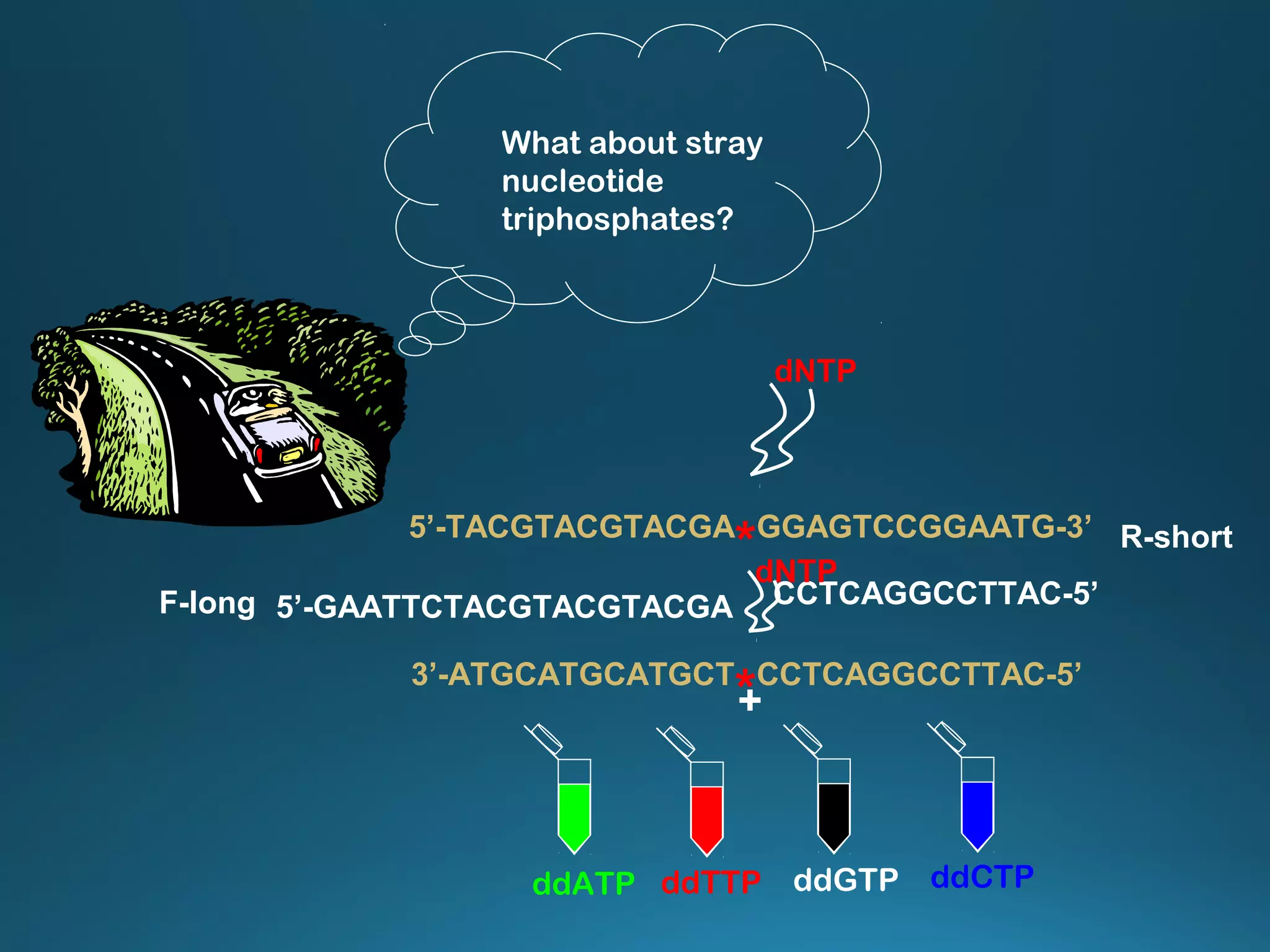
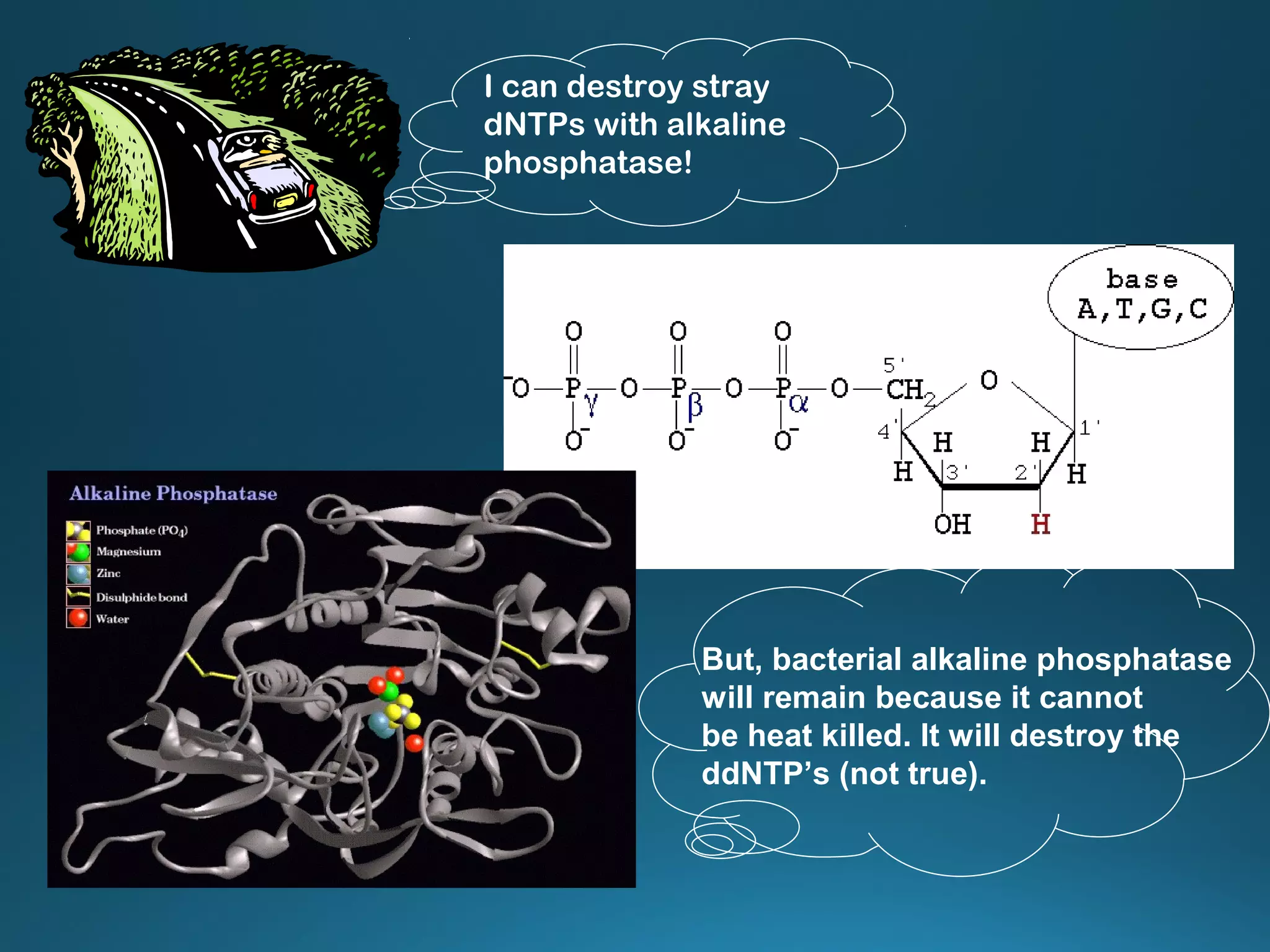
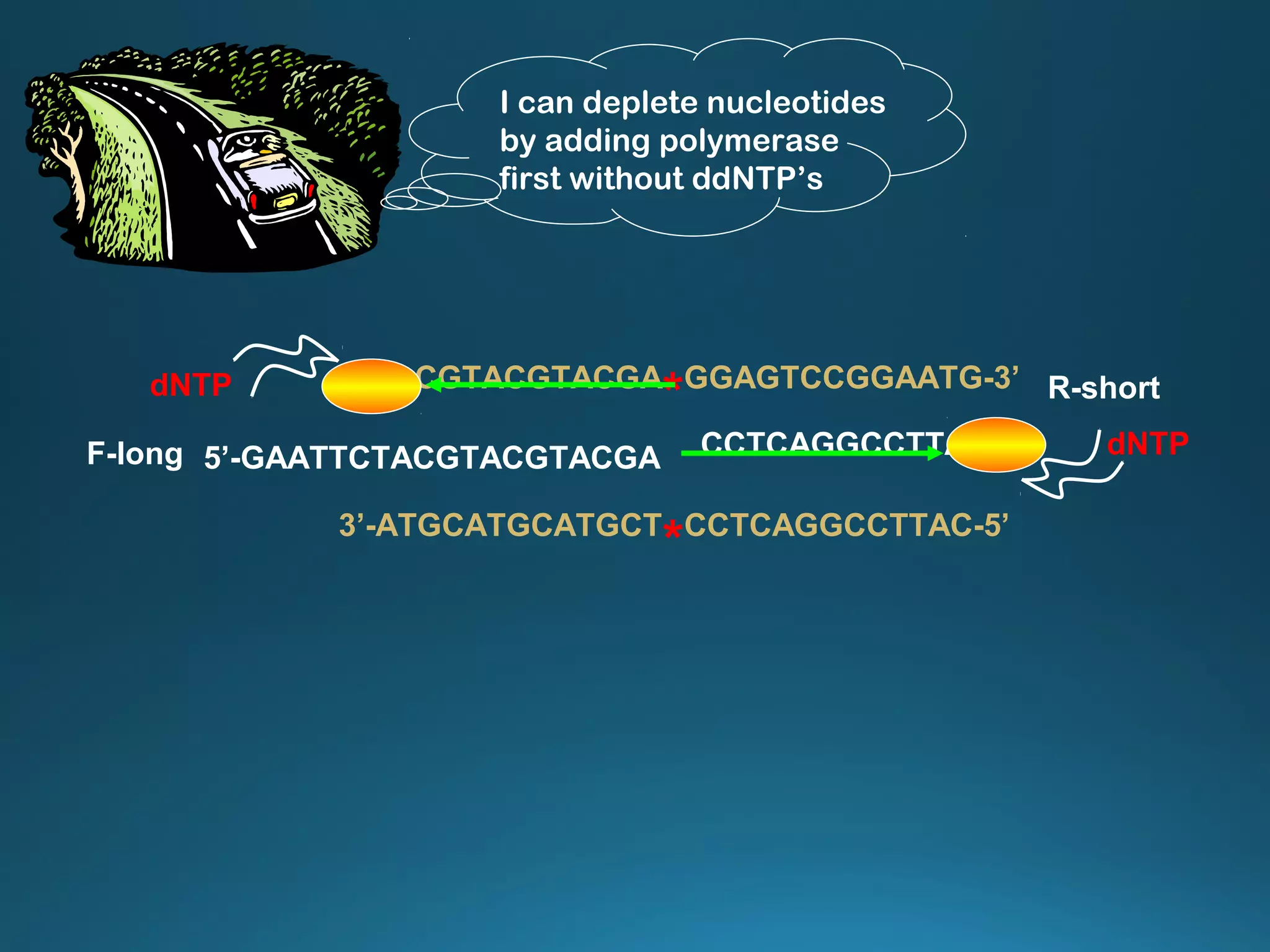
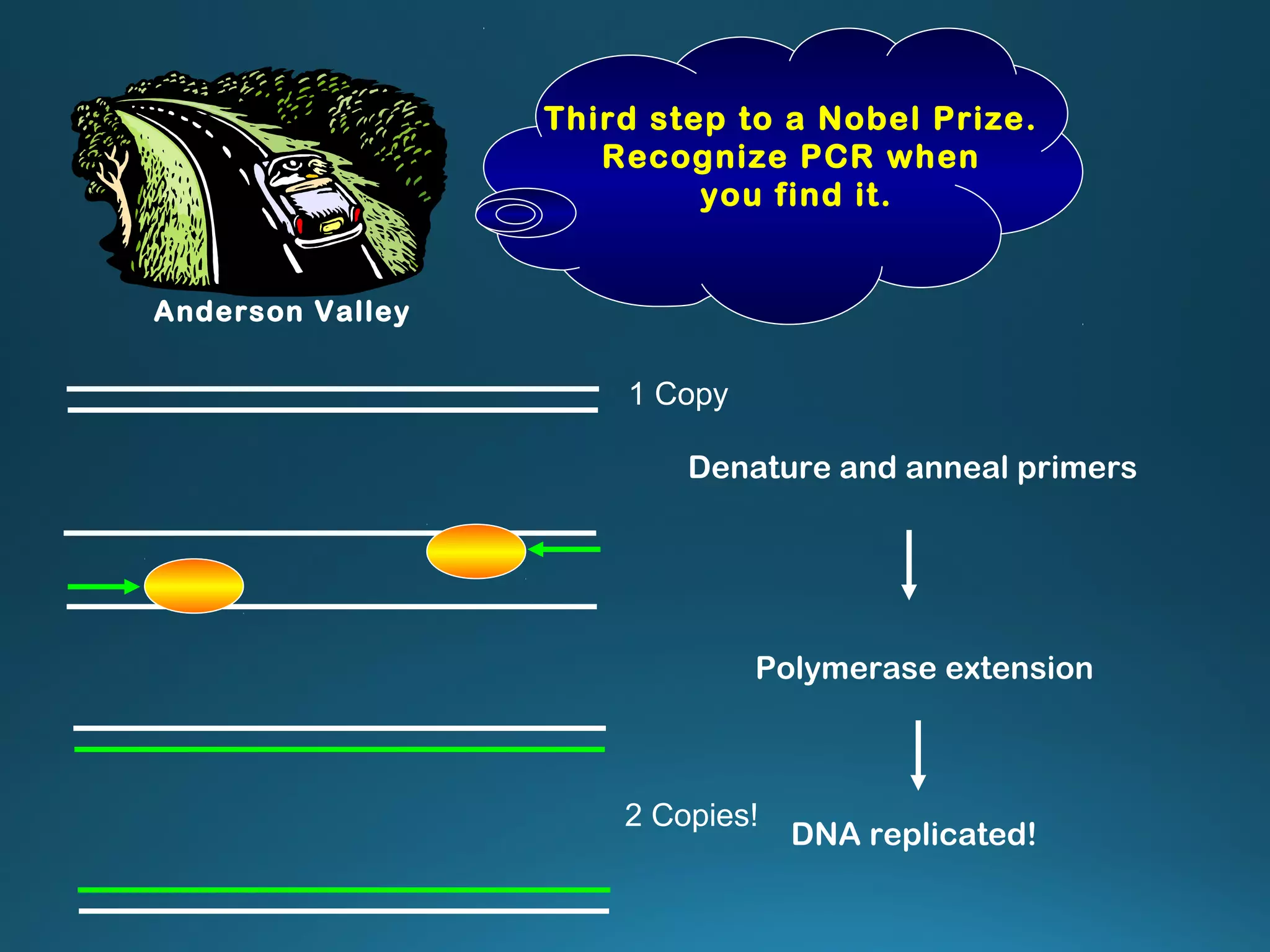
The document discusses the development and methodology of Polymerase Chain Reaction (PCR) and its relation to DNA sequencing, particularly the Sanger dideoxy sequencing method. It details the processes of denaturation, annealing of primers, and extension by DNA polymerase, explaining the significance of chain termination in DNA replication. The document also highlights the contributions of key individuals and the implications of PCR in accessing genetic risk factors for diseases.


















![Loci for Type 2 Diabetes and Triglyceride Levels
GCAGCTCACCTCCAGCTTTAGTTTTC[C/T]CATGACAGTAAGTCTATTACCCTCC
Risk allele](https://image.slidesharecdn.com/analysis-of-gene-expression-using-rtpcr-160211143334/75/Analysis-of-gene-expression-using-rt-pcr-38-2048.jpg)