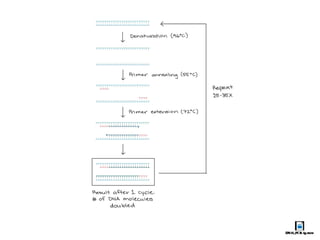
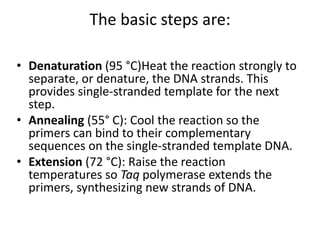
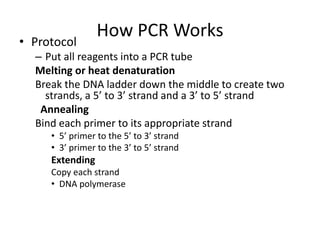
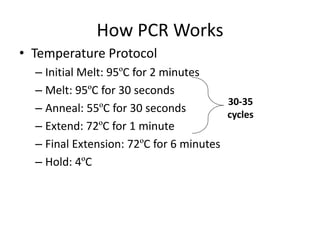
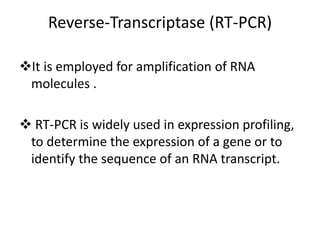
The document summarizes polymerase chain reaction (PCR), including its history, principles, types, and applications. It describes how PCR was invented in 1983 by Kary Mullis, allowing for the amplification of specific DNA sequences. The basic steps of PCR involve denaturation of DNA, annealing of primers, and extension of new strands by DNA polymerase. Various types of PCR are discussed, such as real-time PCR, reverse transcriptase PCR, and nested PCR. The document explains that PCR has many applications, including diagnosis of infectious diseases and detection of genetic variations.