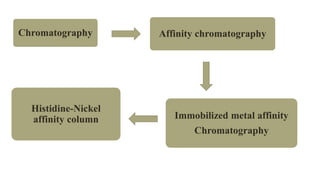
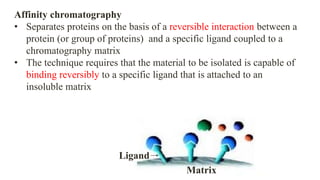
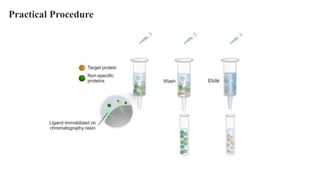
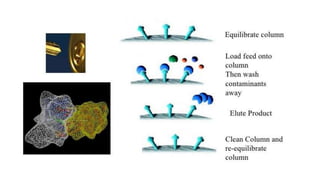
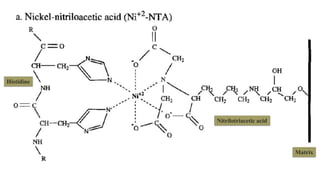
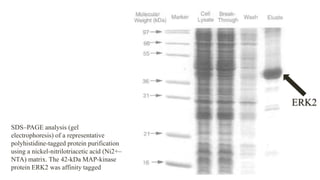
The document discusses histidine-nickel affinity chromatography, a method for separating proteins based on their reversible interaction with ligands on a chromatography matrix. It details the characteristics of an ideal matrix, the role of ligands and spacer arms, and outlines practical procedures for protein purification. The use of immobilized metal affinity chromatography with histidine residues is emphasized for its efficiency in purifying tagged proteins.