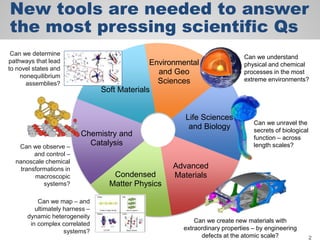
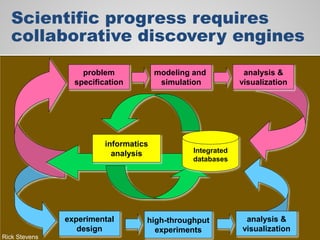
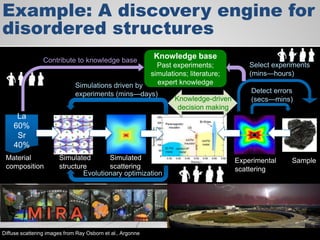
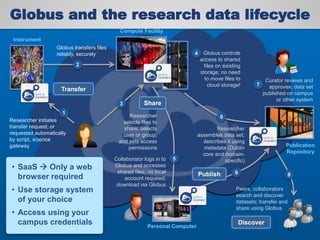
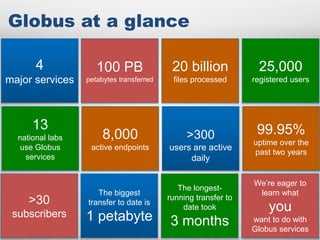
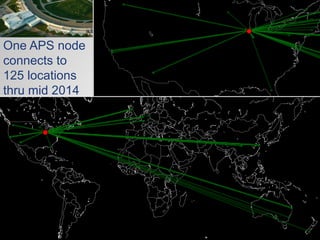
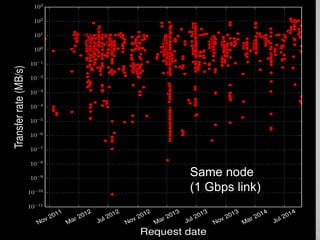
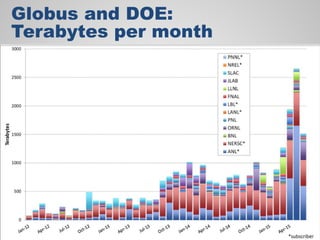
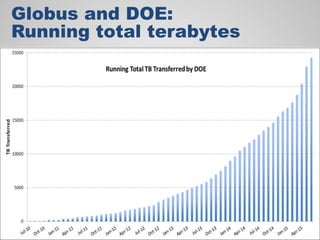
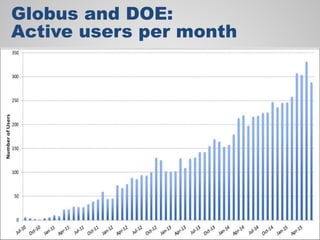
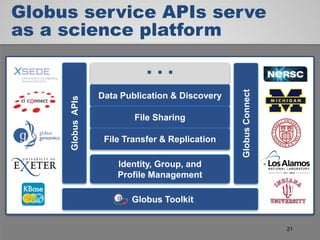
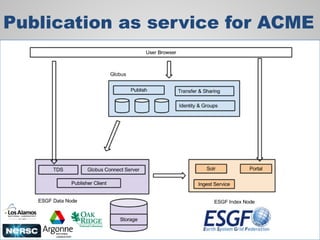
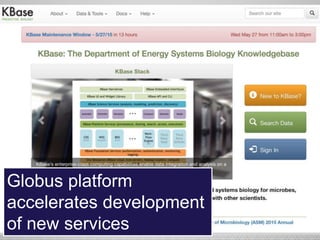
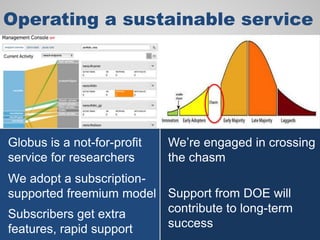
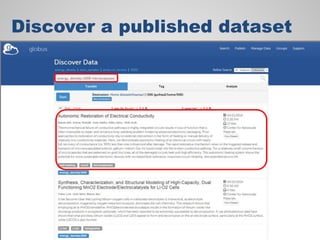
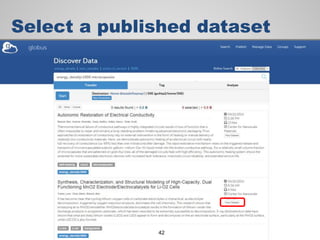
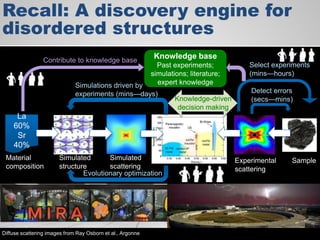
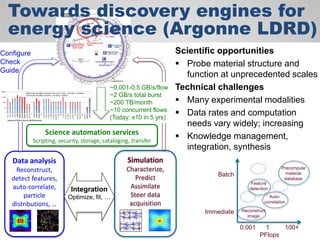
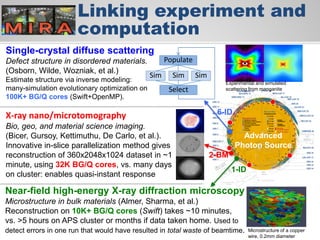
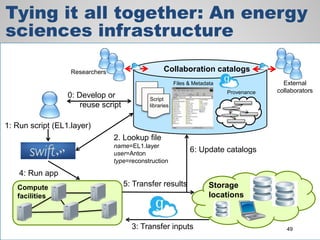
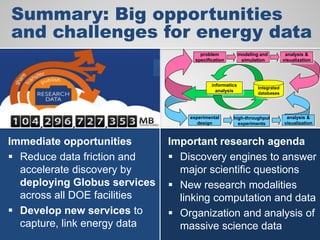
The document discusses the evolution of data-driven discovery in energy science, emphasizing the development of new tools and collaborative research methodologies to enhance data access and sharing. It highlights the importance of eliminating data friction through services like Globus, which streamline data transfer, analysis, and publication processes. Additionally, it outlines the need for discovery engines to facilitate the integration of simulation and experimental data in order to address complex scientific questions.