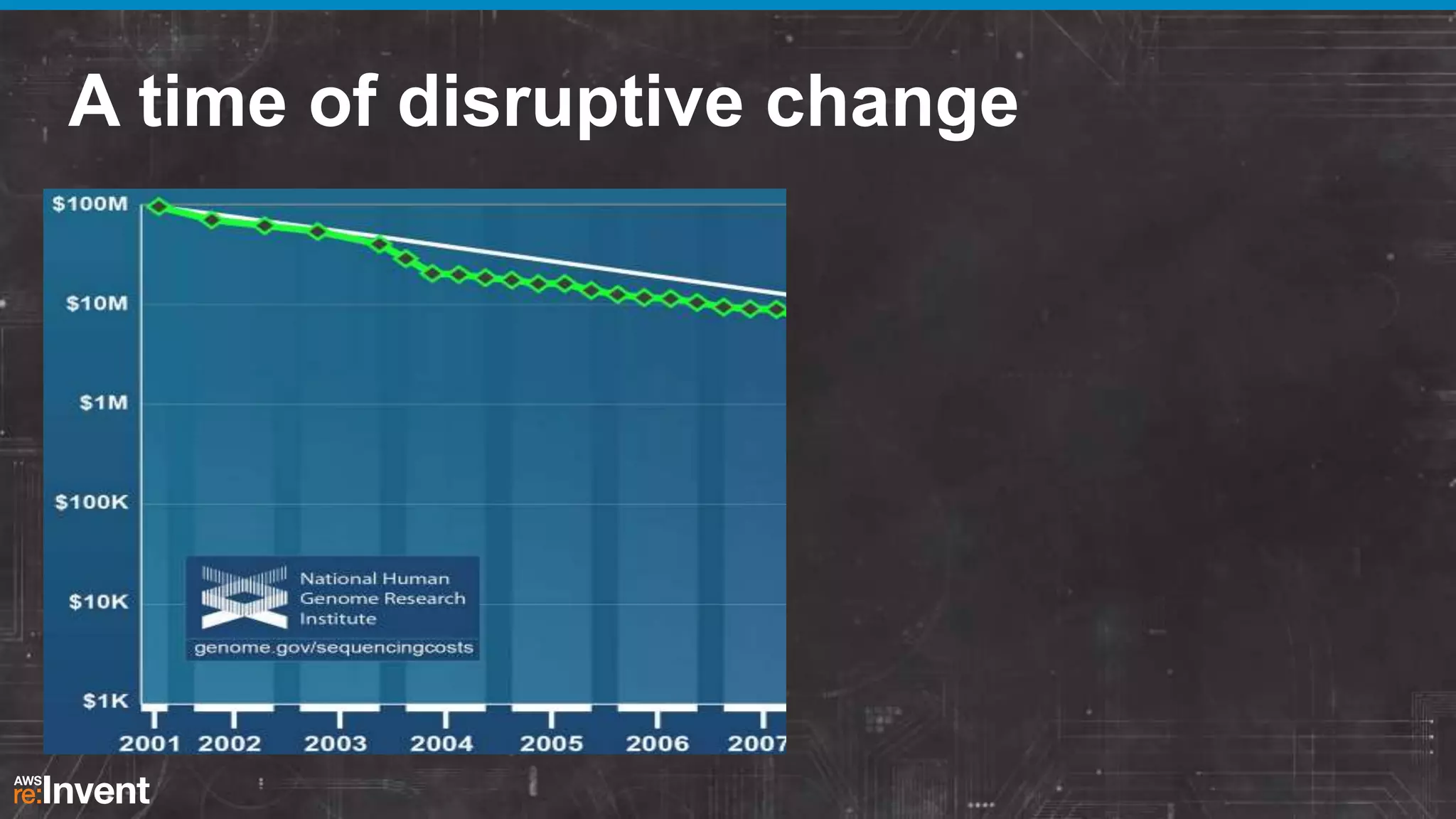
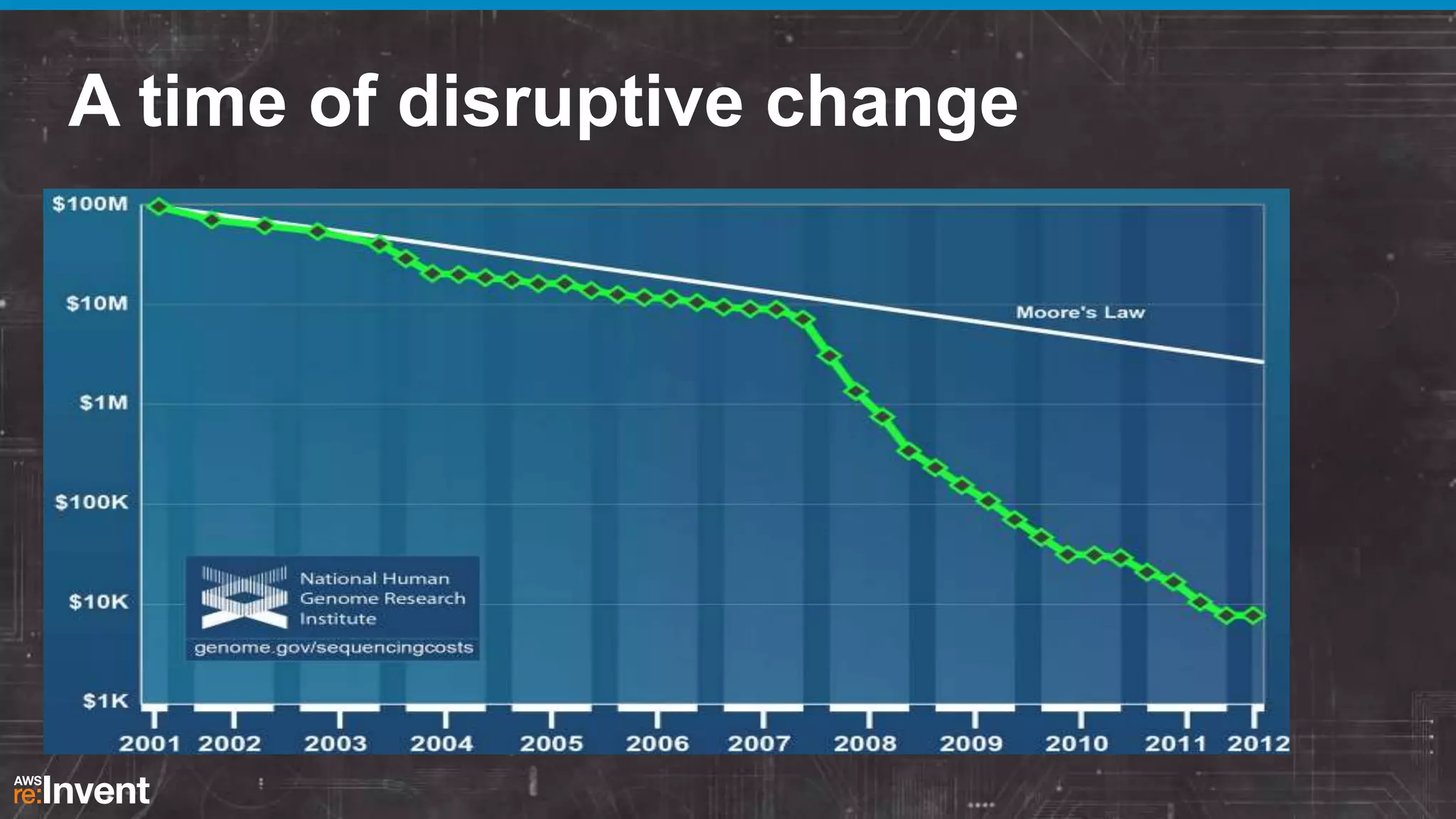
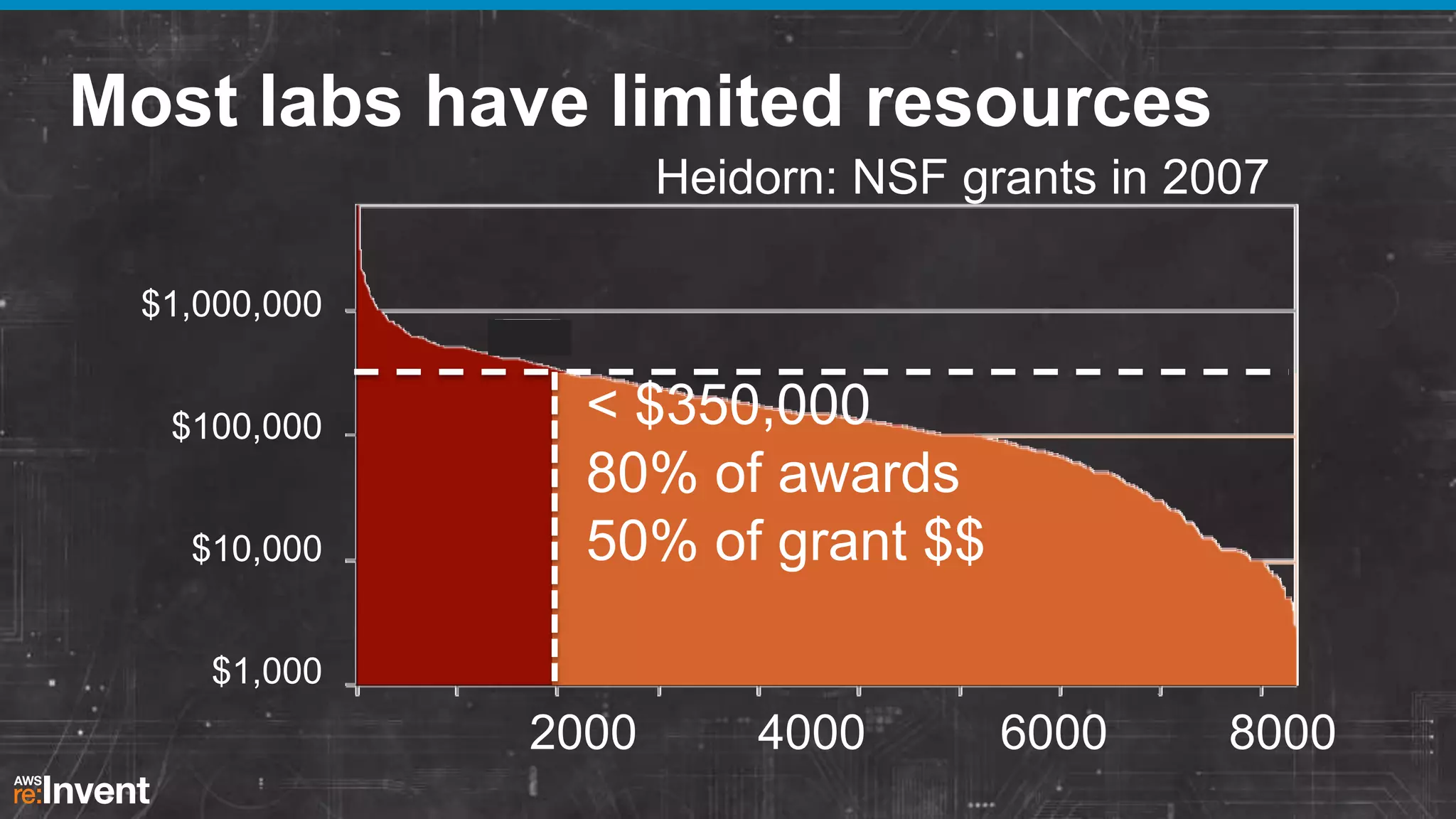
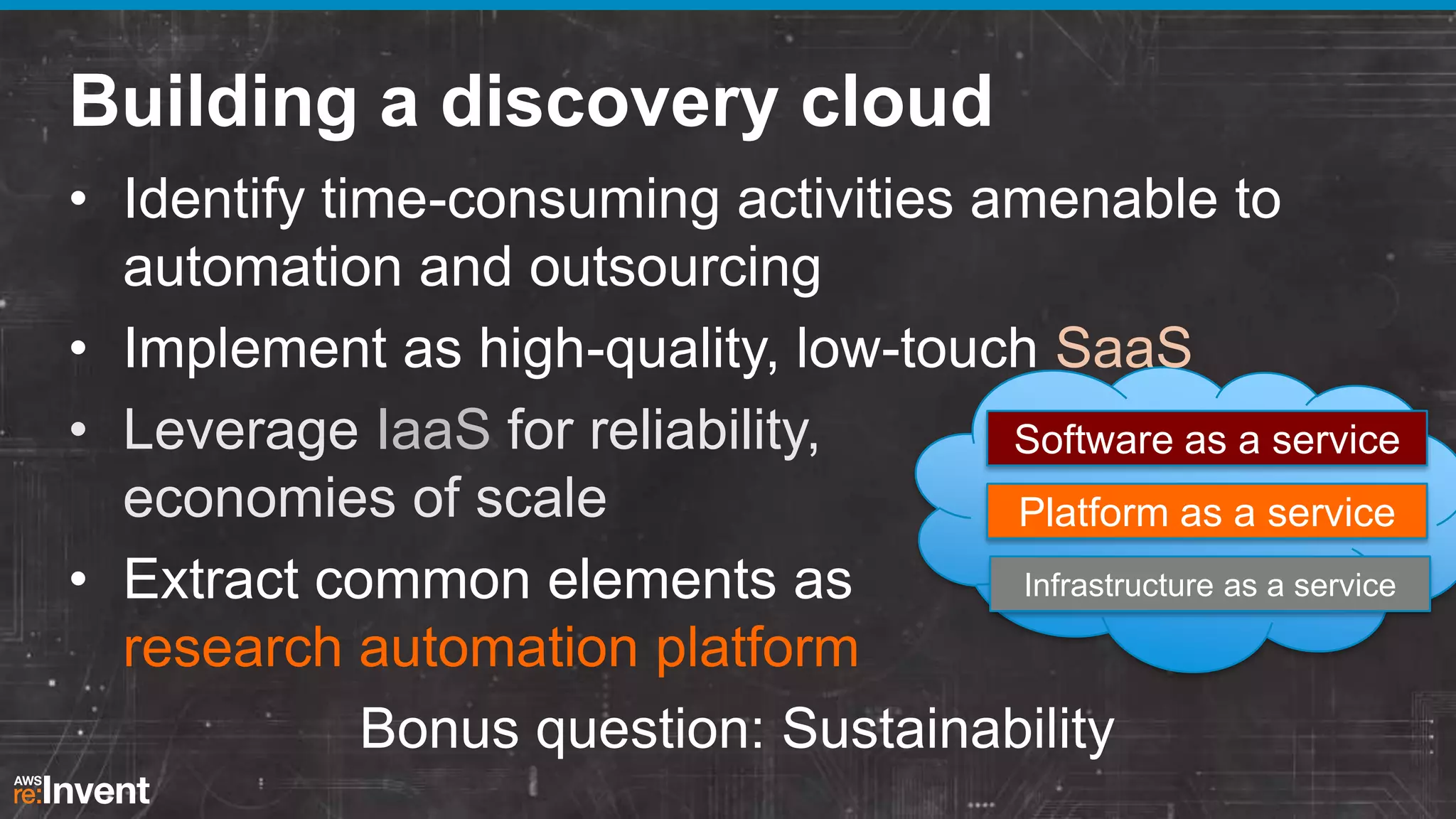
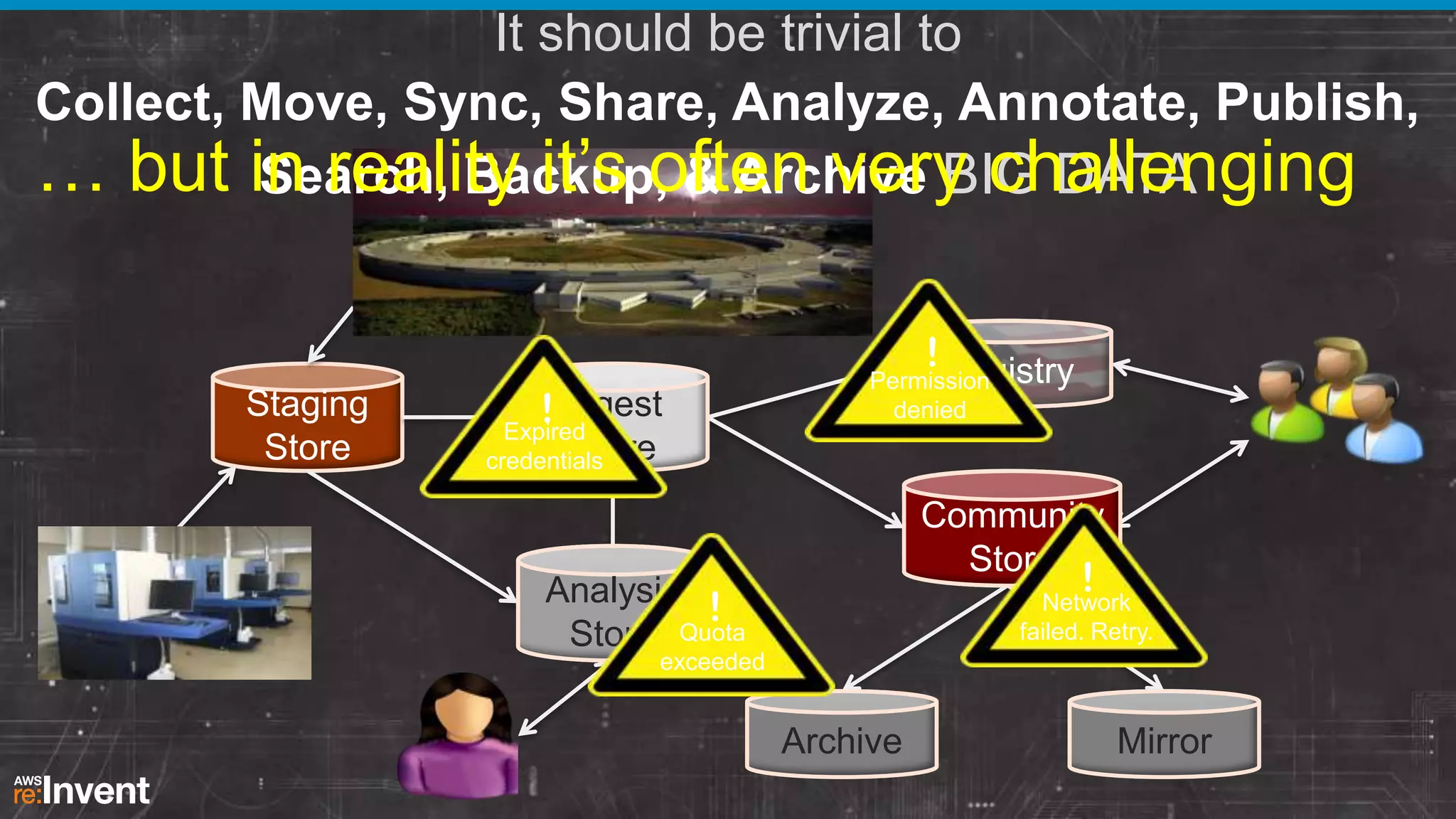
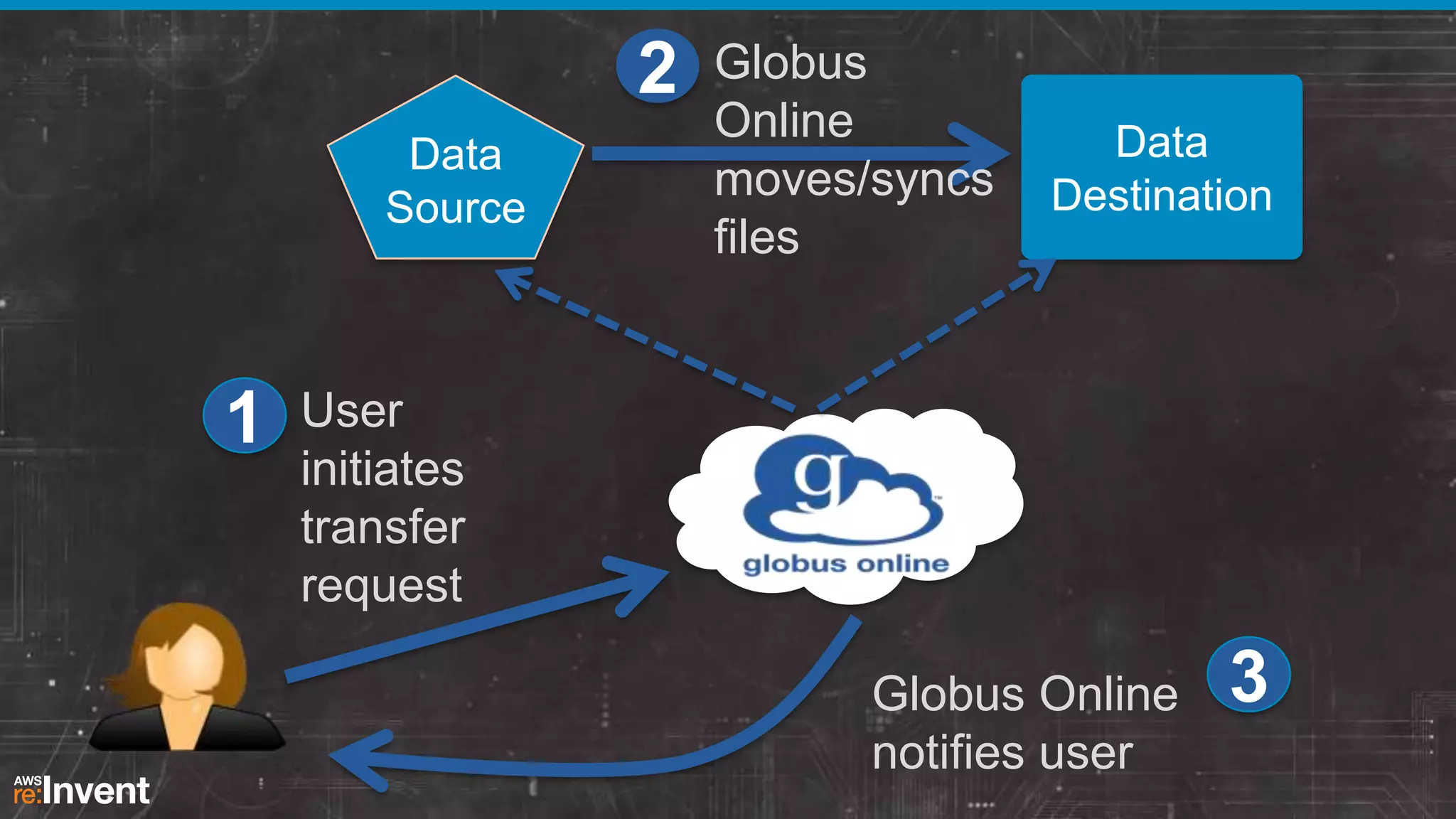
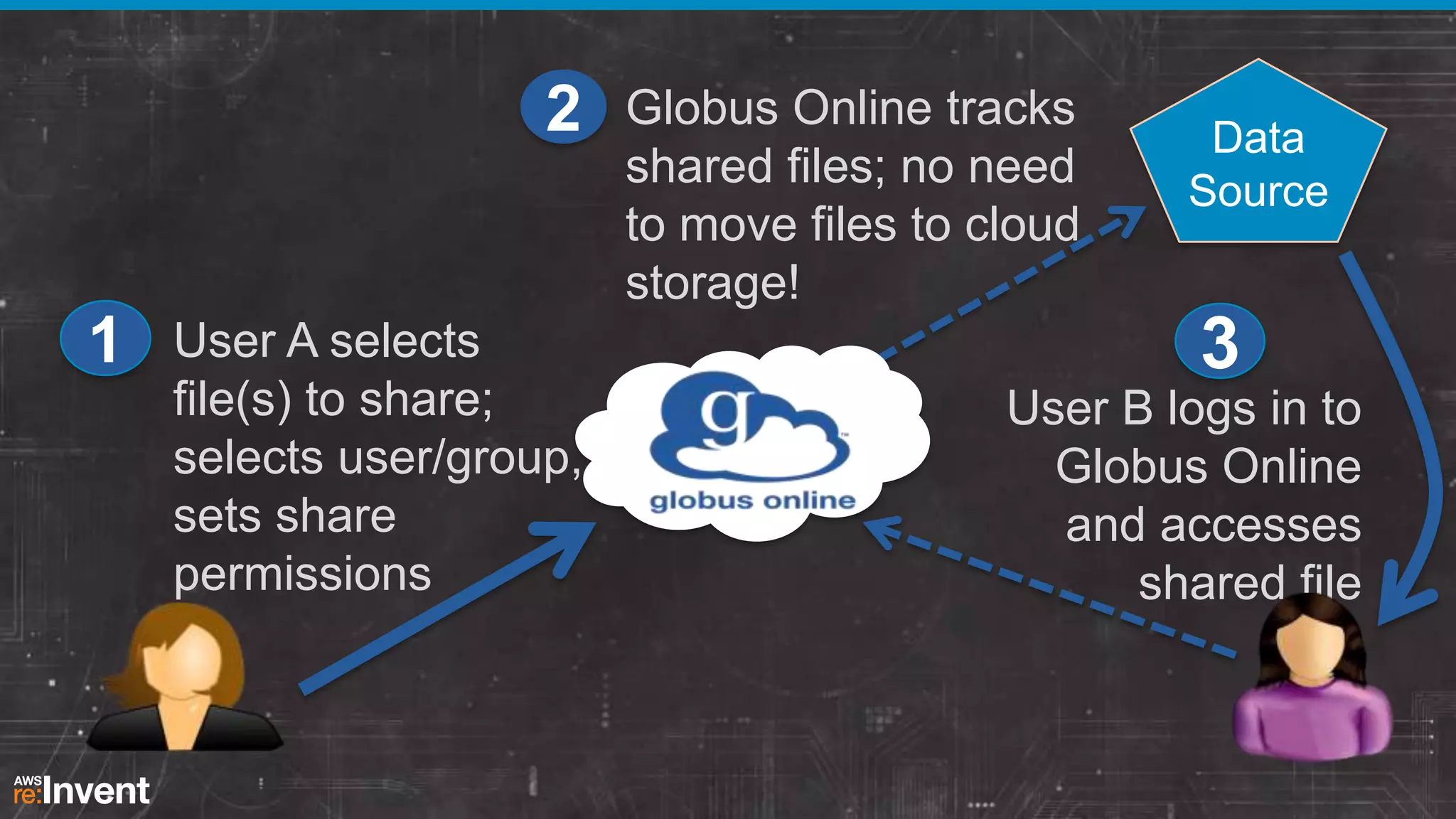
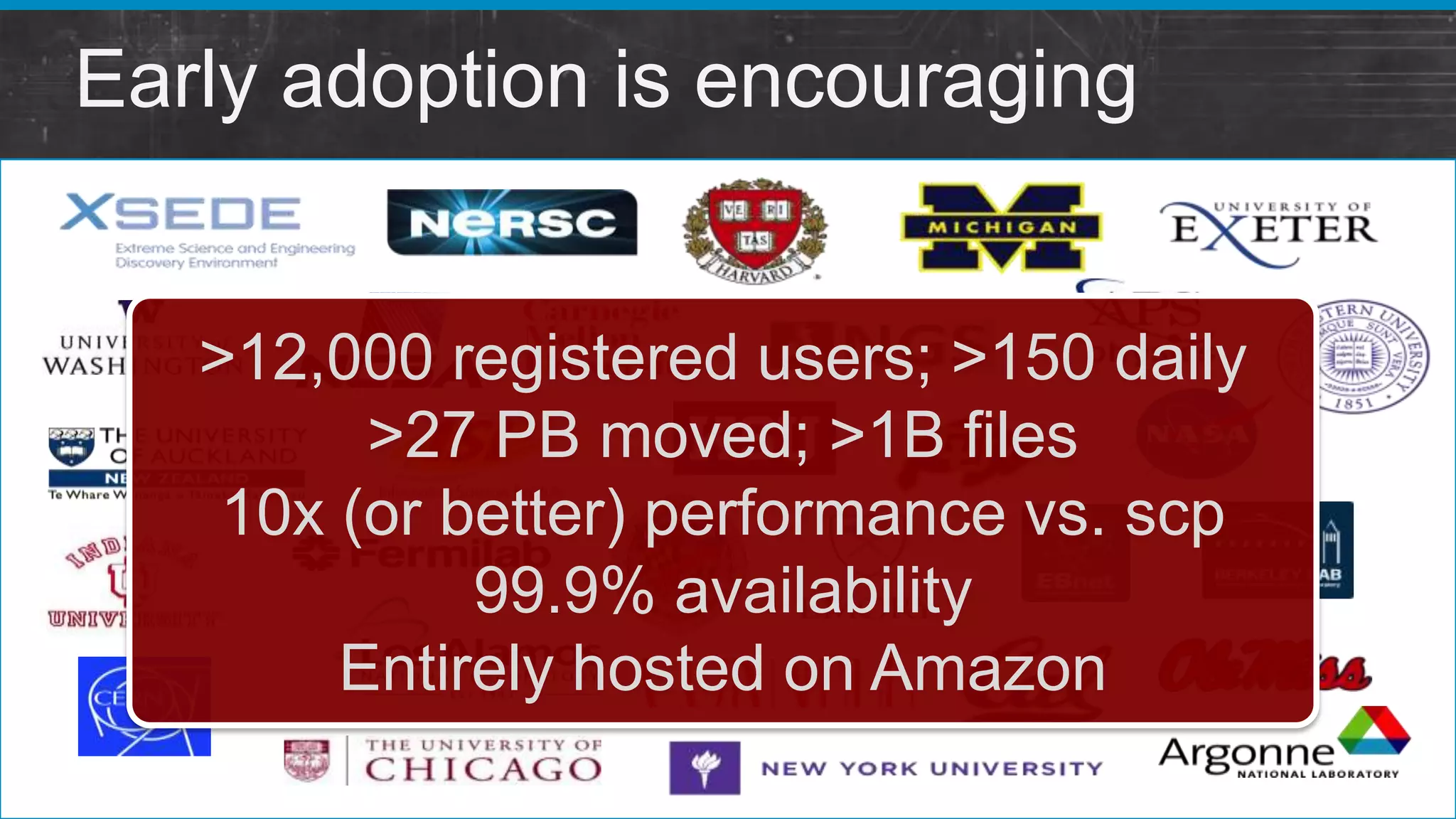
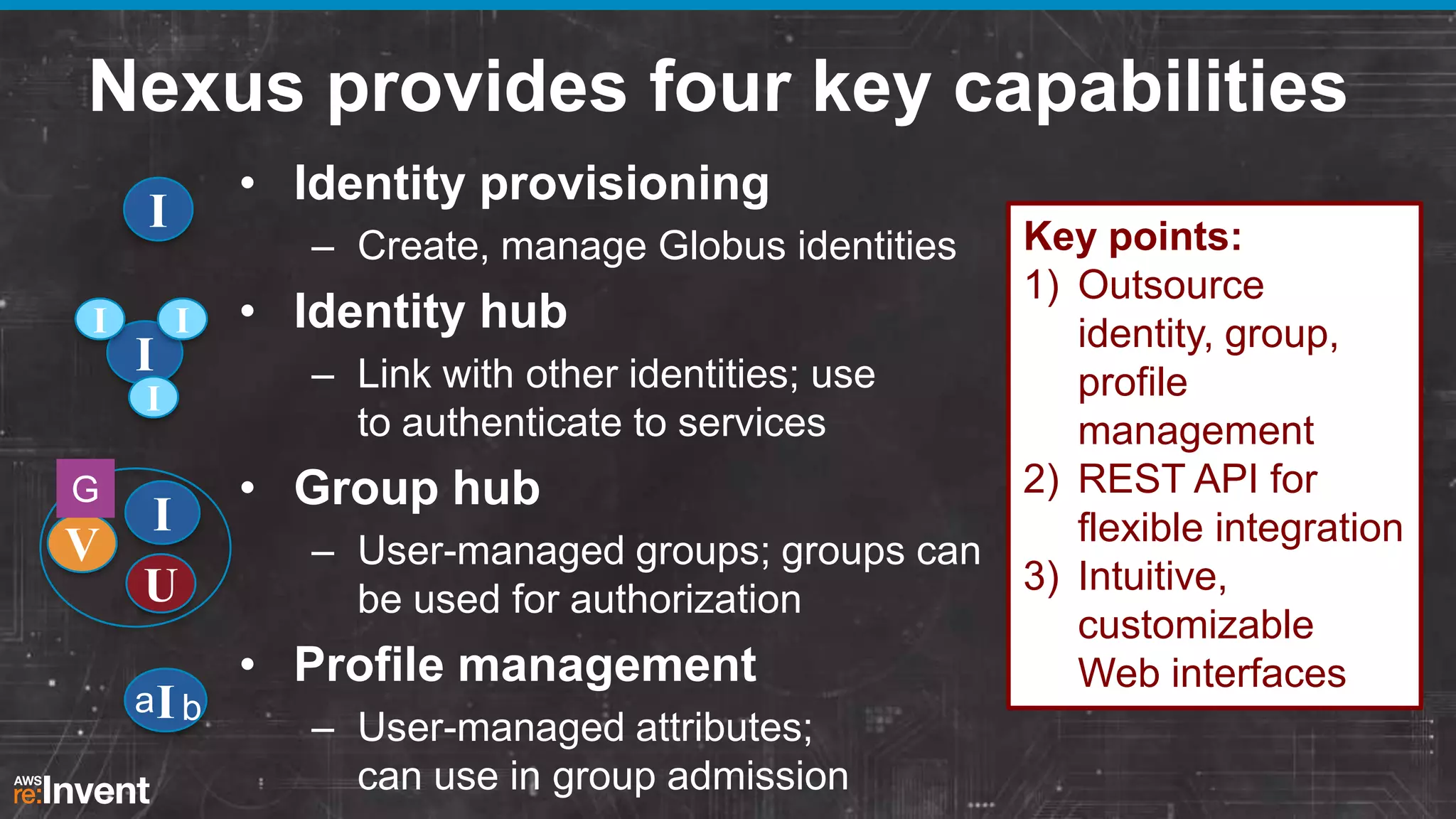
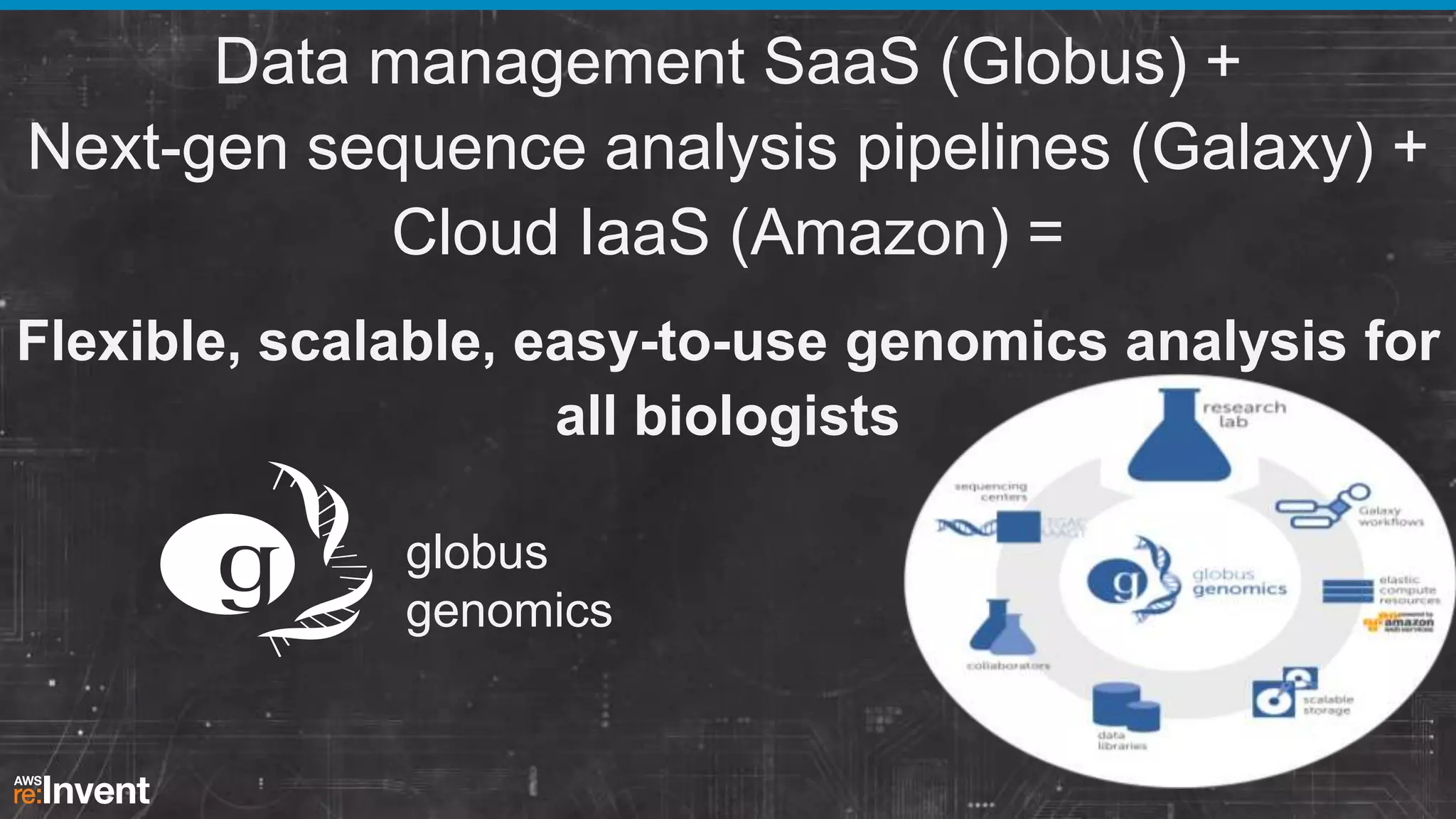
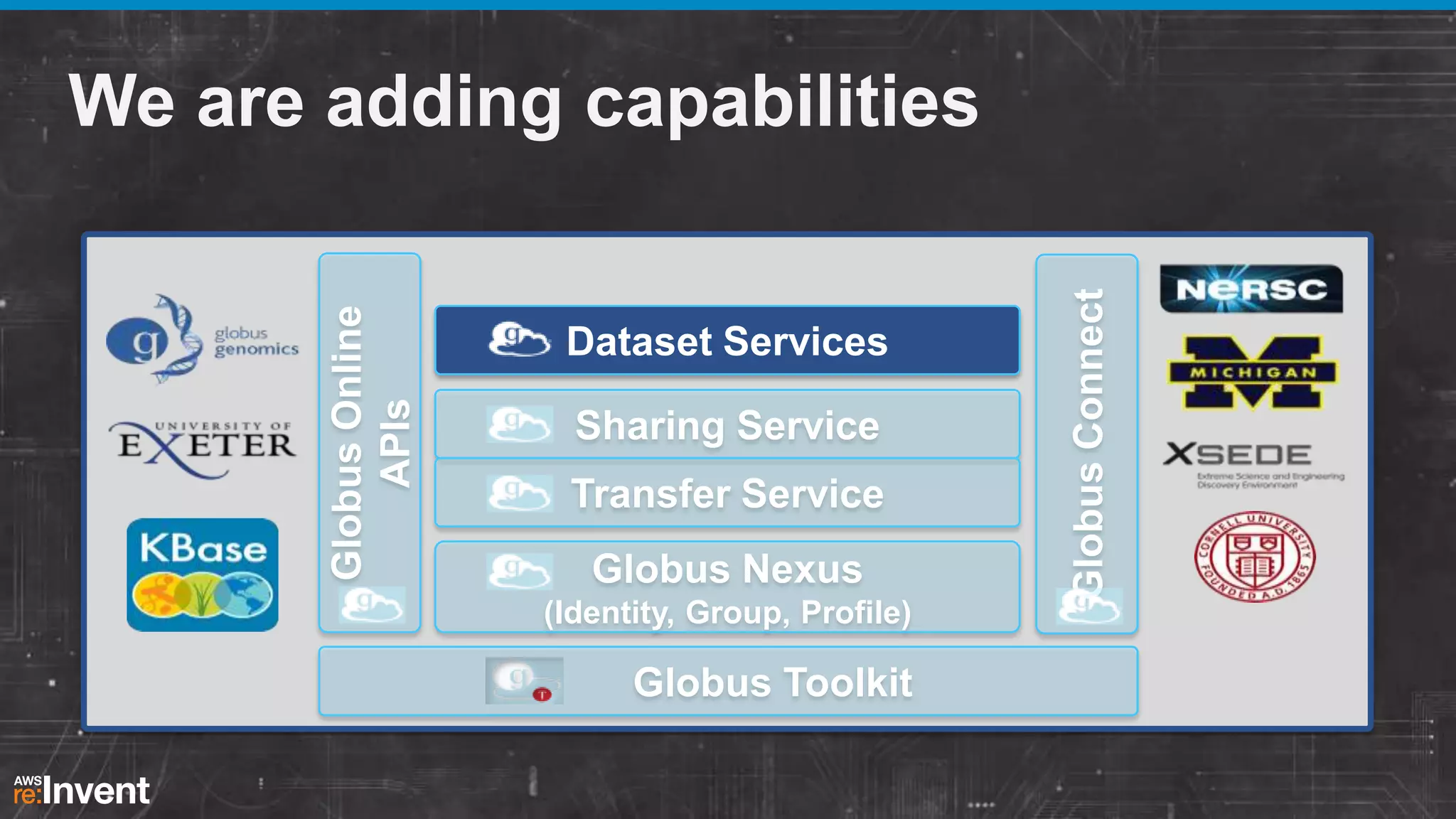
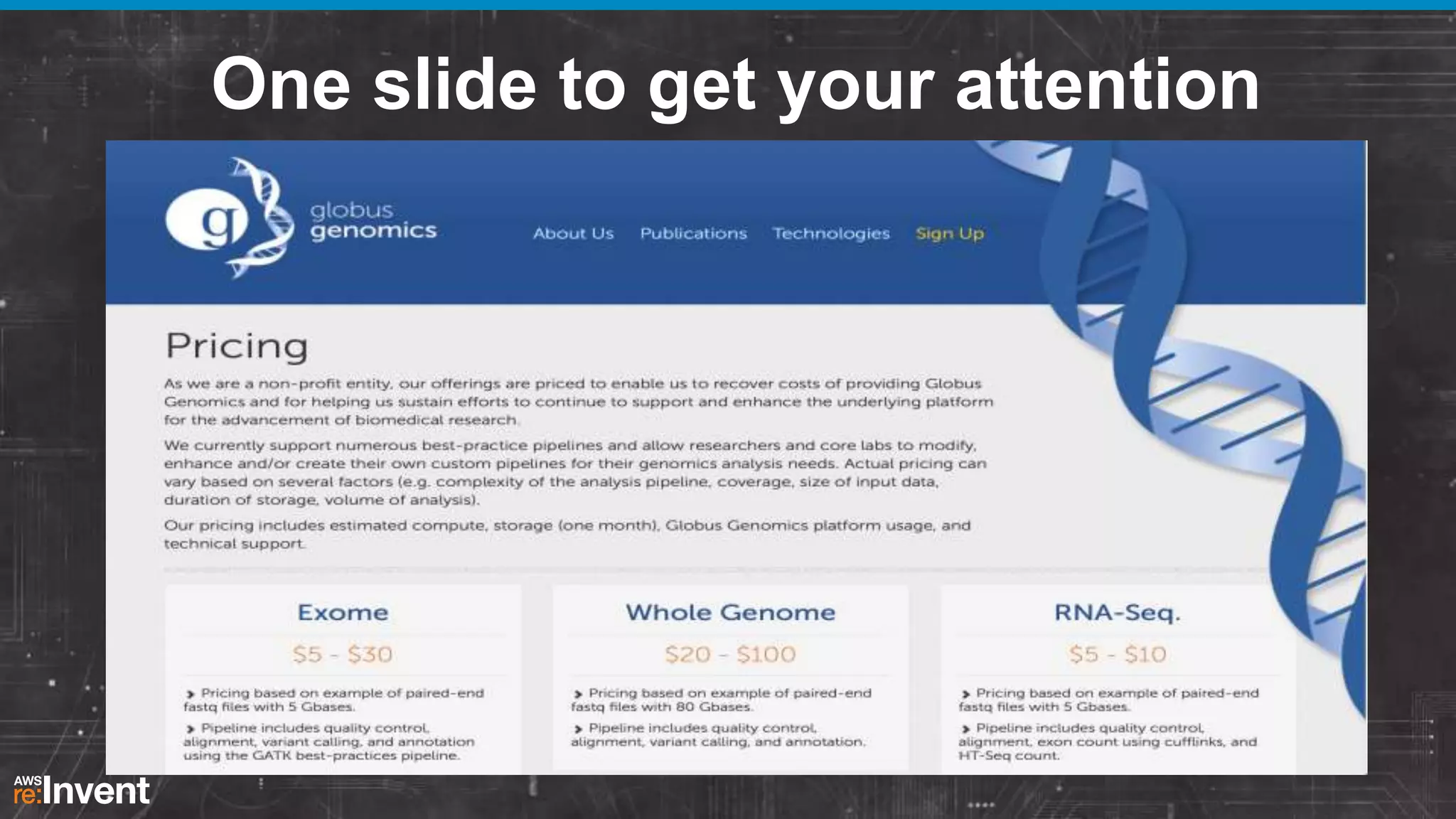
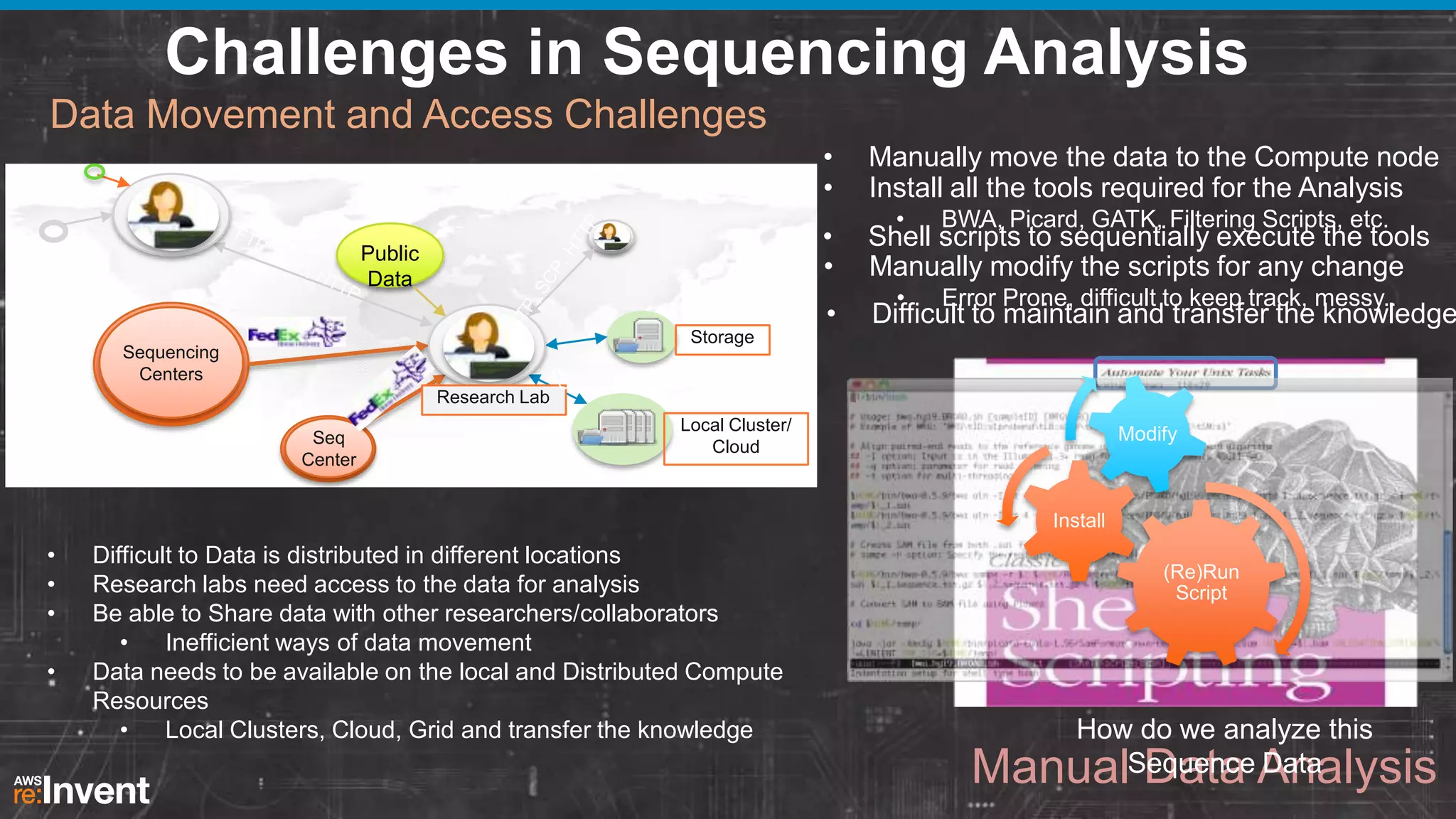
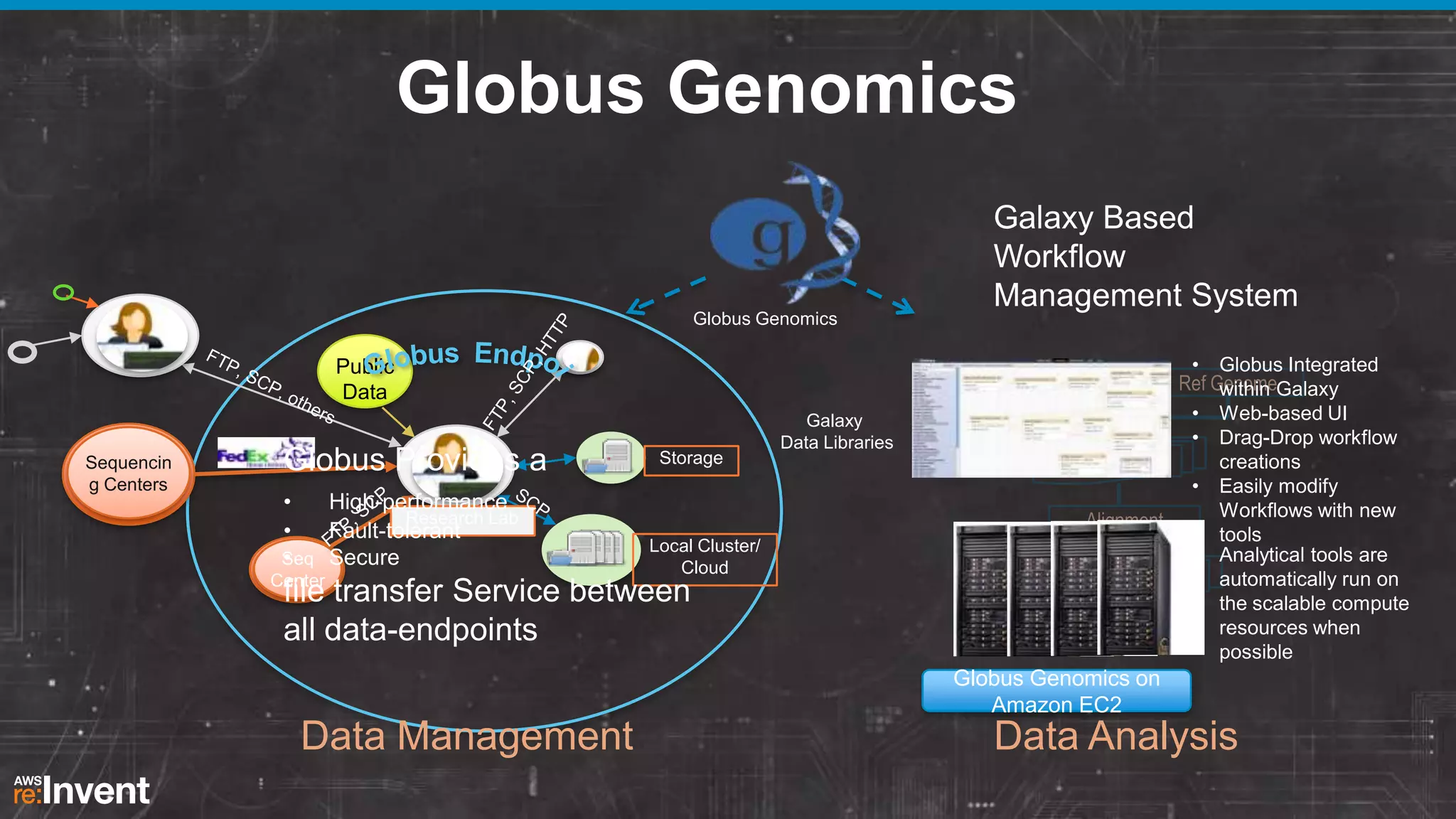
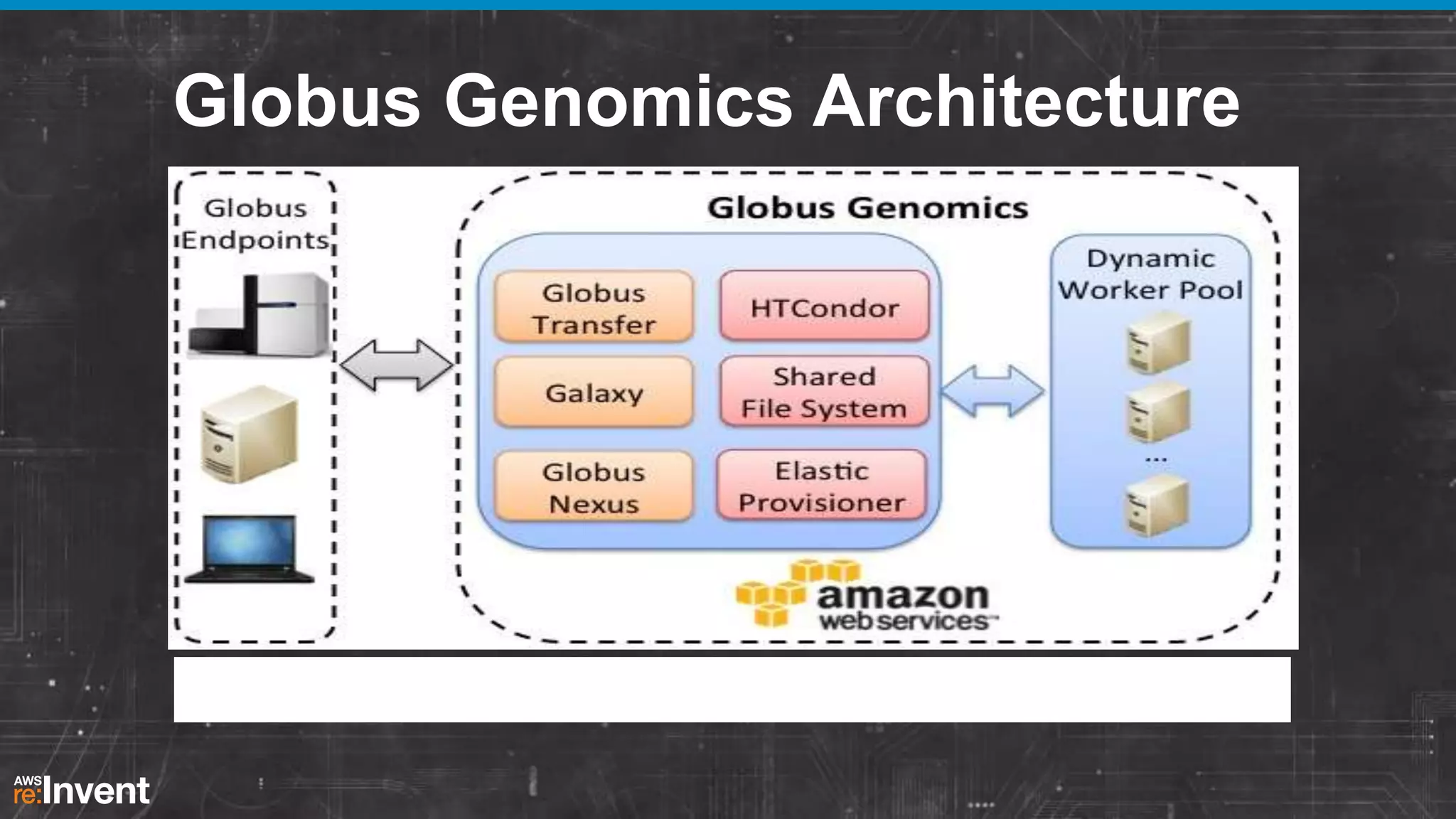
This document summarizes a presentation about providing next-generation sequencing analysis capabilities using Globus Genomics. It outlines challenges with current manual approaches to sequencing data analysis, including difficulties moving large datasets between locations and maintaining complex analysis scripts. The presentation introduces Globus Genomics, which uses Globus data transfer services integrated with Galaxy to provide a workflow-based system for sequencing analysis without requiring local installation or configuration. Key benefits include on-demand access to scalable cloud resources, ability to easily modify and reuse analysis workflows, and integration with data sources. The system aims to accelerate genomic research by automating and simplifying analysis.