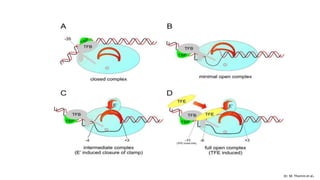
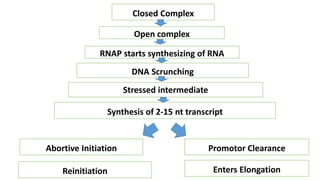
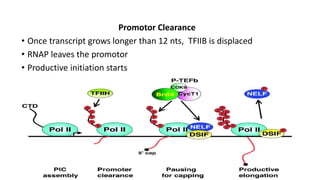
This document discusses abortive initiation, which is the repetitive synthesis and release of short nascent RNA transcripts between 2-15 nucleotides long by RNA polymerase during transcription initiation. It involves three steps - formation of a closed preinitiation complex, formation of an open complex, and transitioning from initiation to elongation through promoter escape. During DNA scrunching, the polymerase remains stationary while the downstream DNA passes through, leading to unwinding and compaction. This stressed intermediate can either abort transcription or continue through promoter clearance once the transcript grows past 12 nucleotides. The fate of these short abortive transcripts is that they are very short-lived and quickly degraded, though their biological functions are unknown.