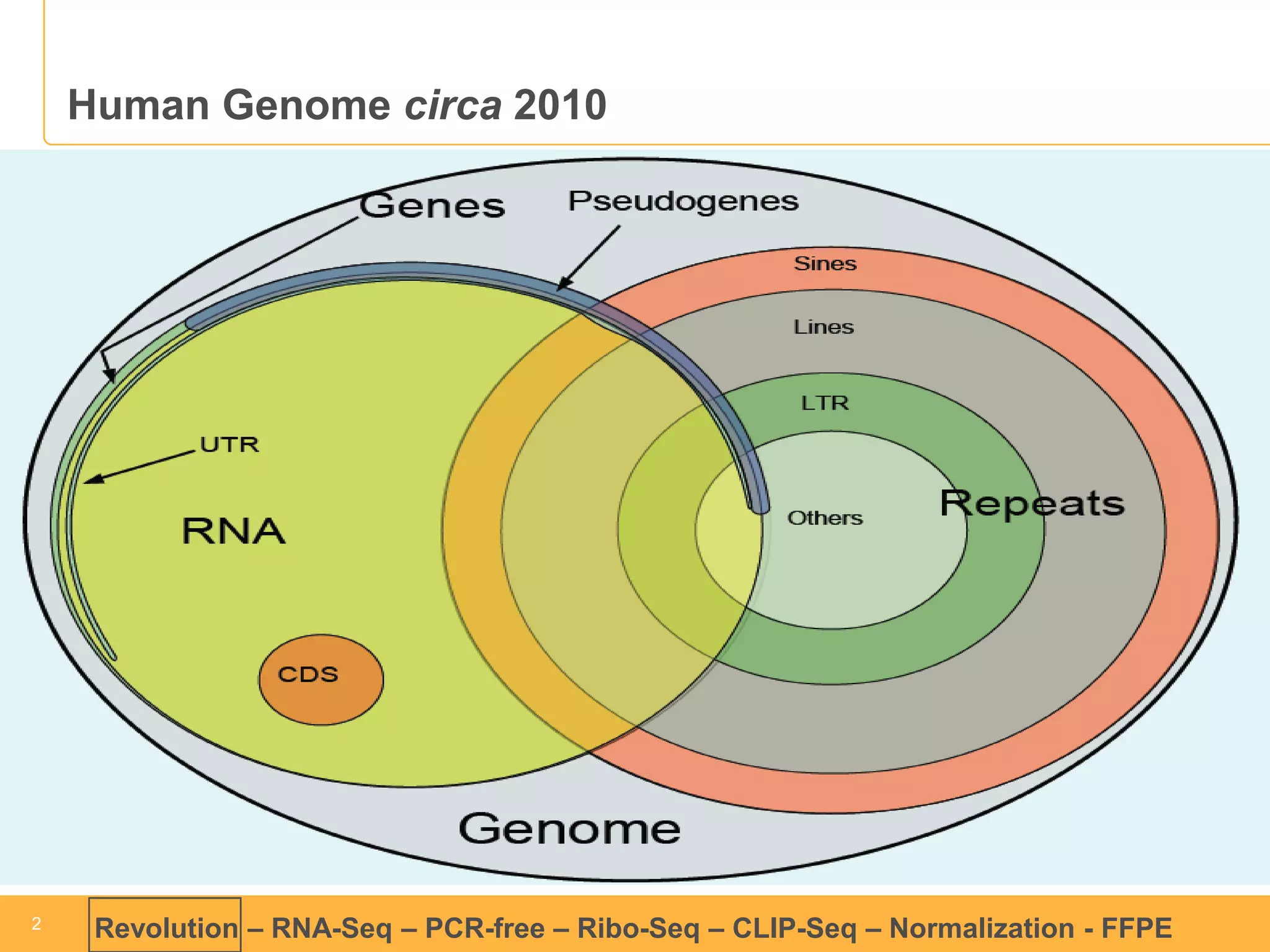
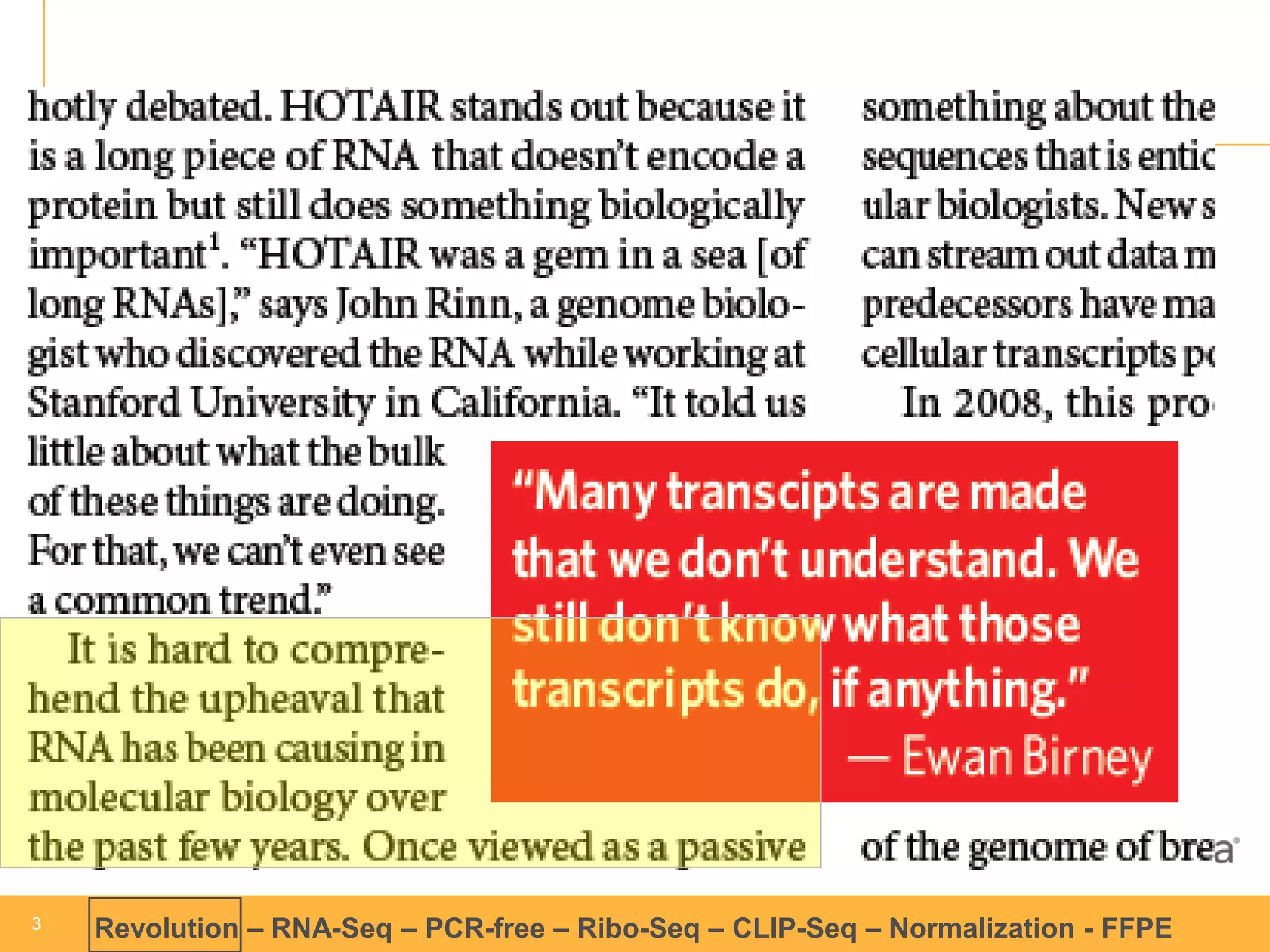
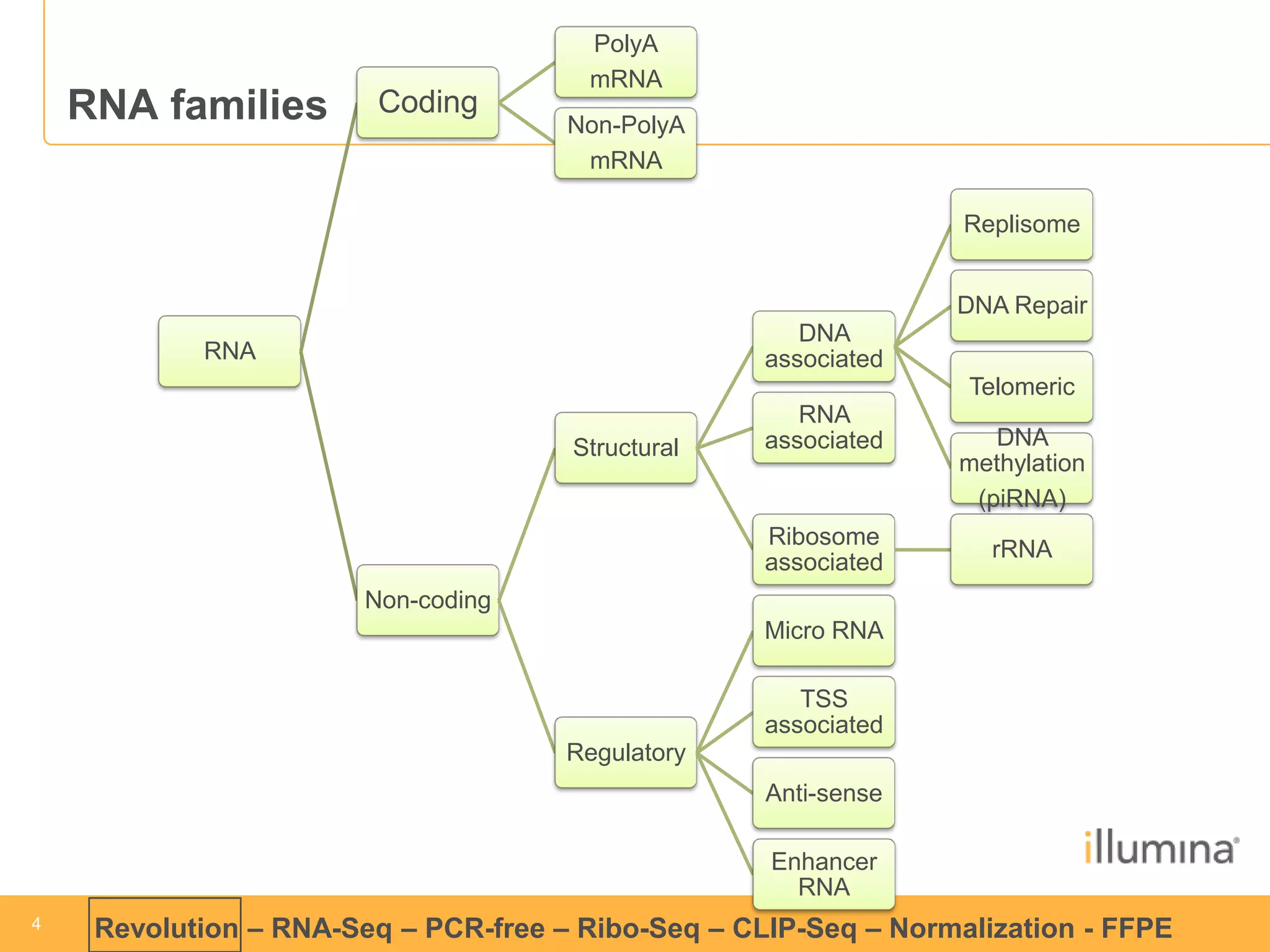
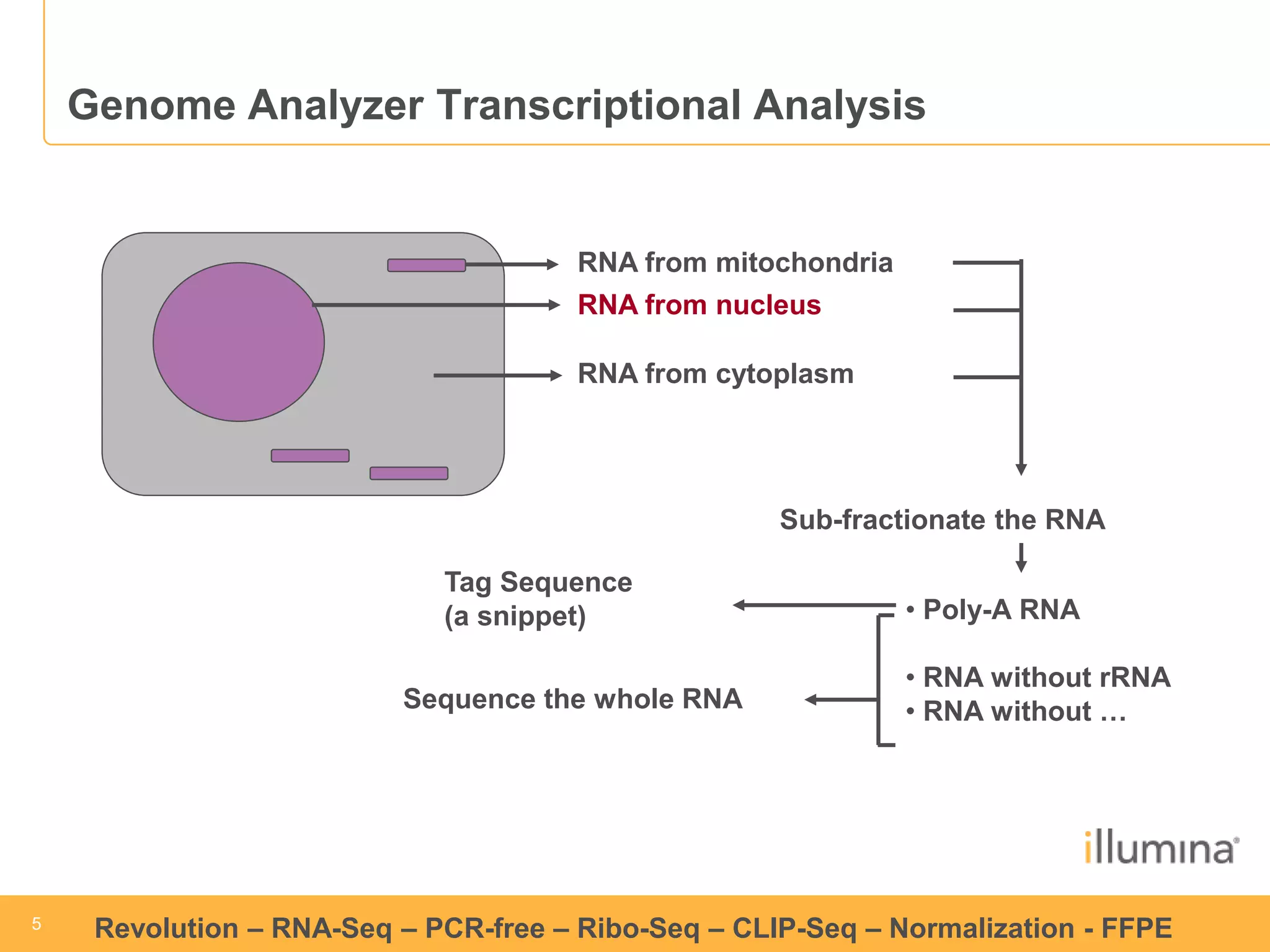
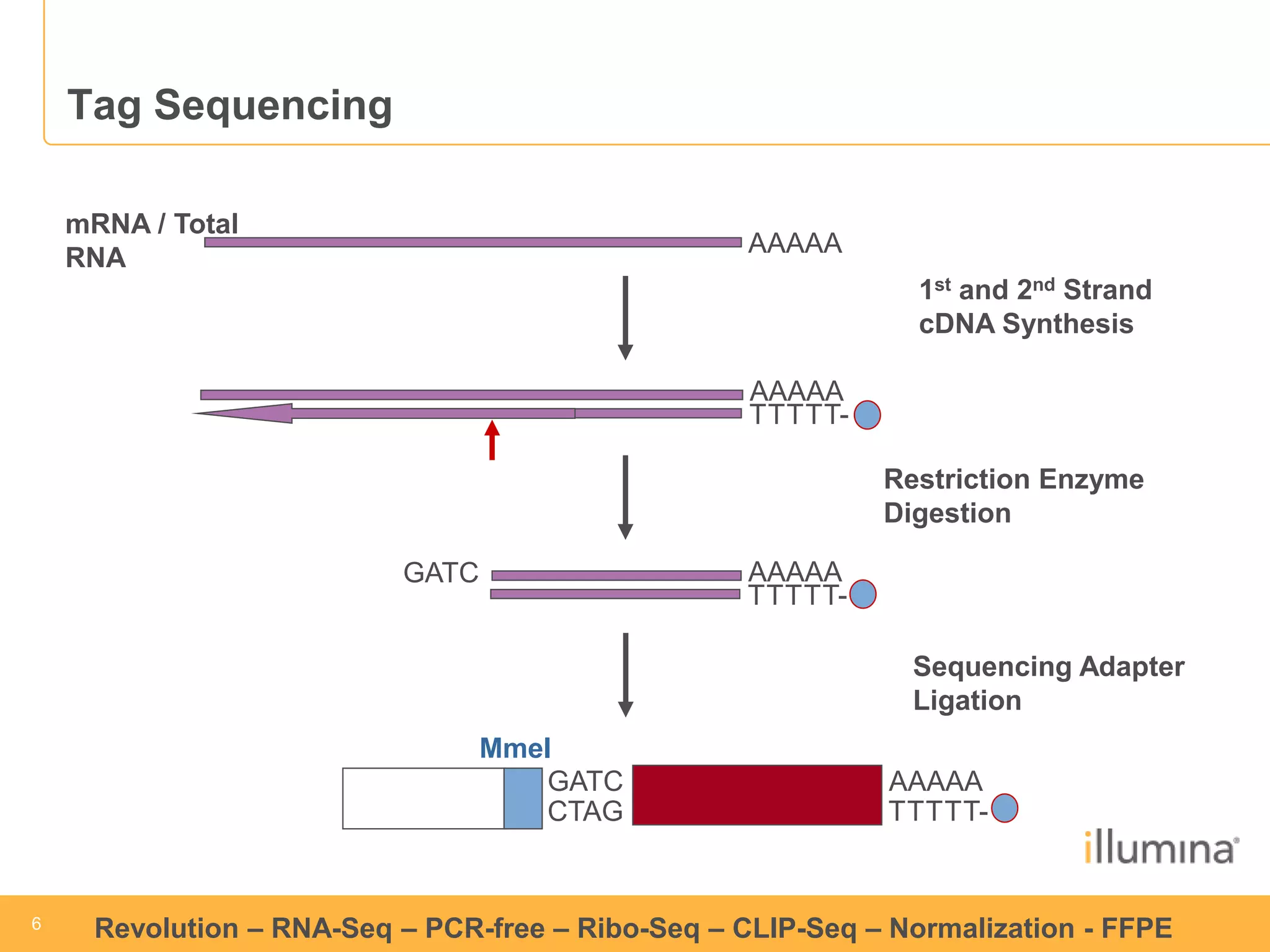
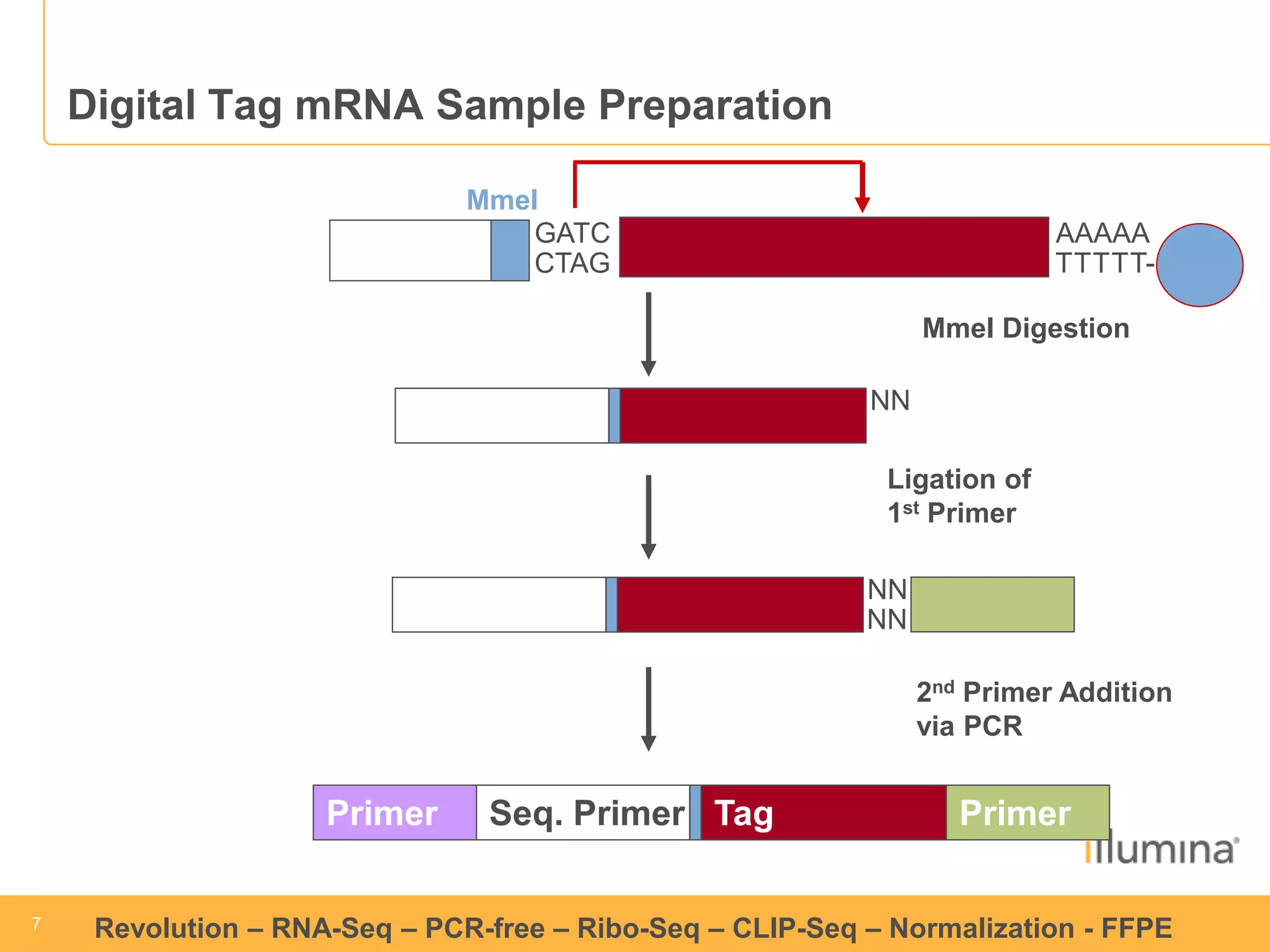
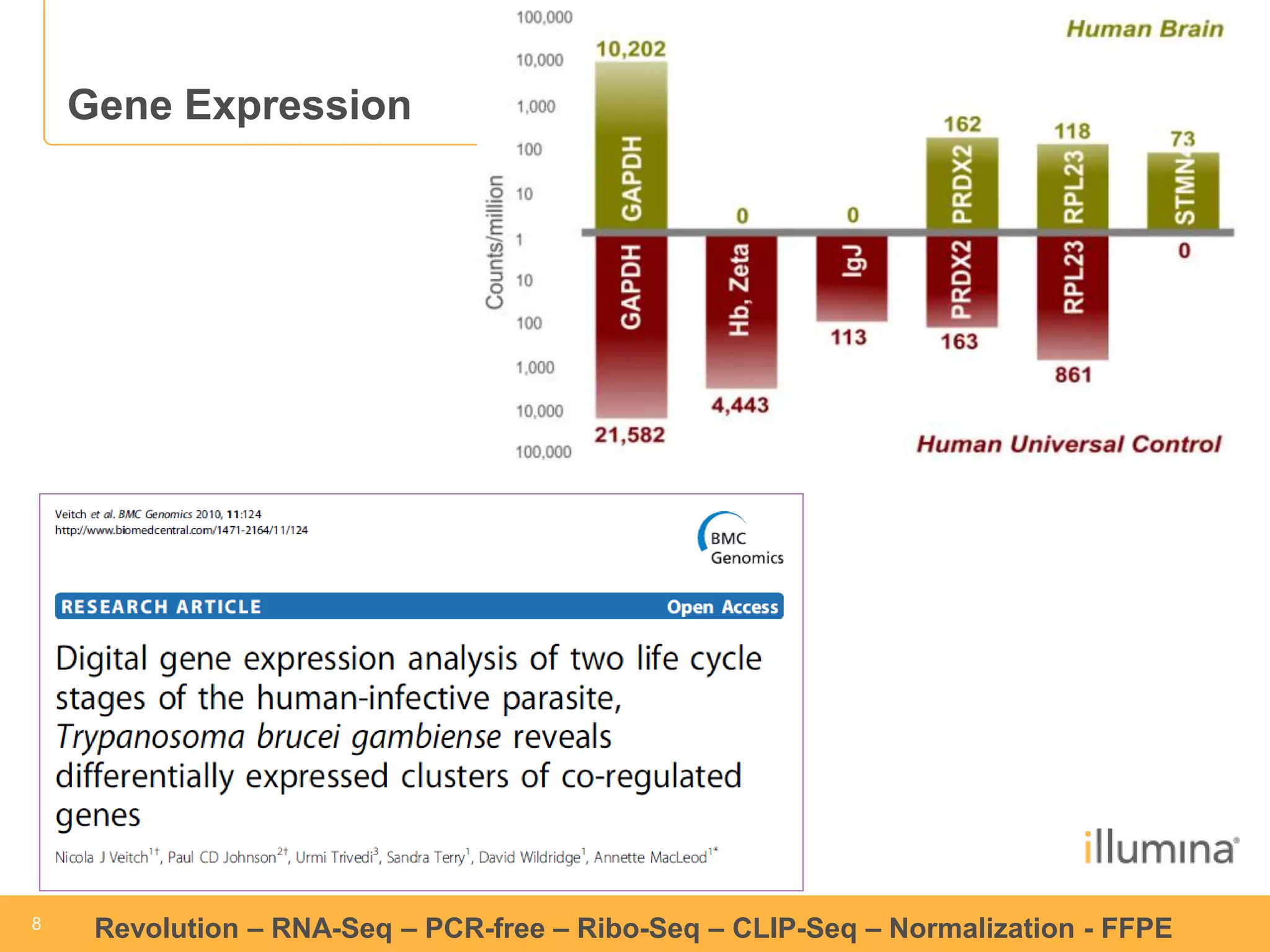
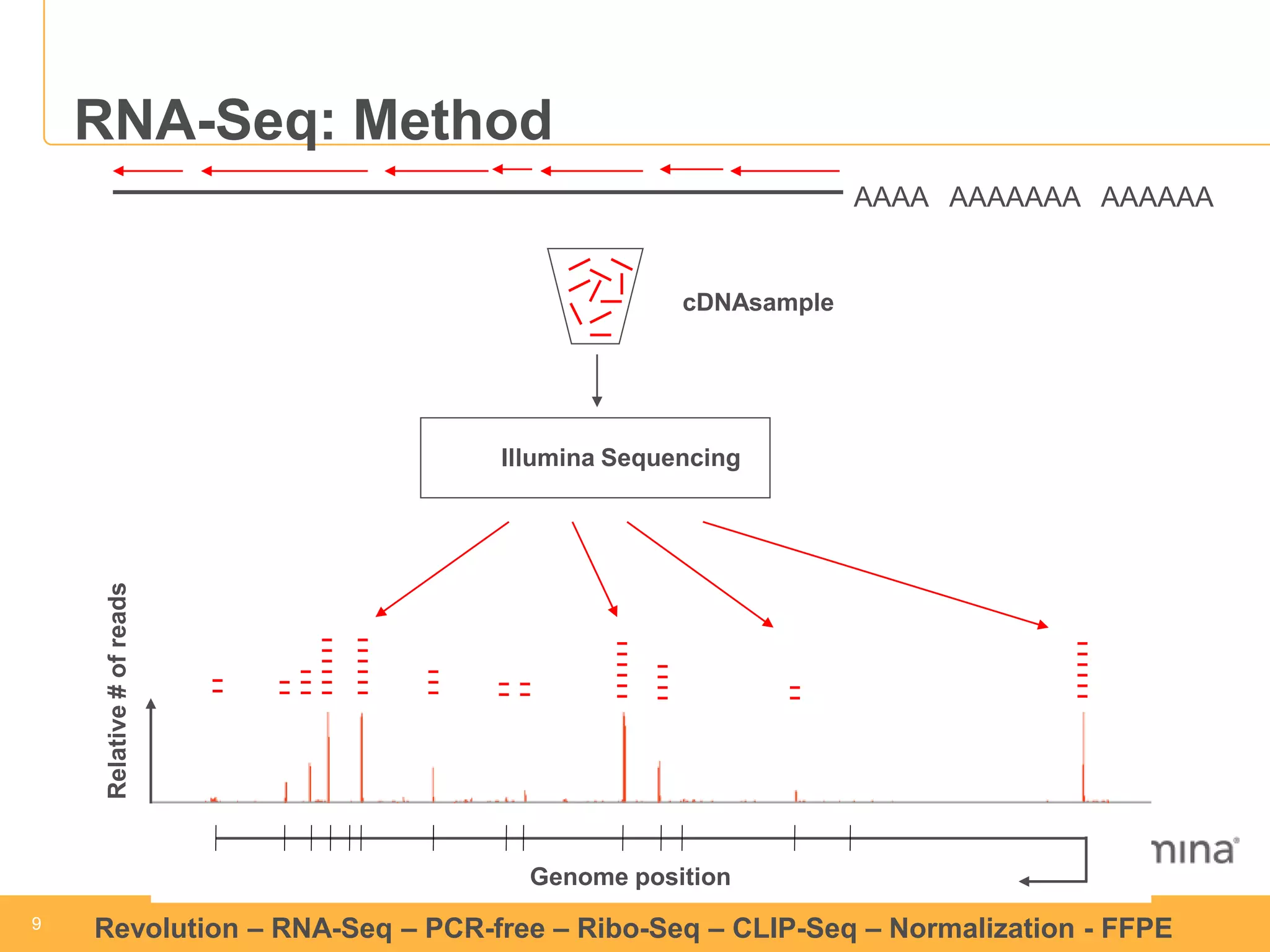
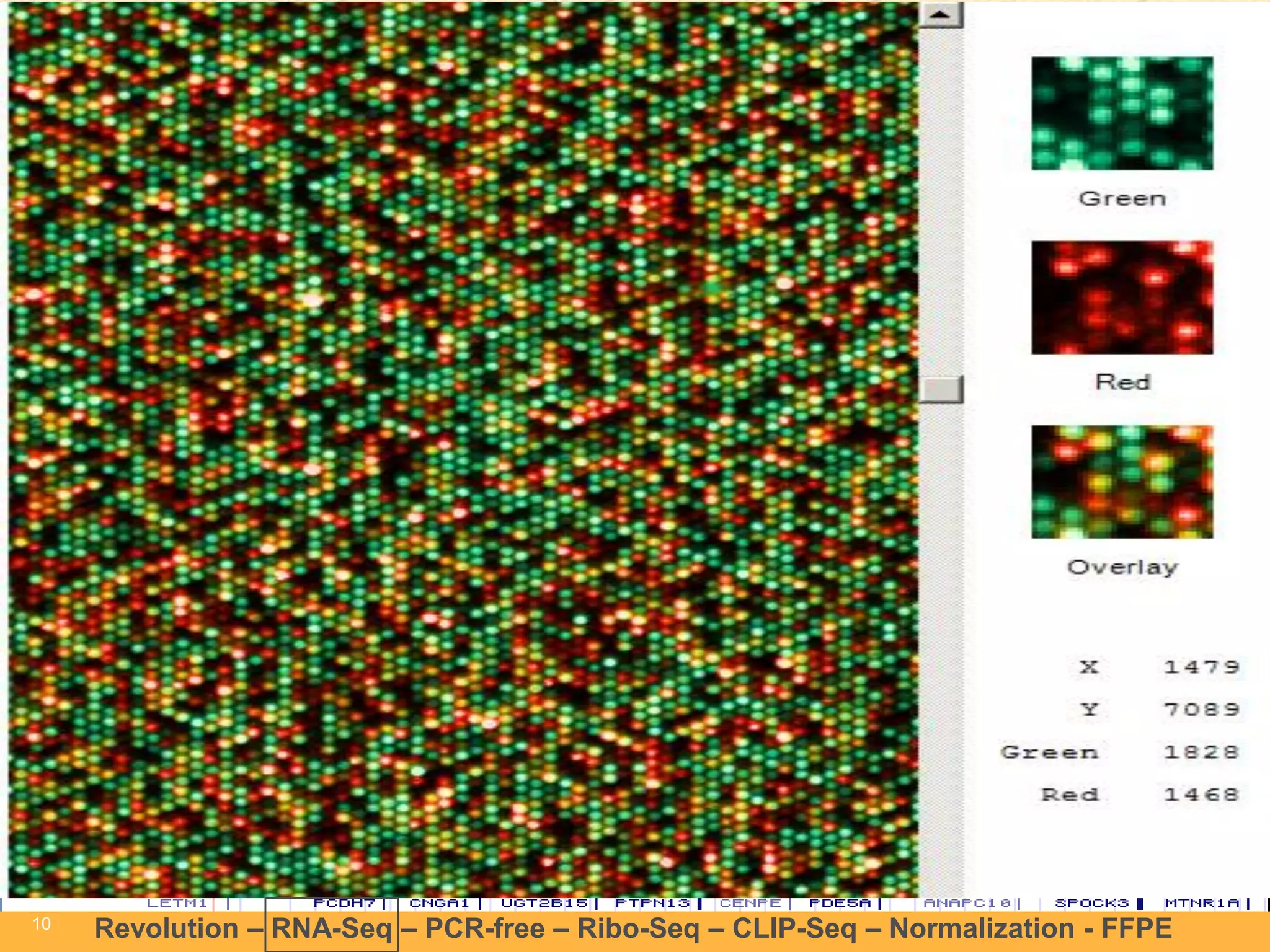
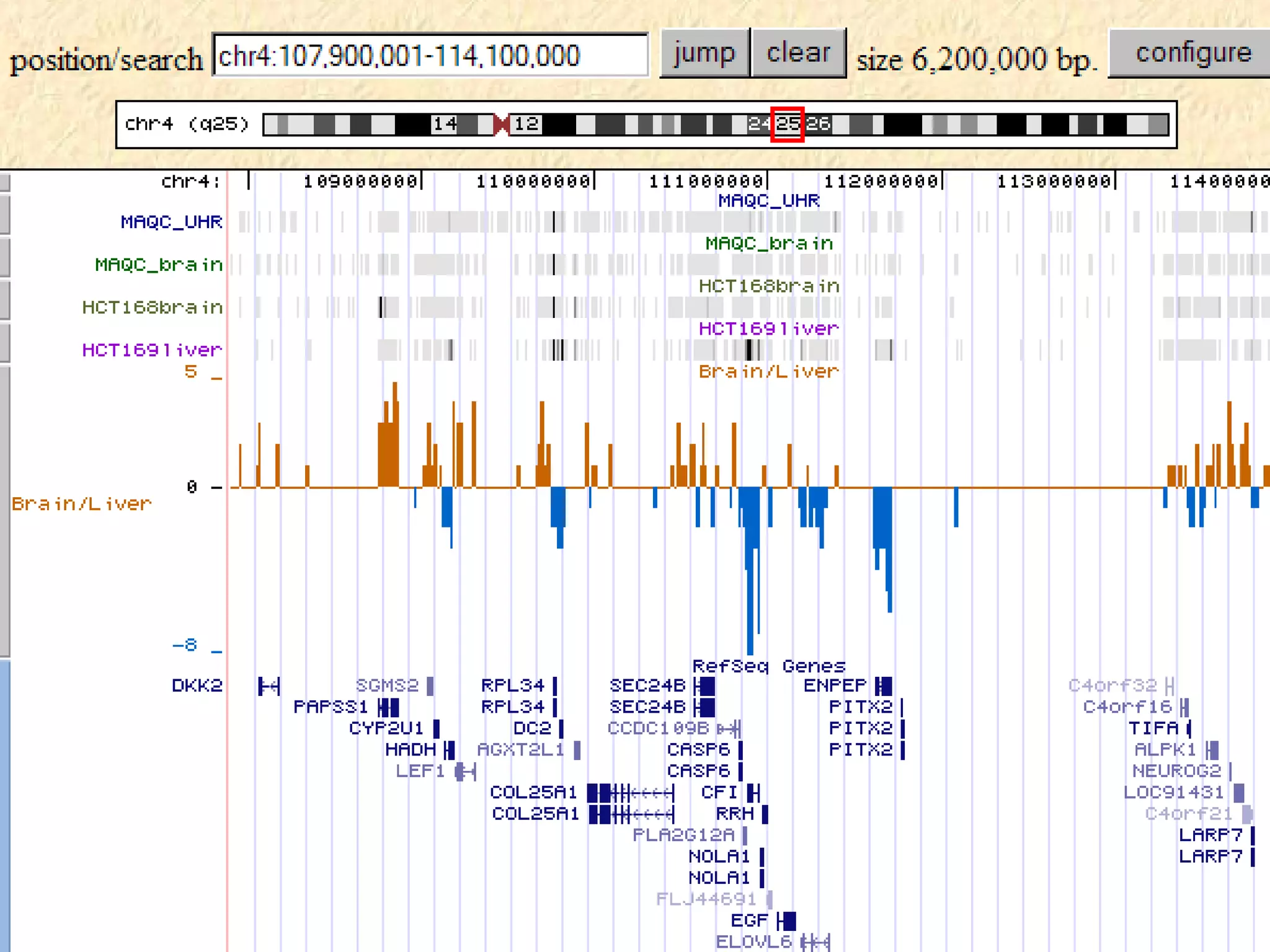
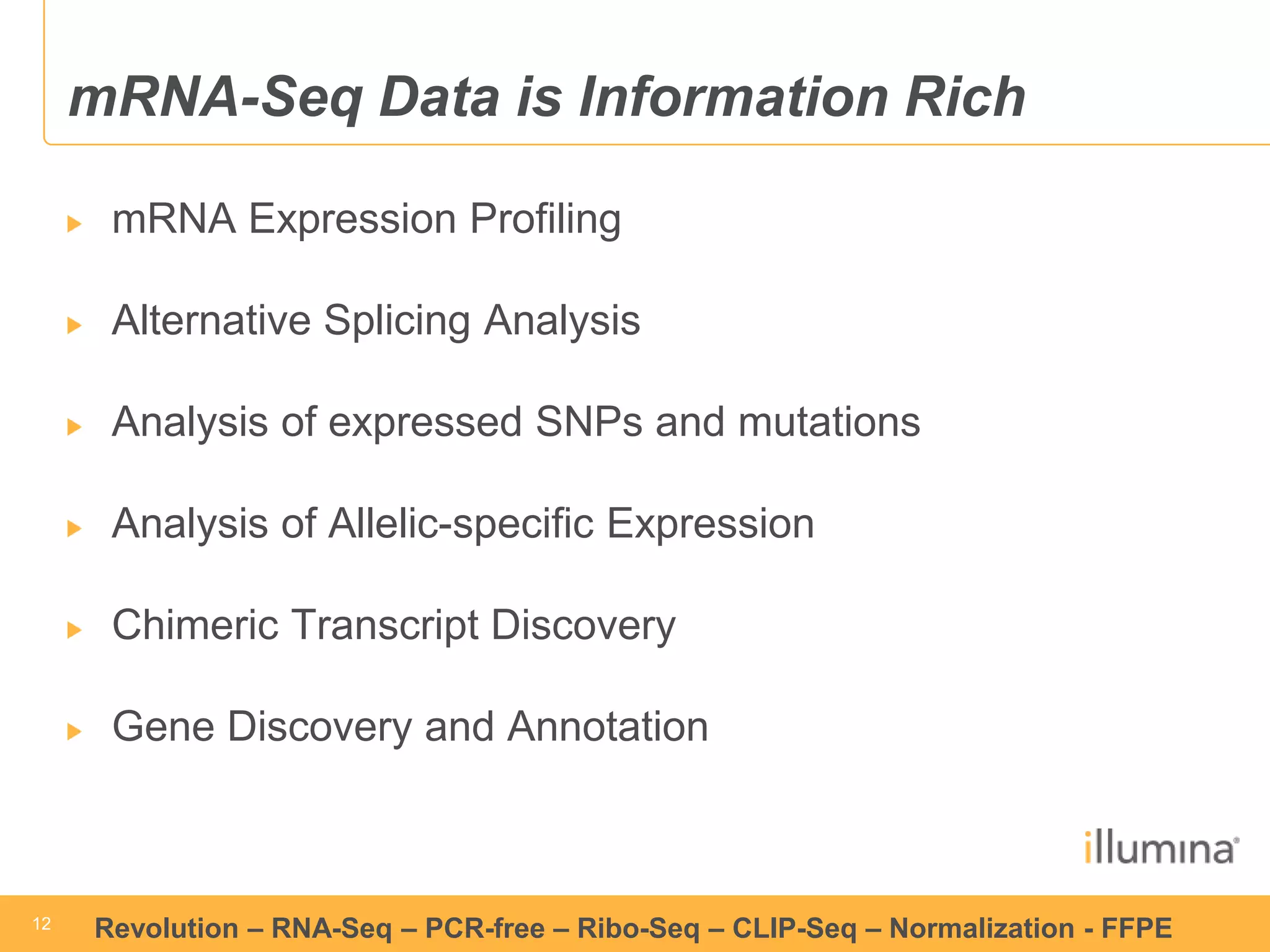
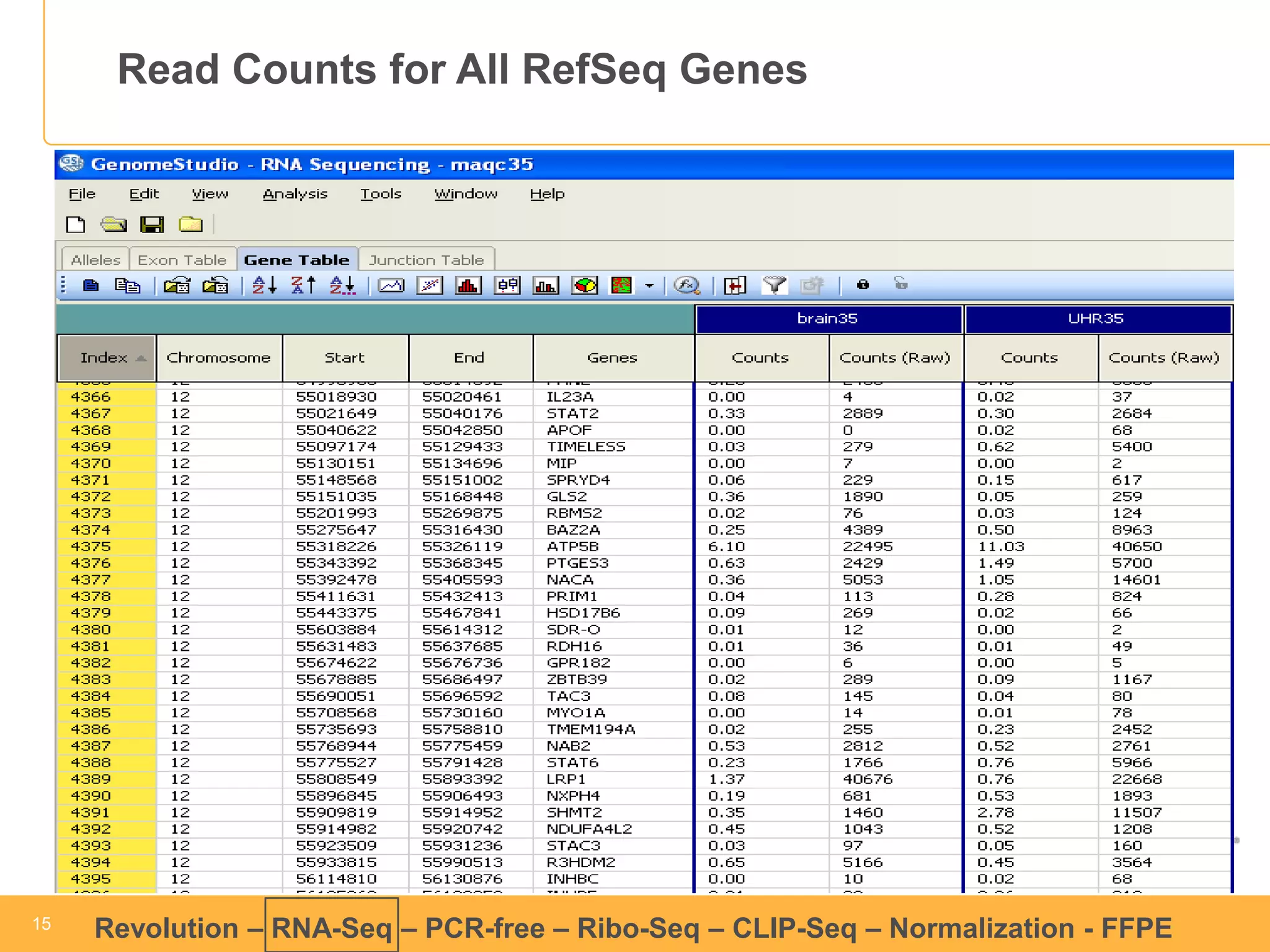
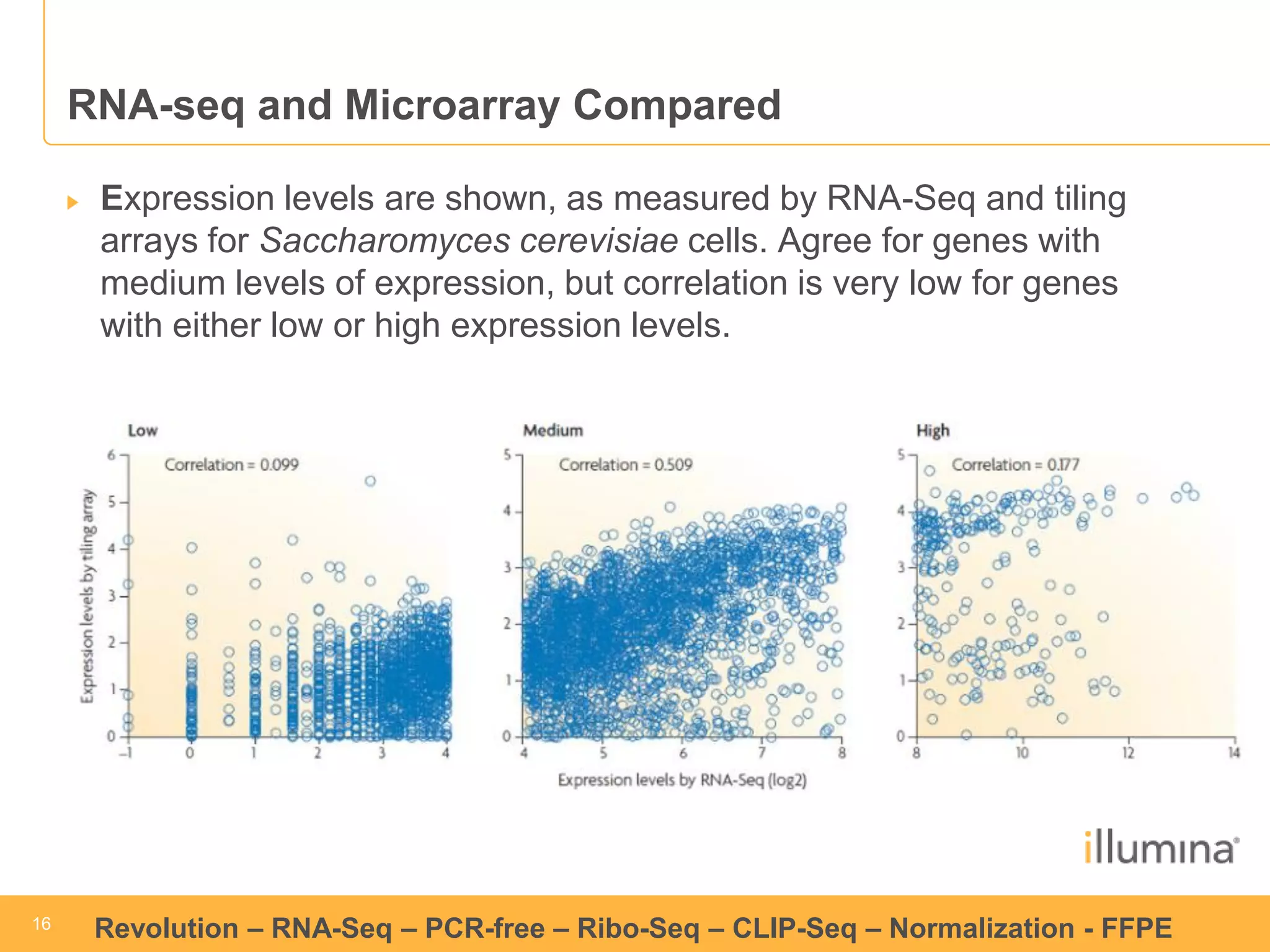
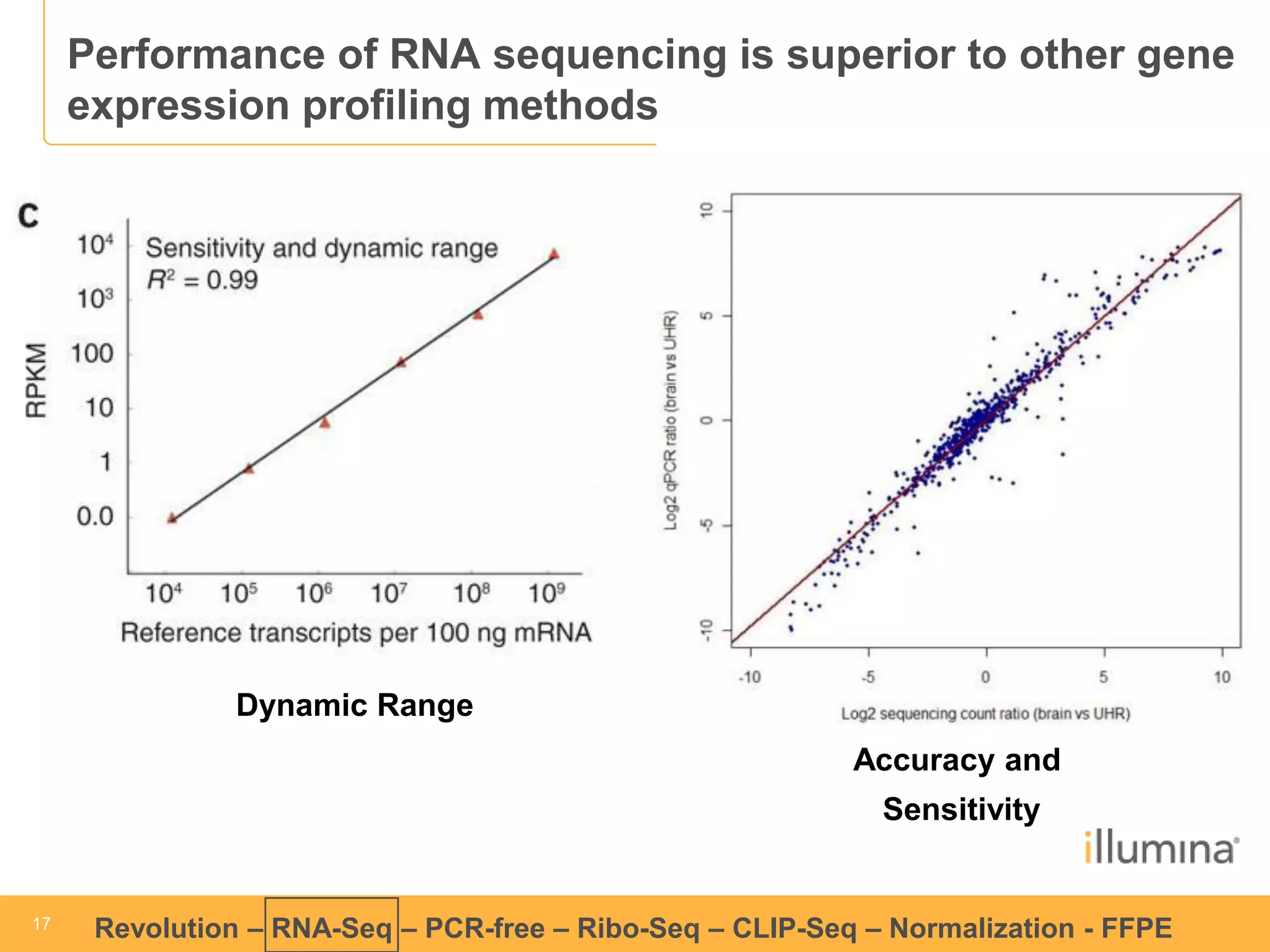
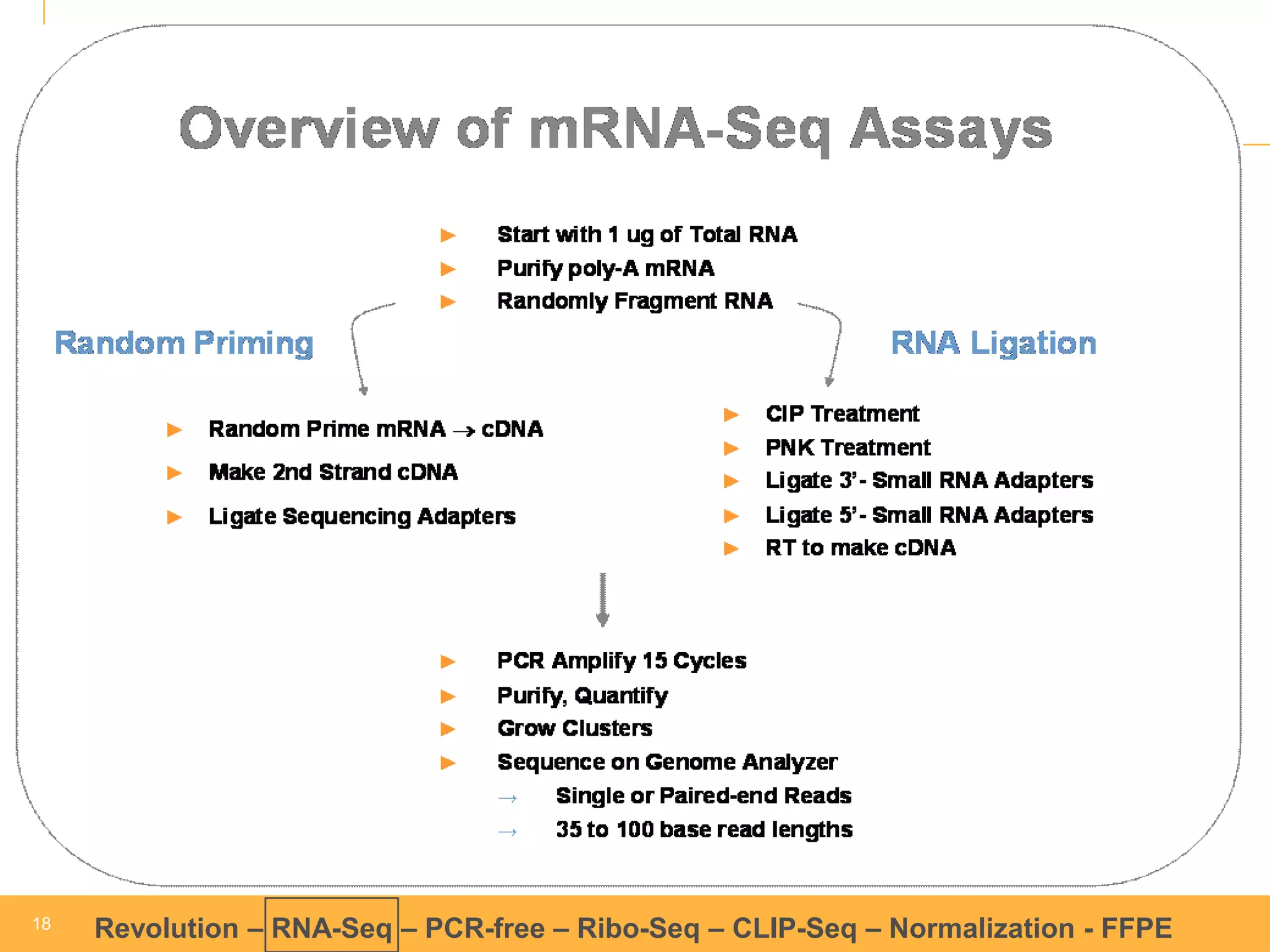
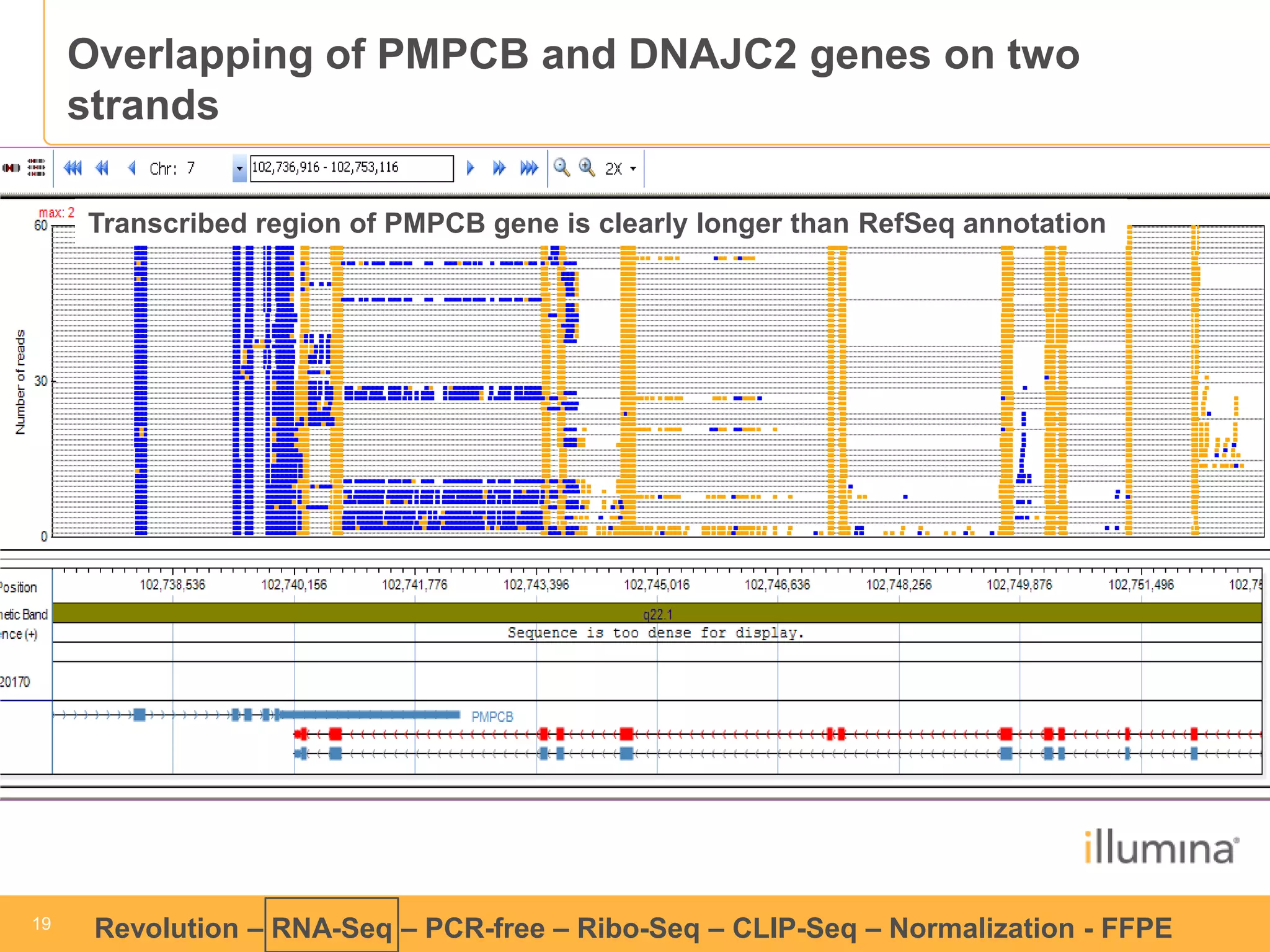
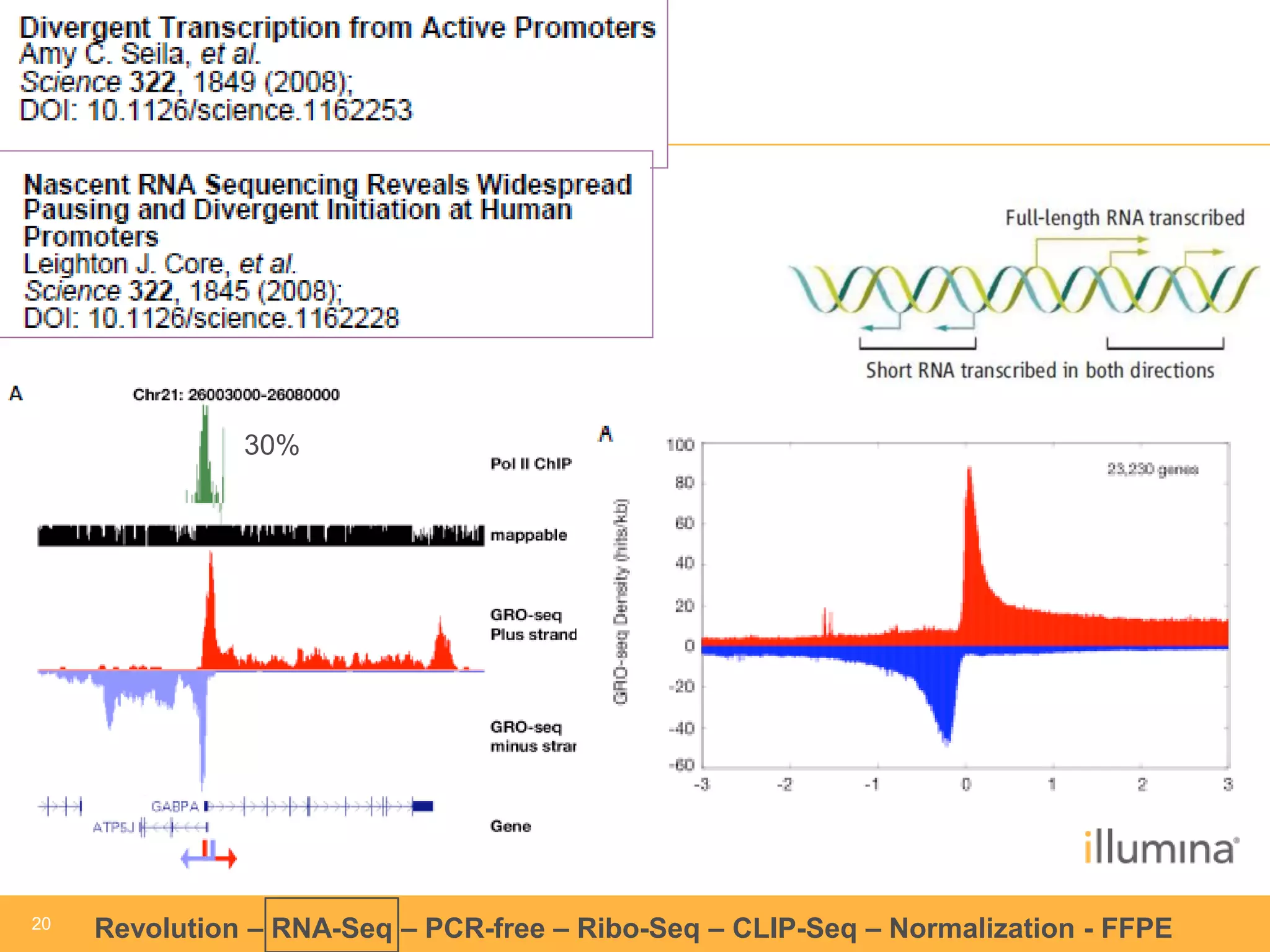
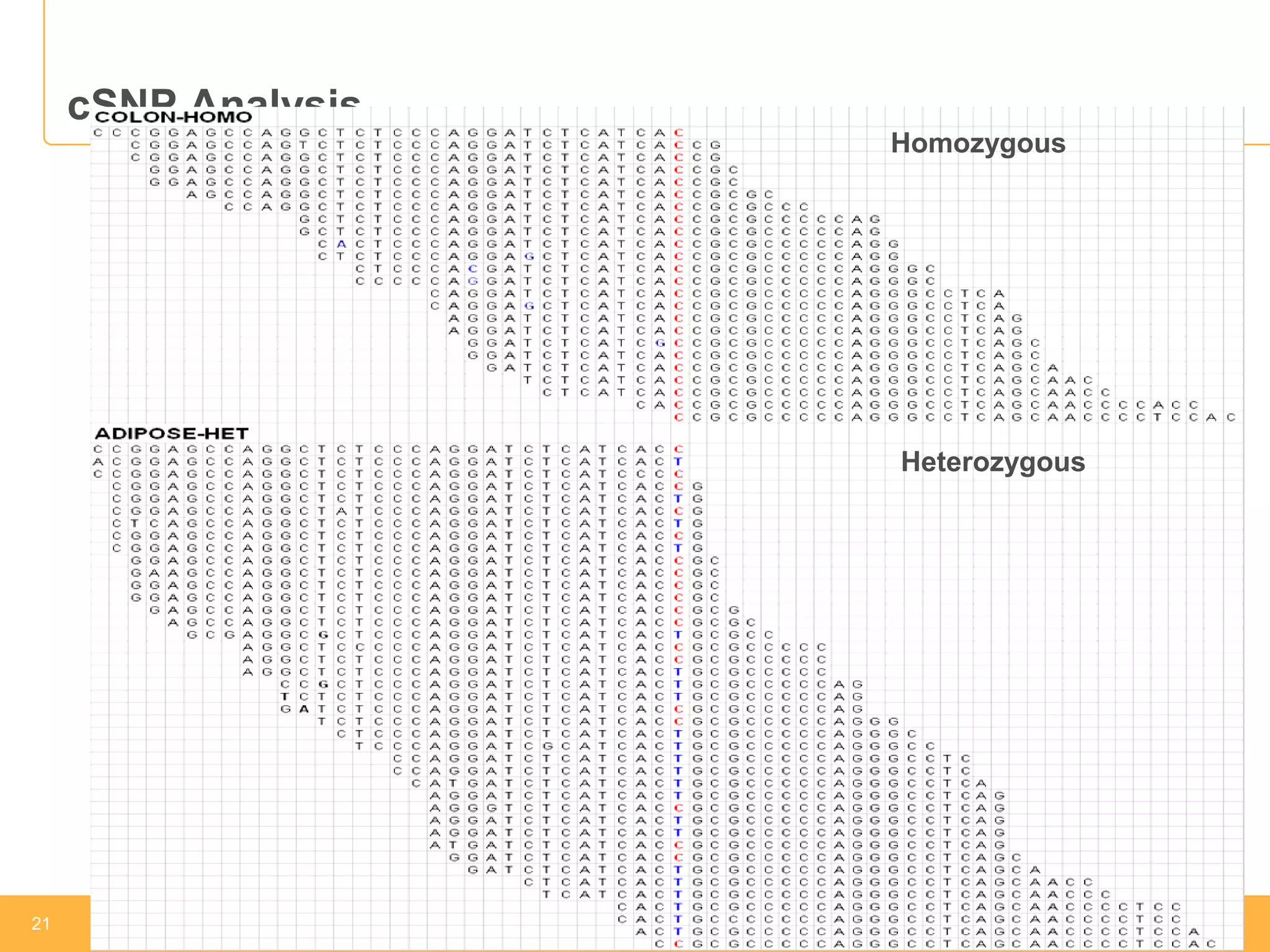
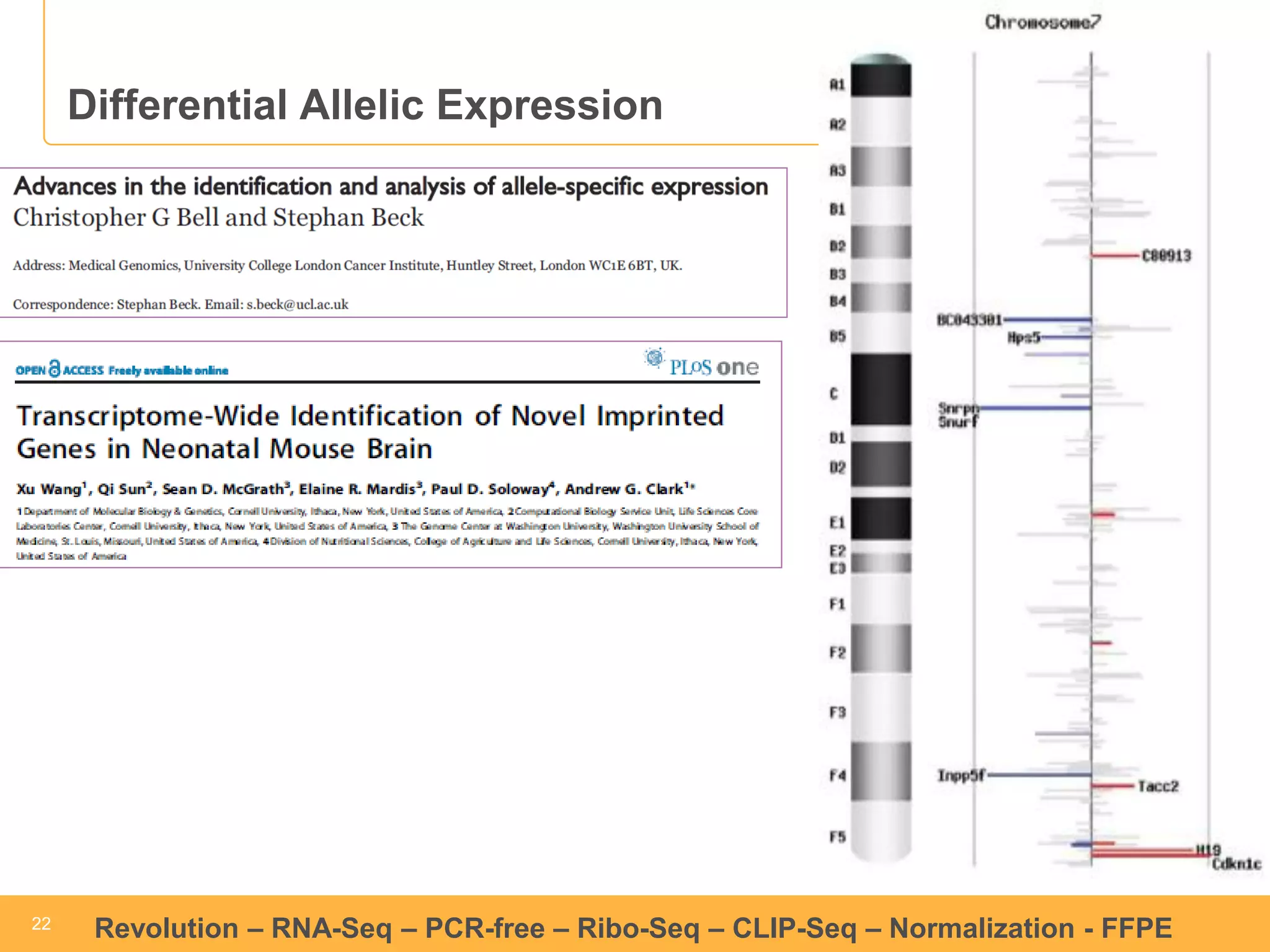
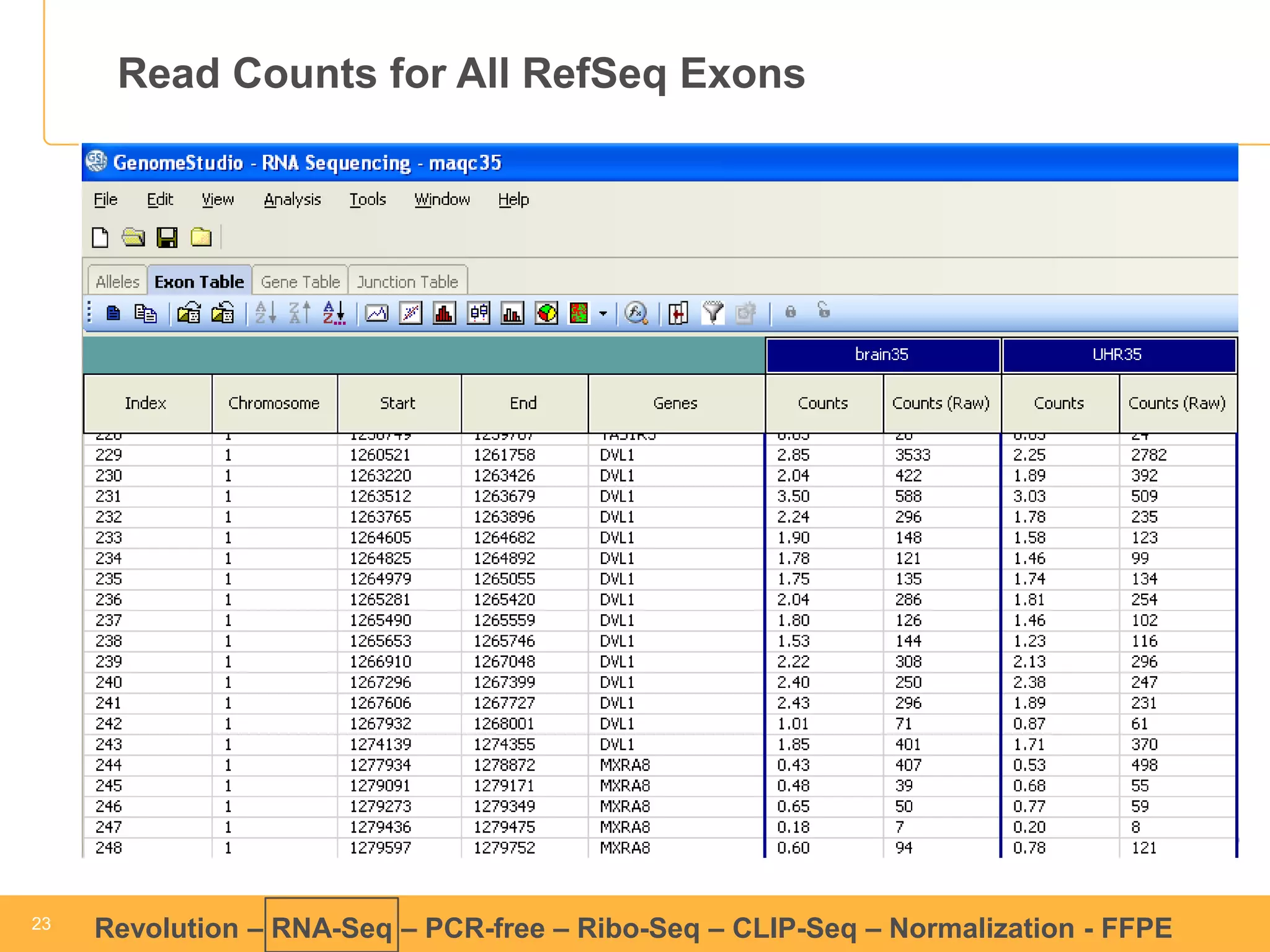
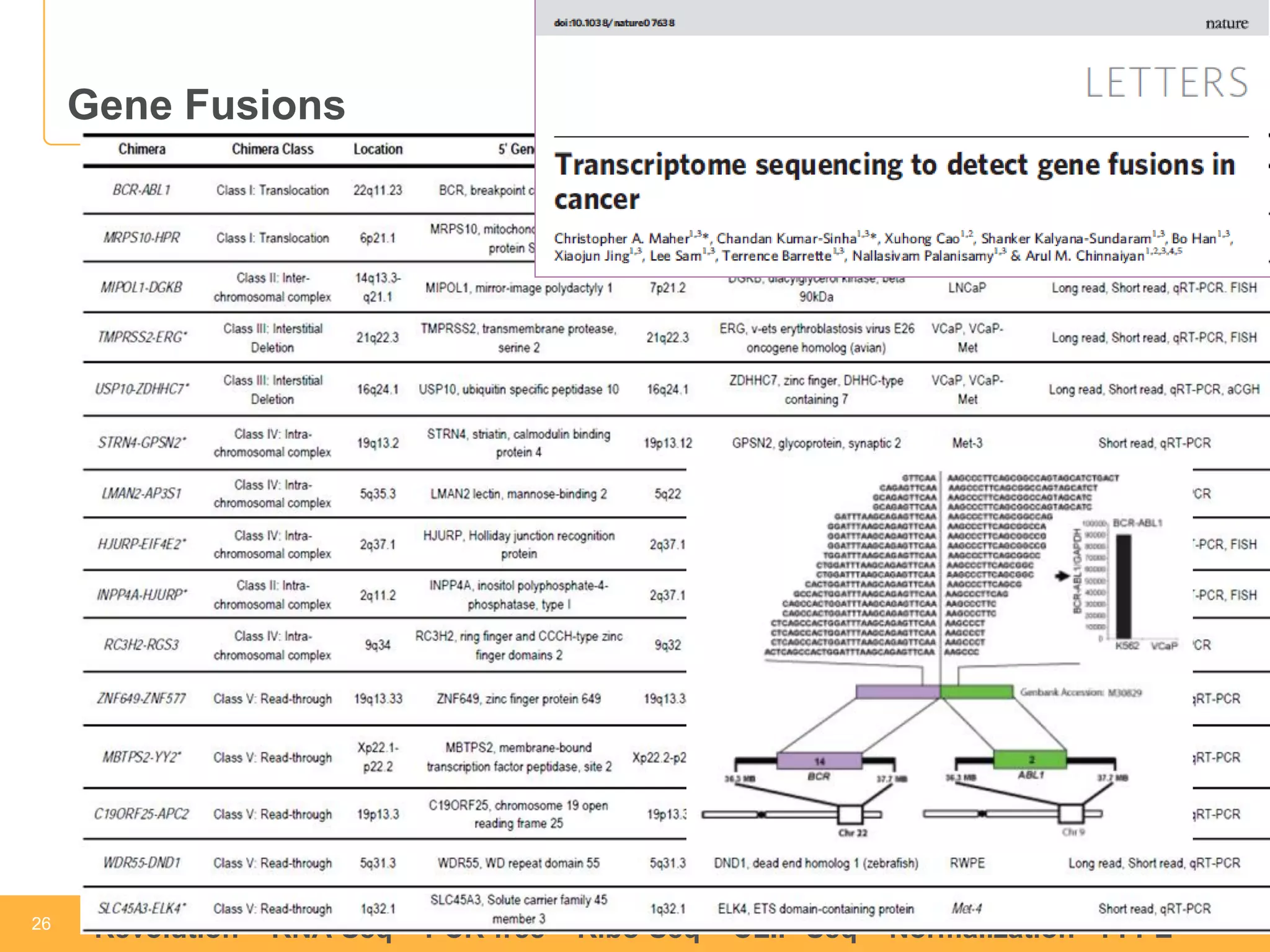
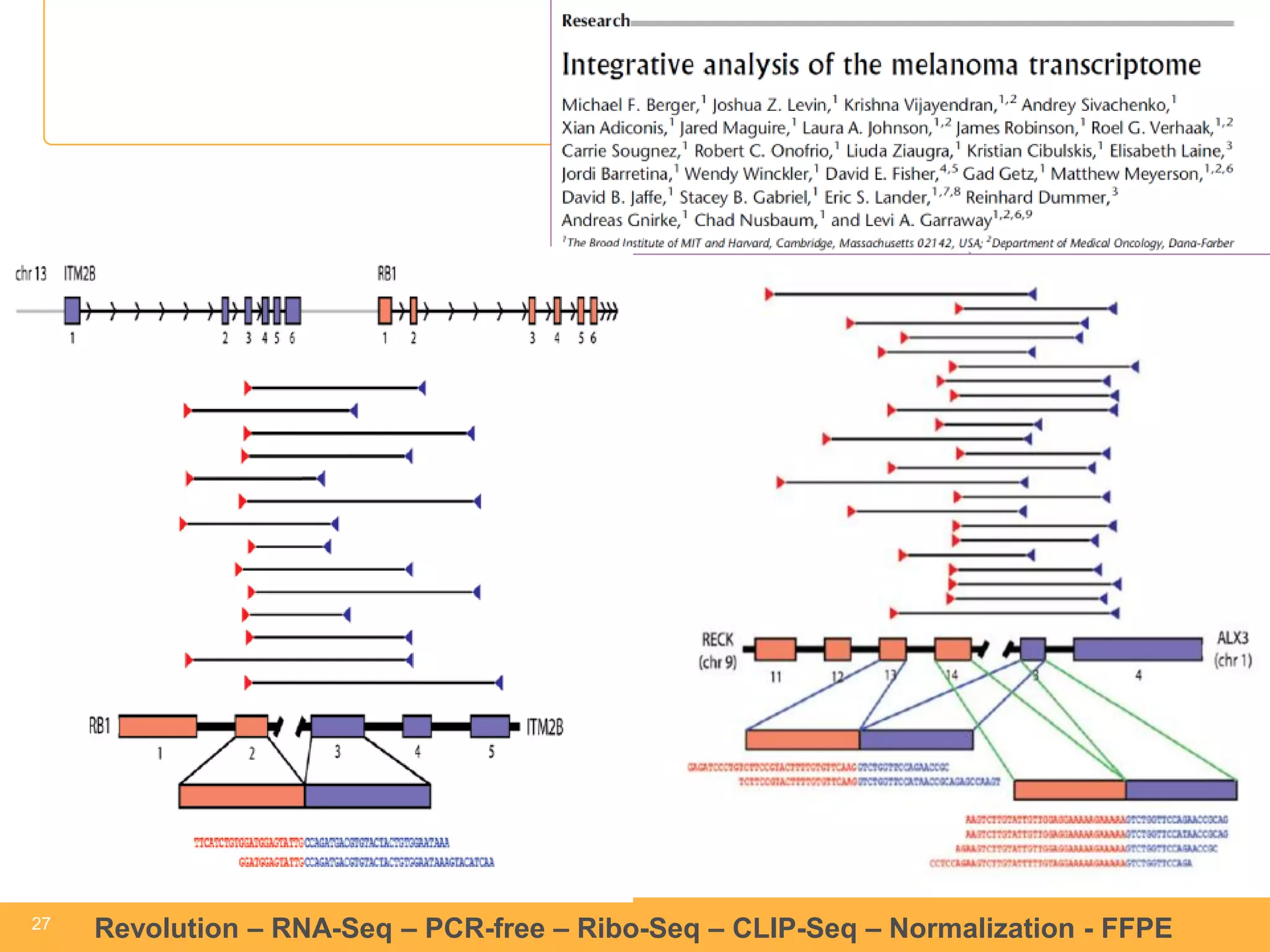
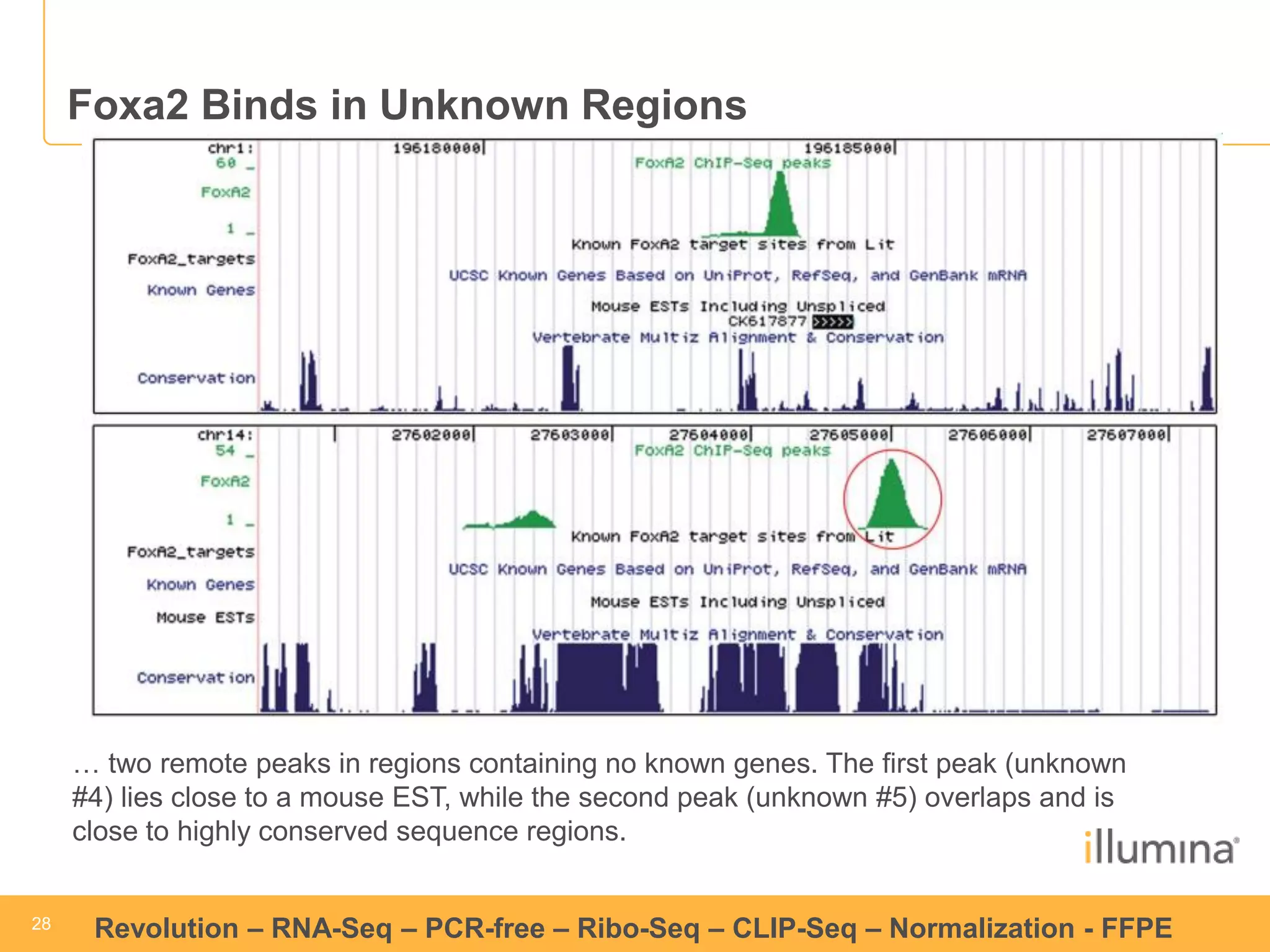
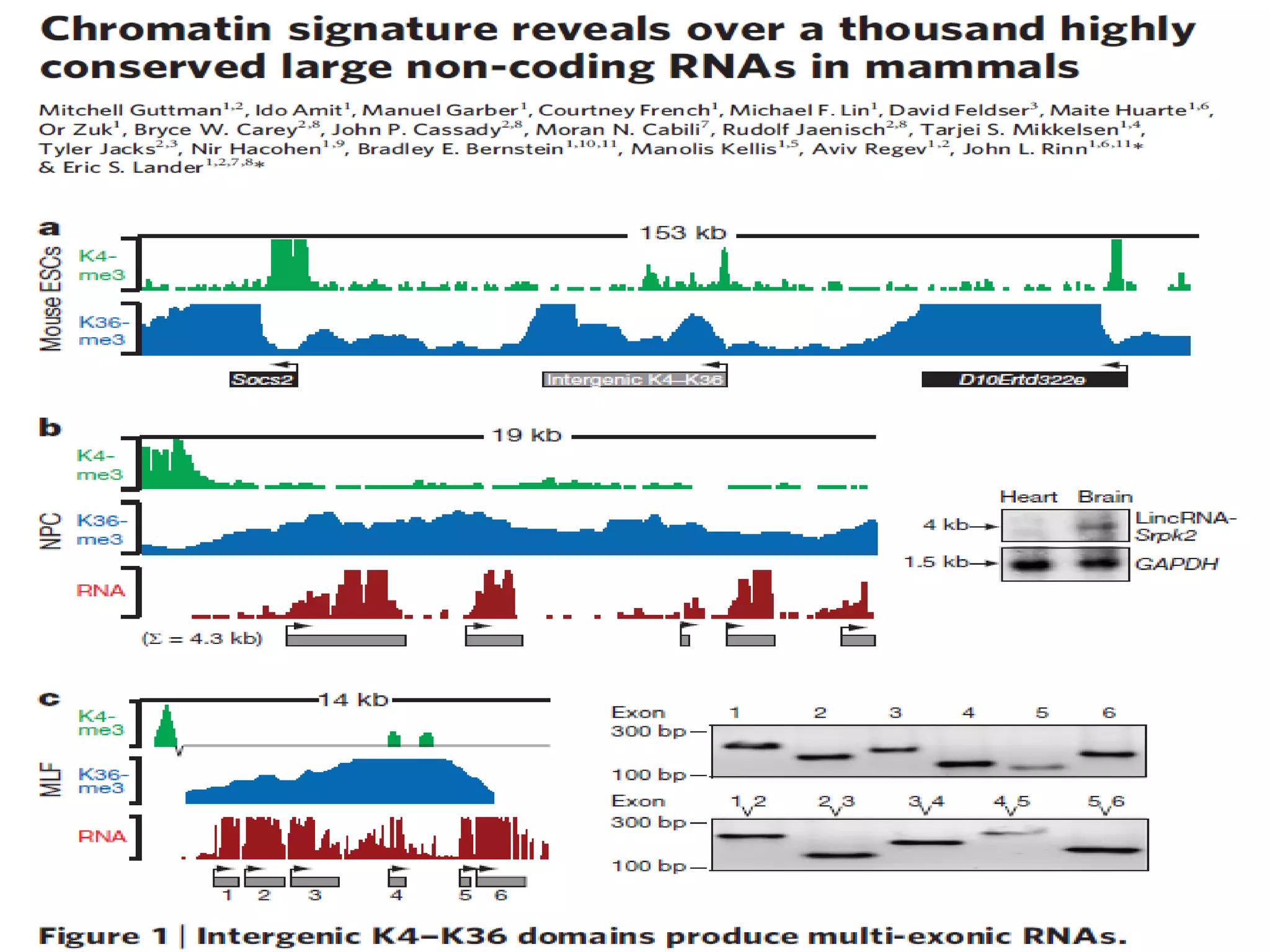
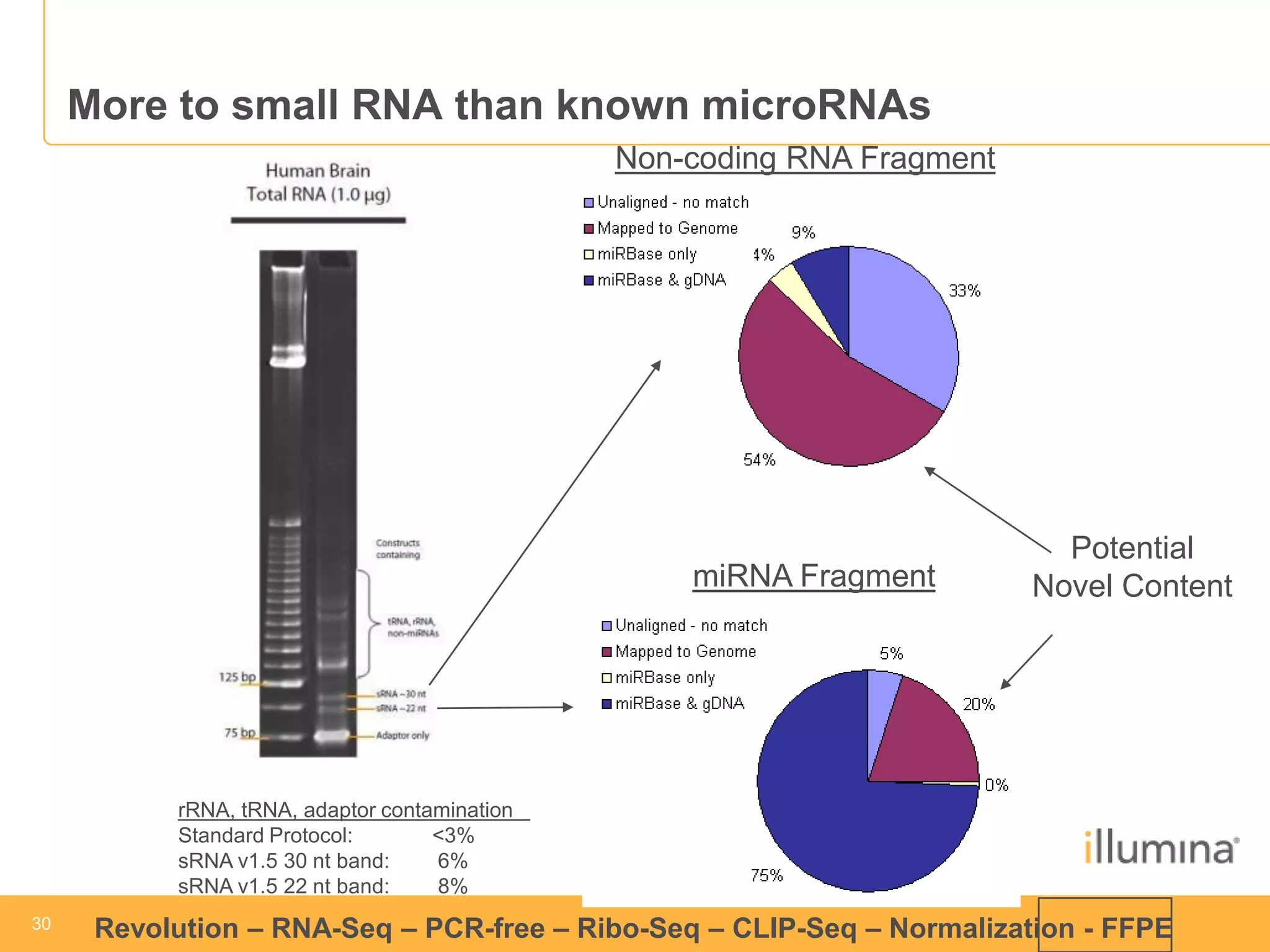
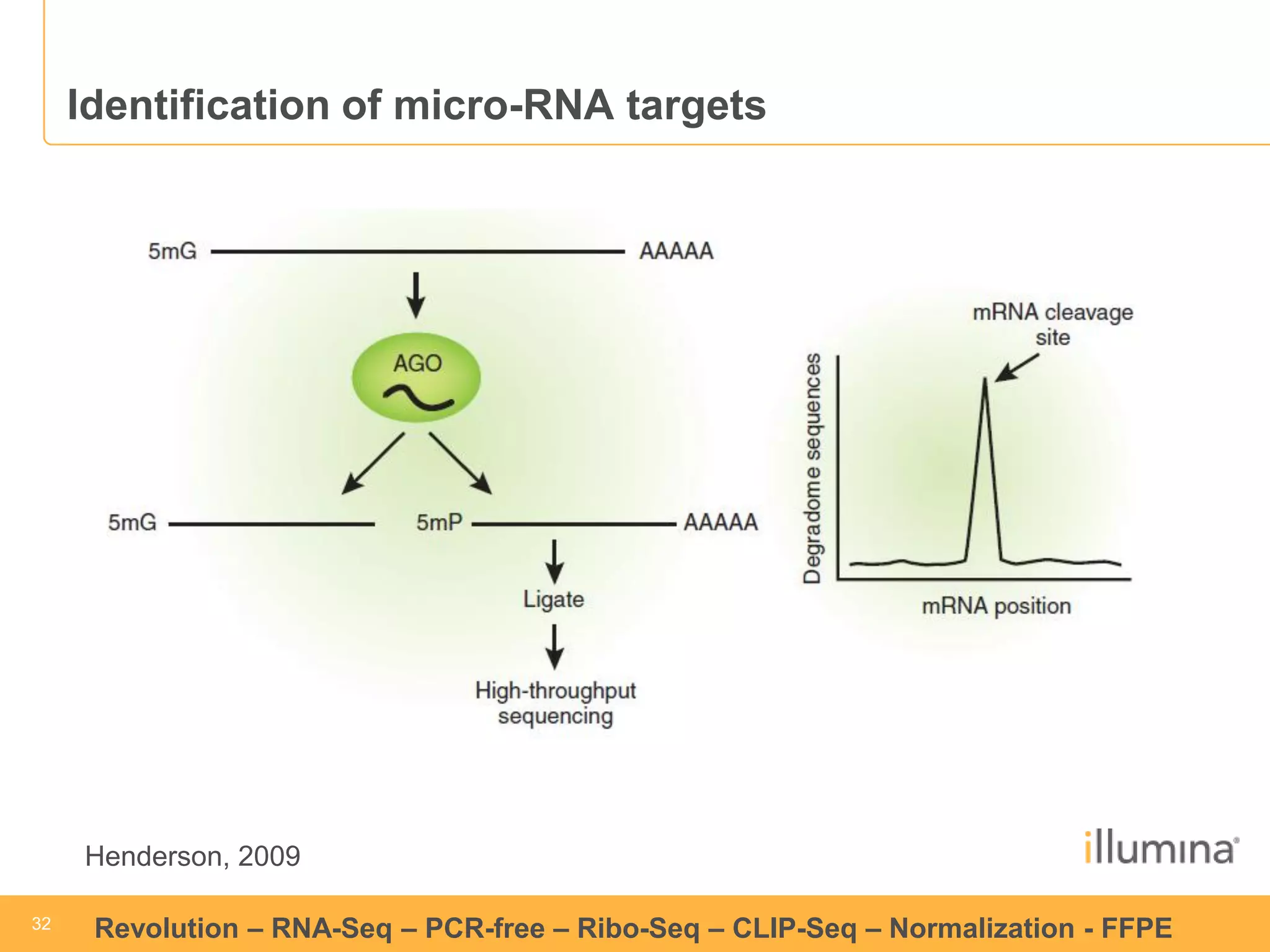
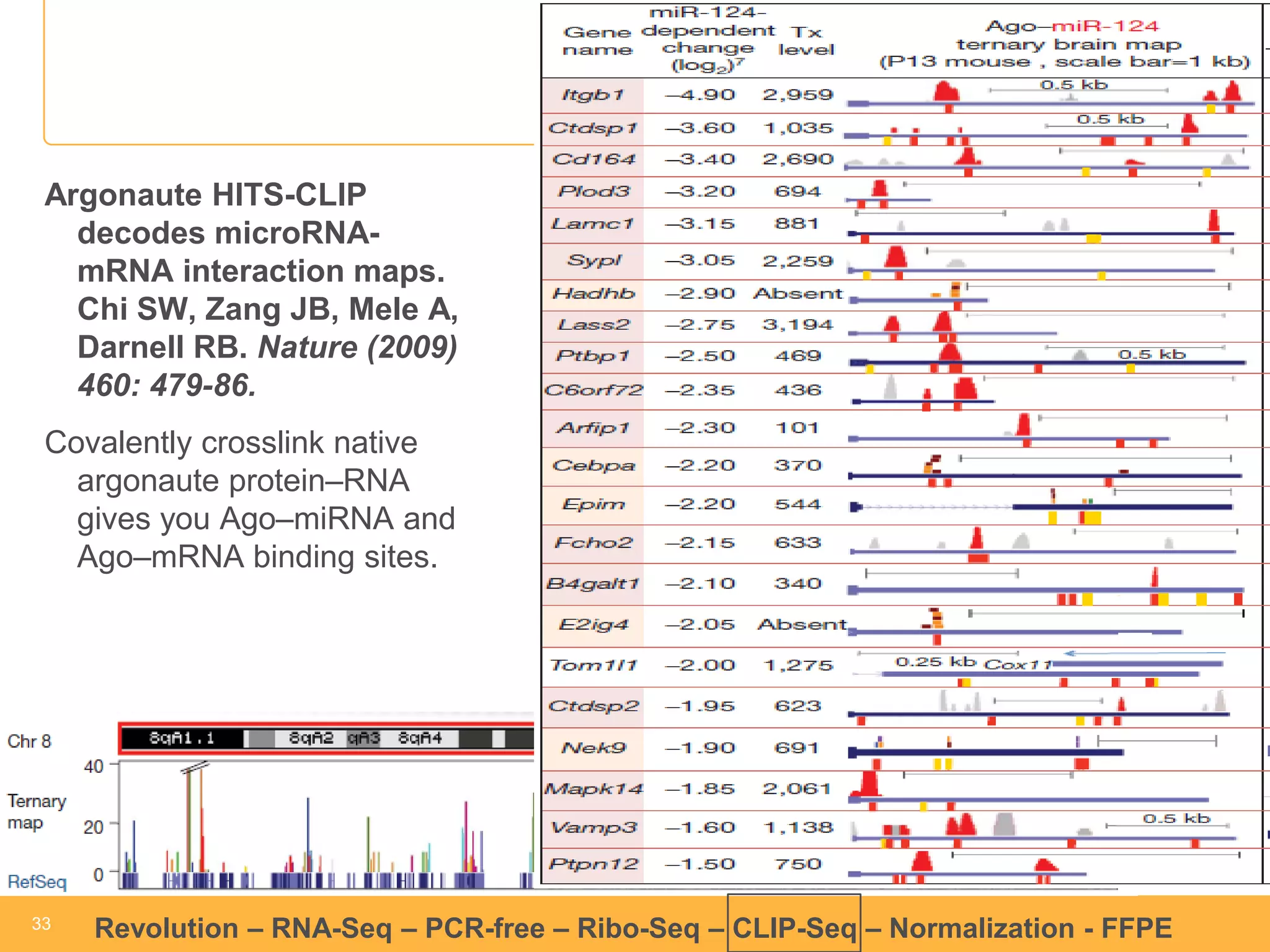
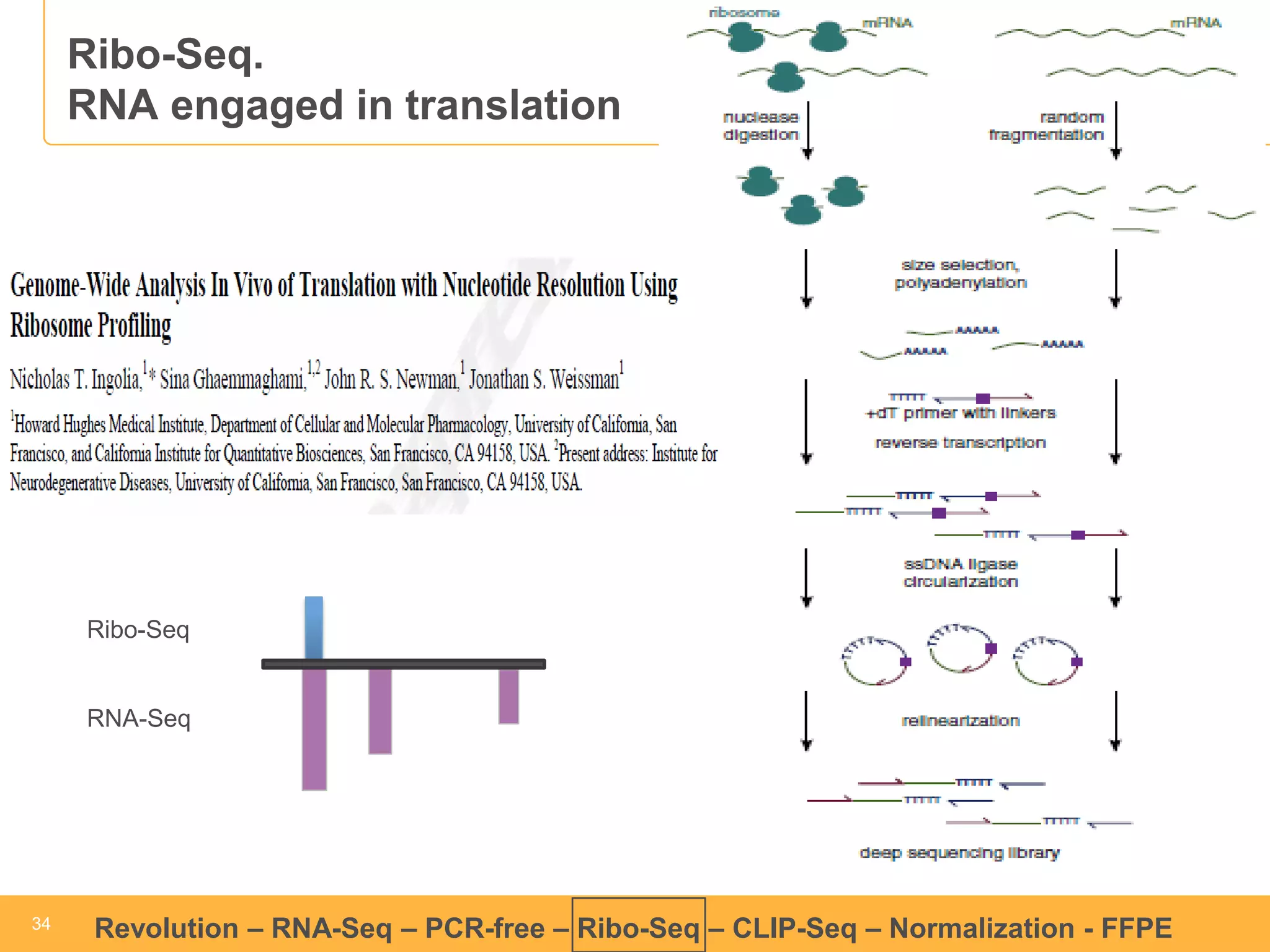
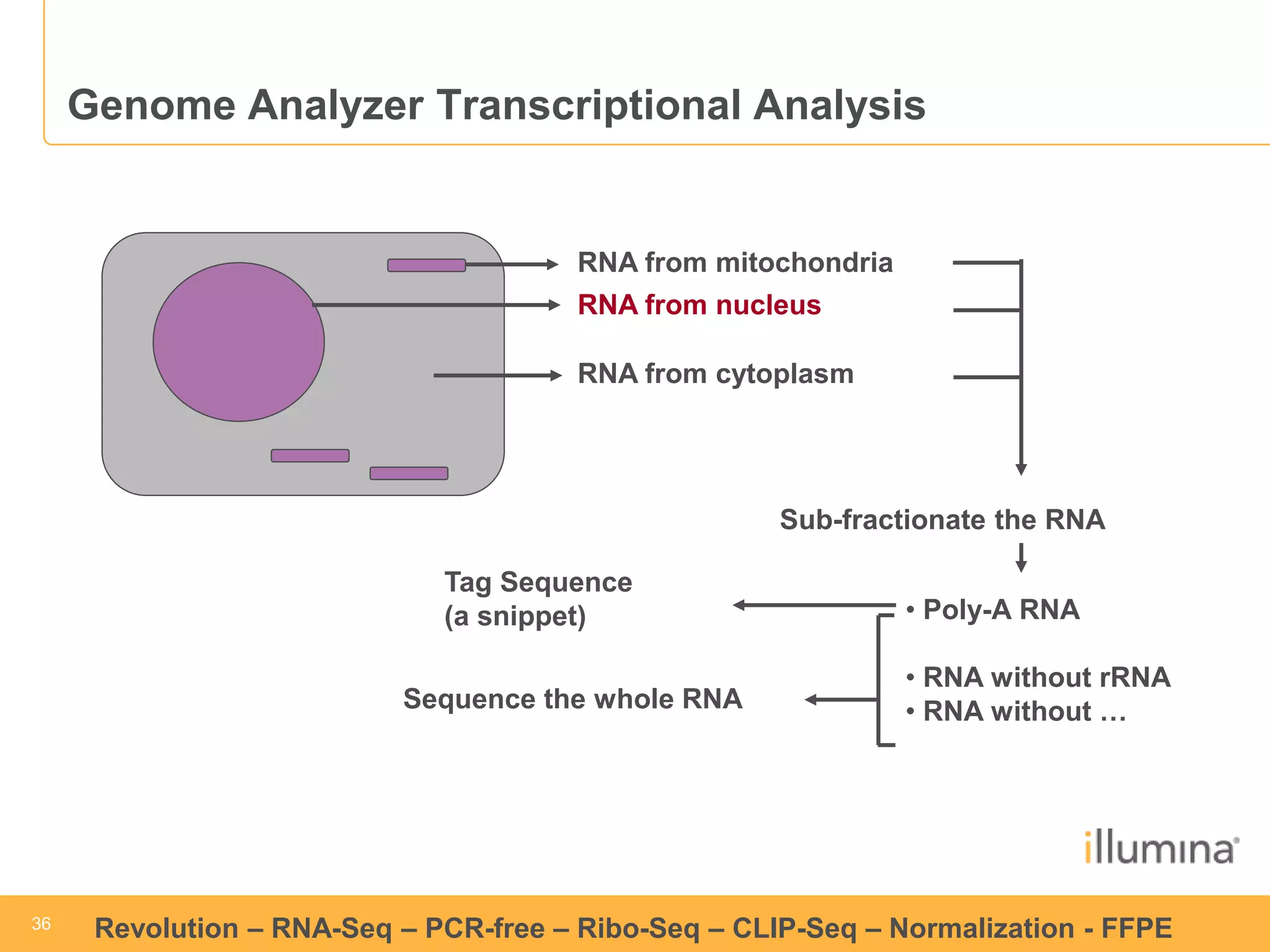
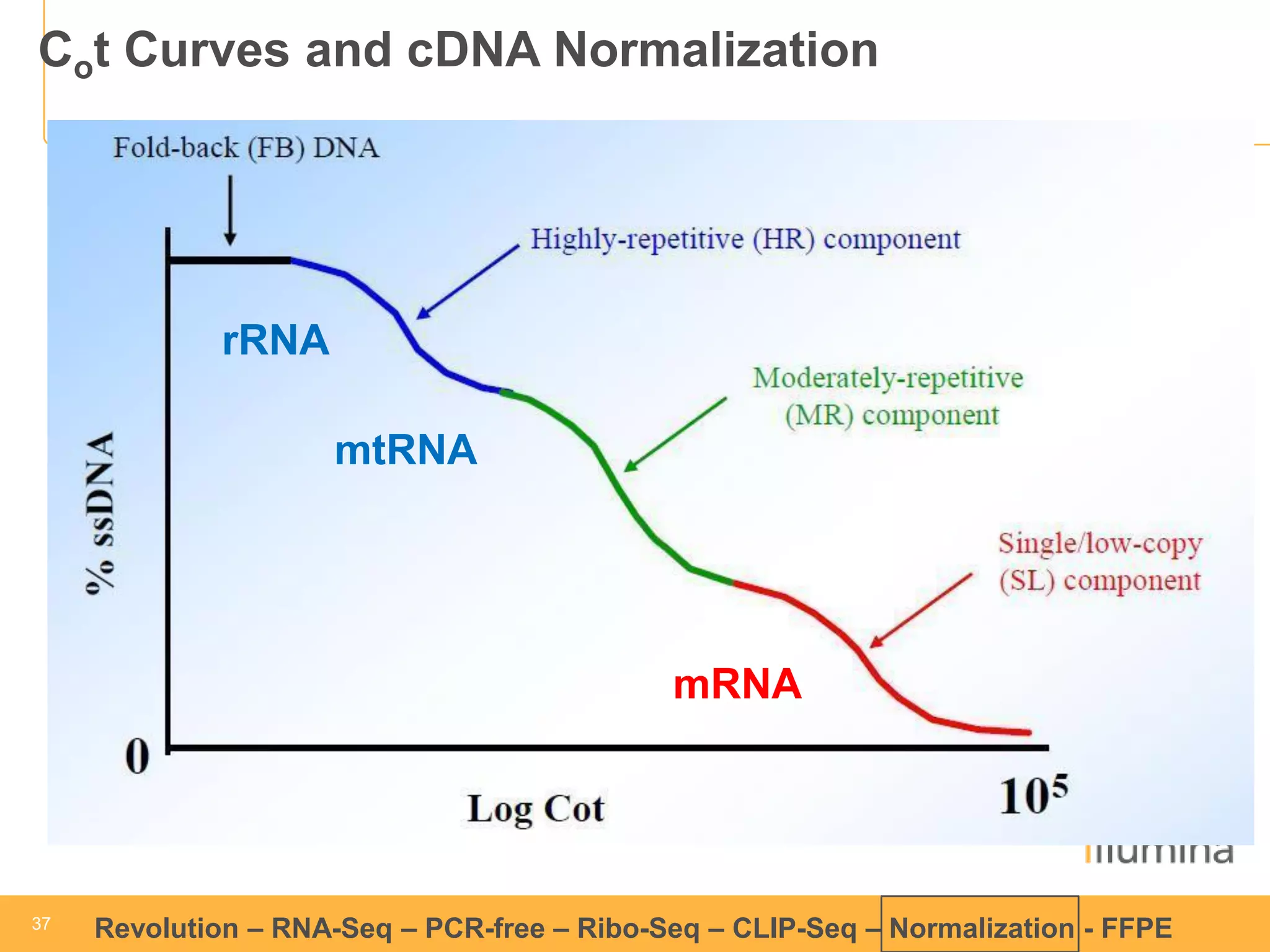
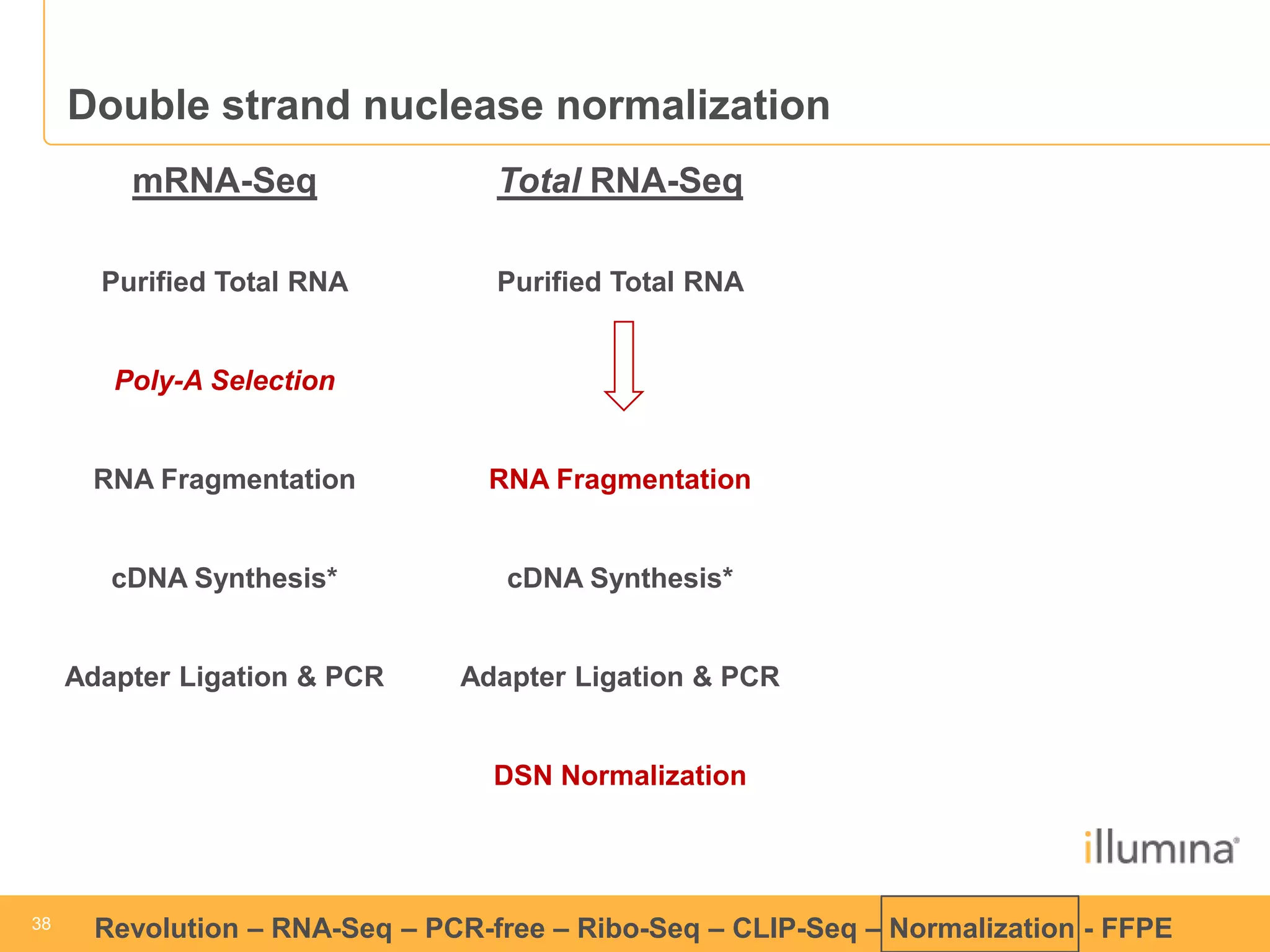
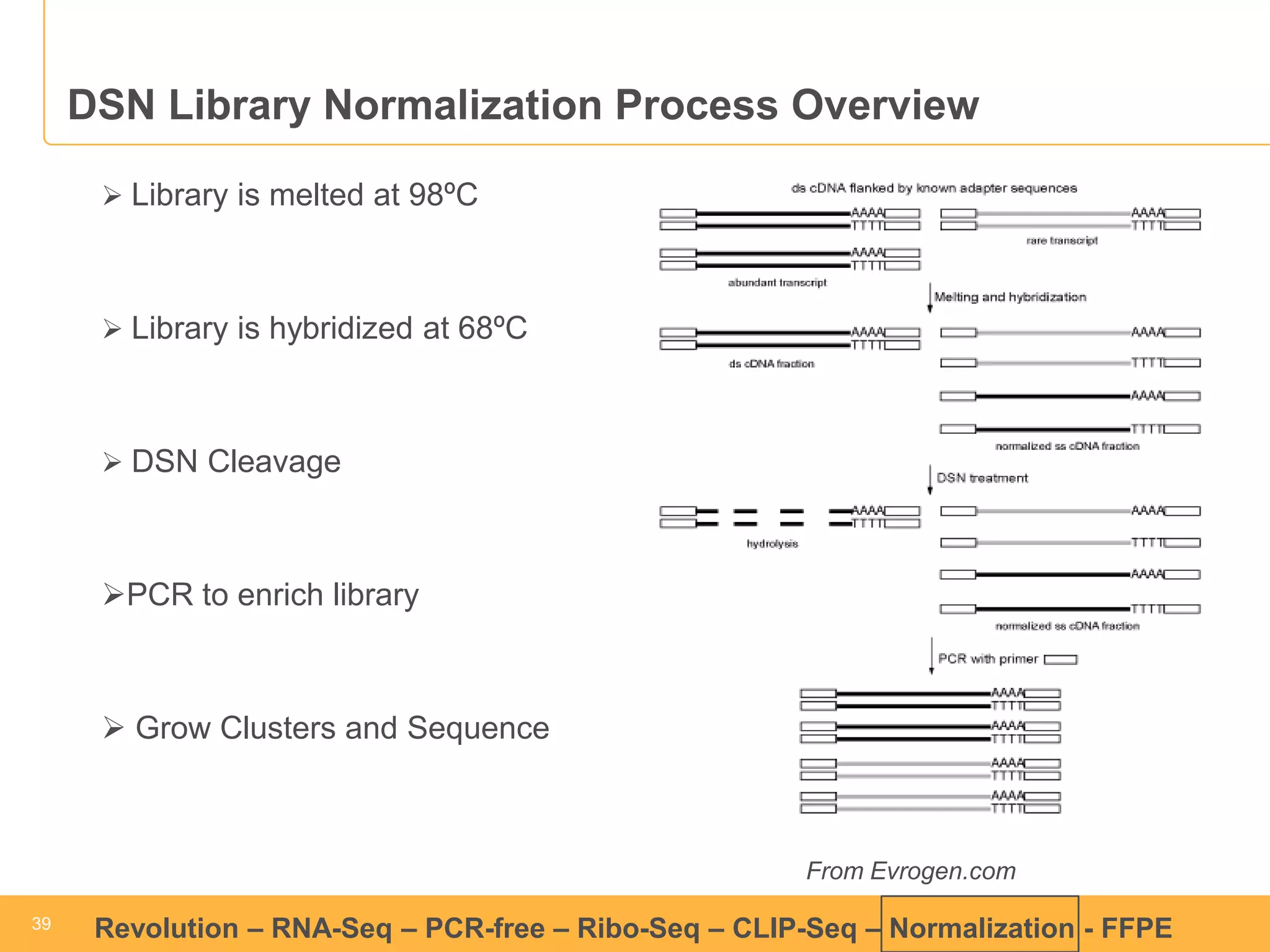
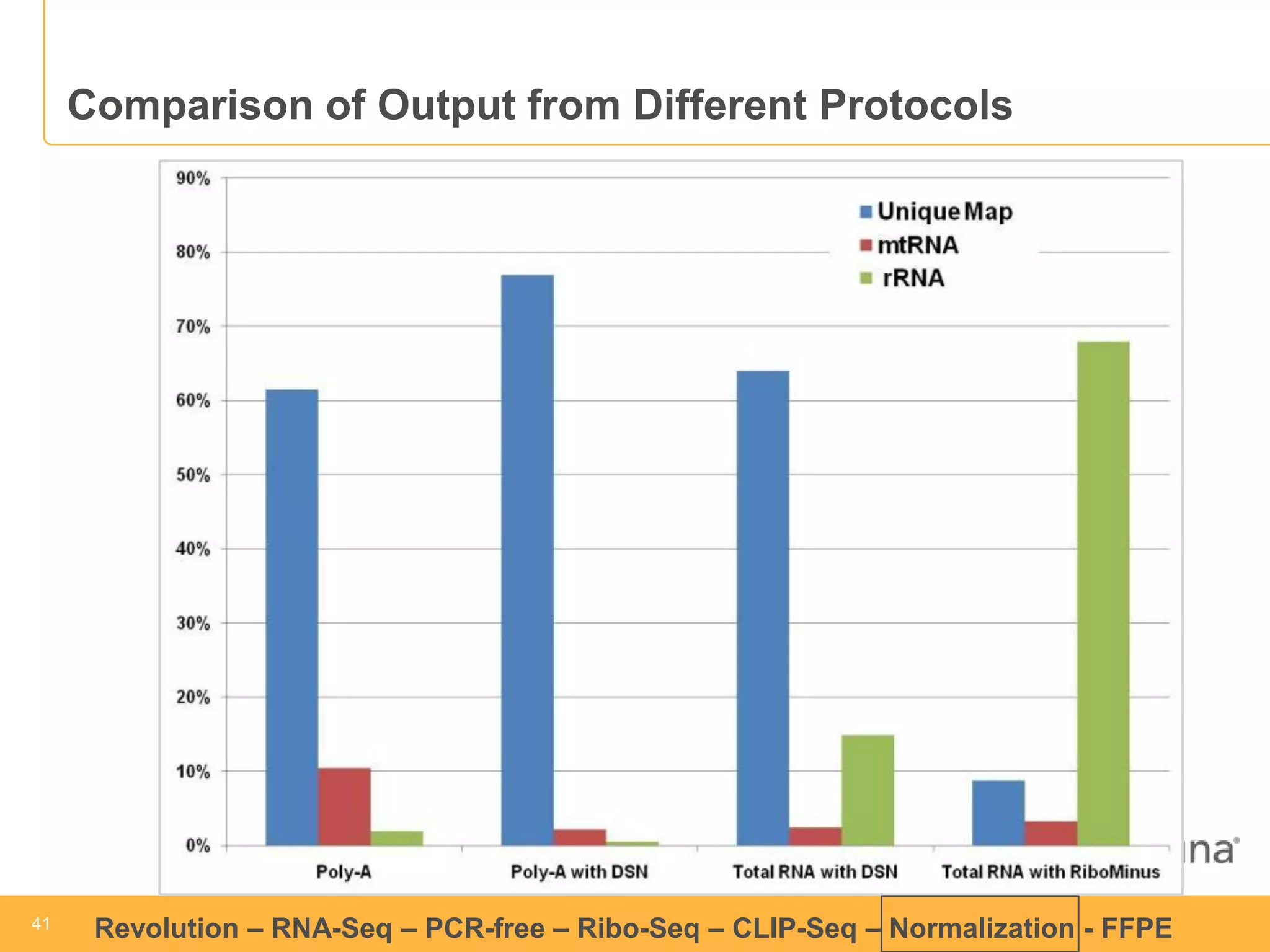
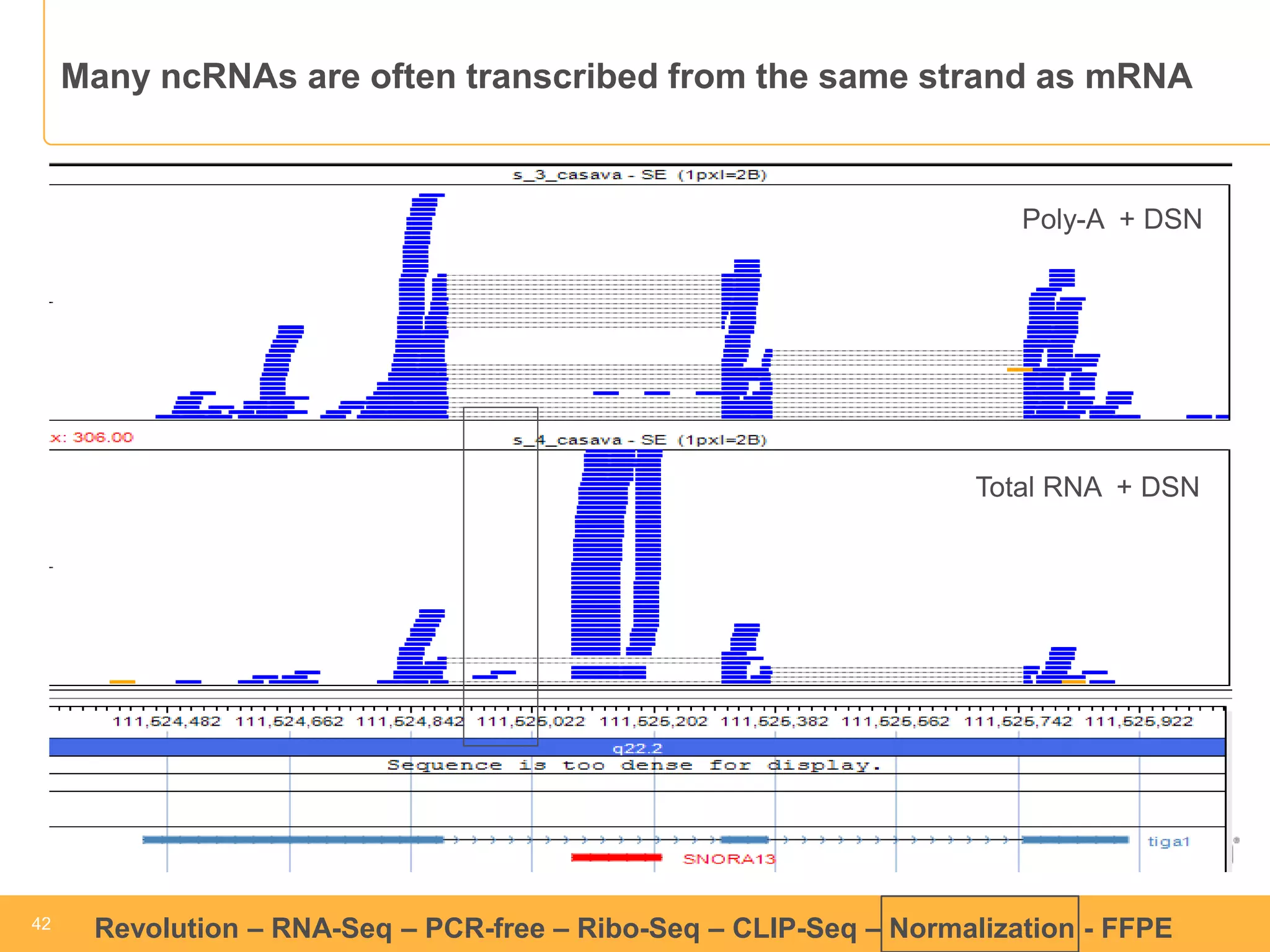
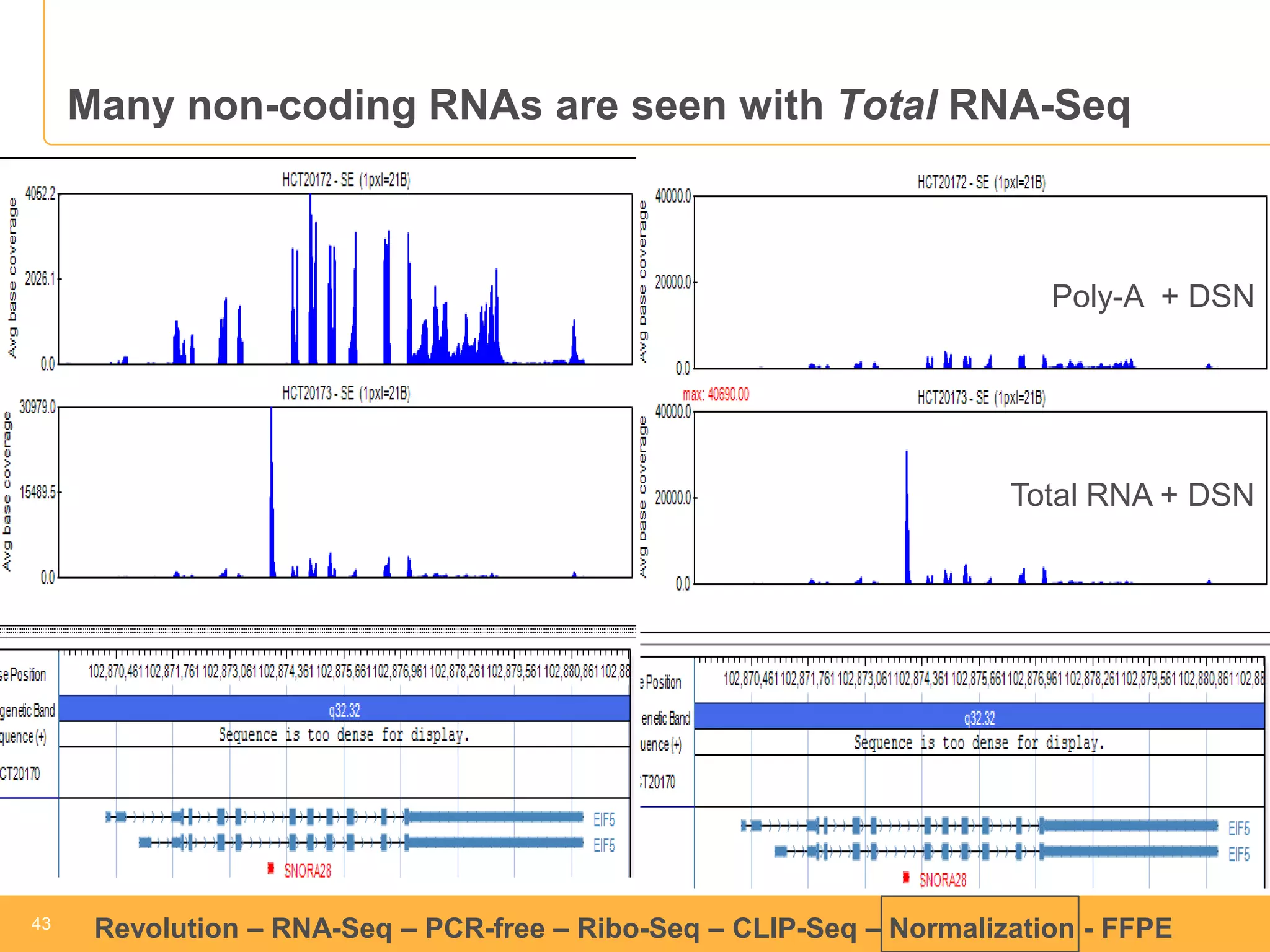
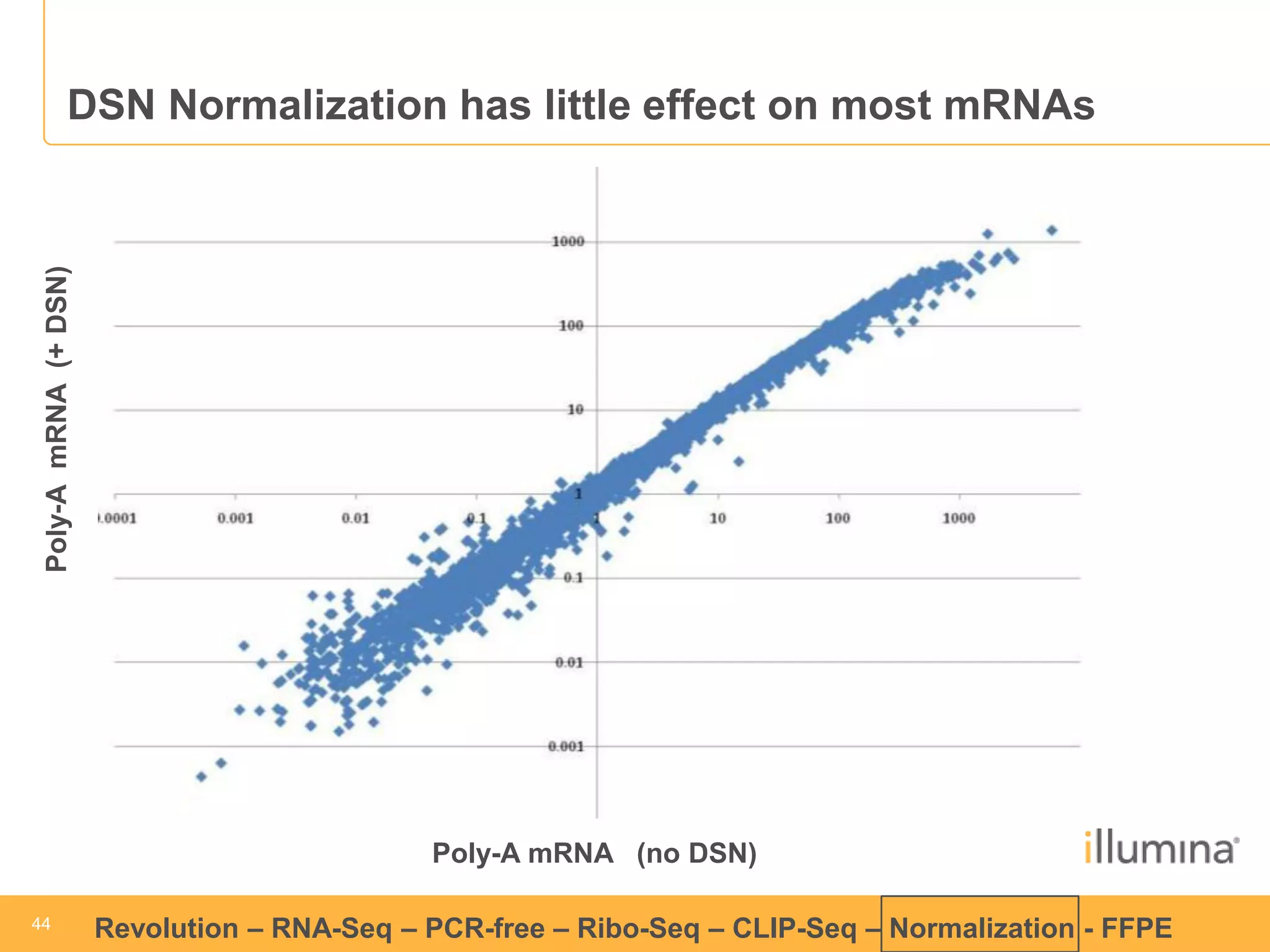
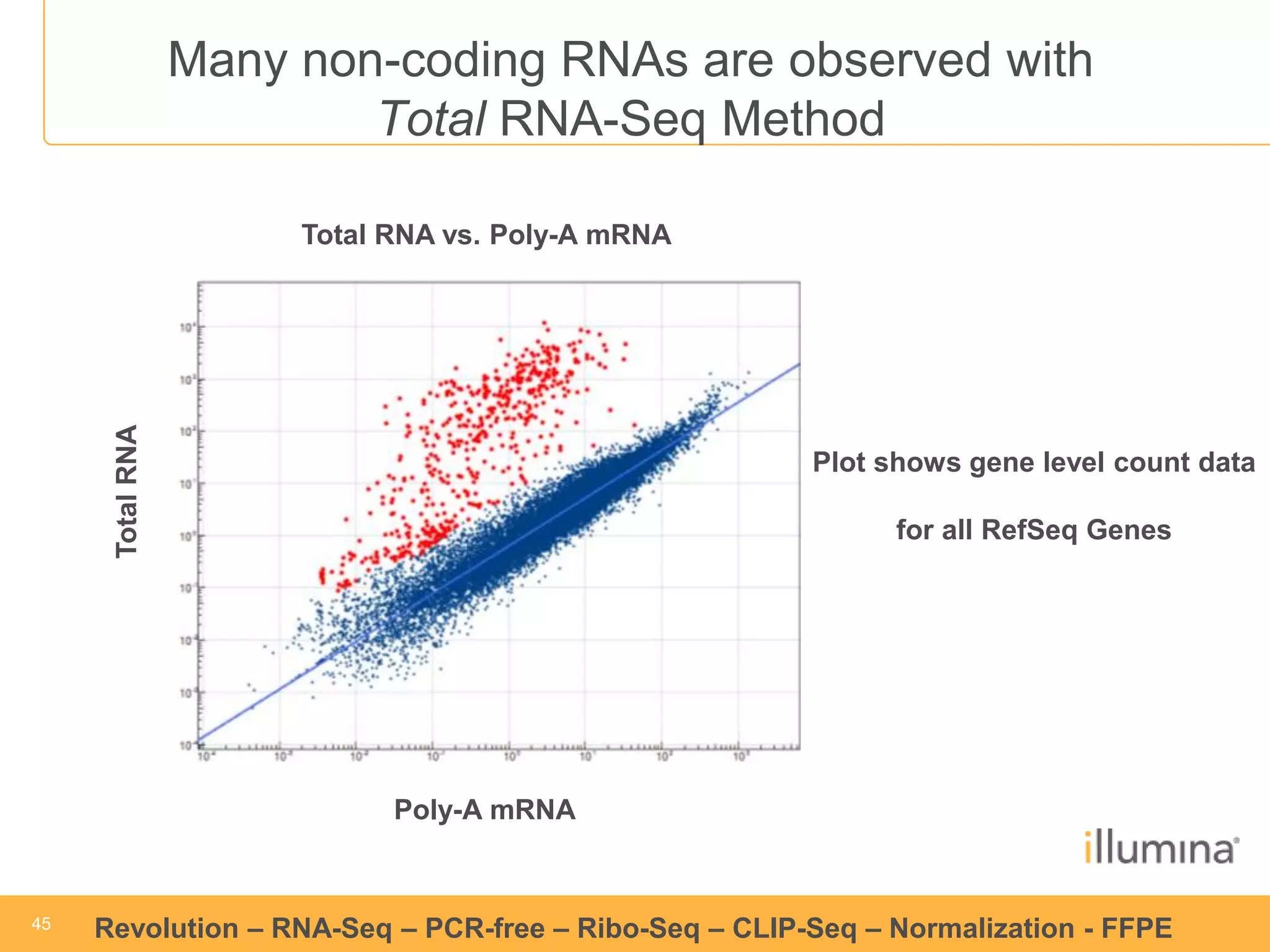
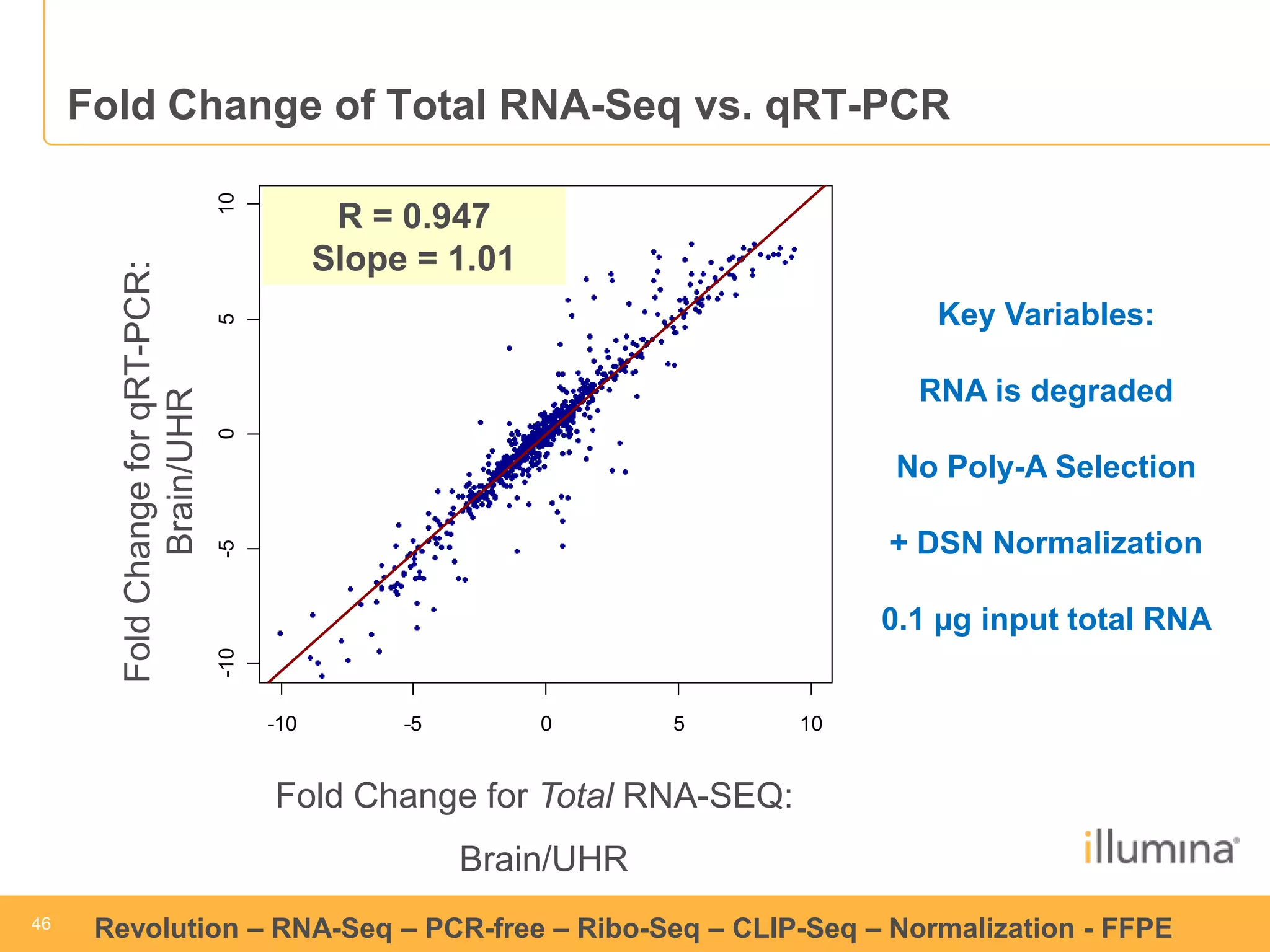
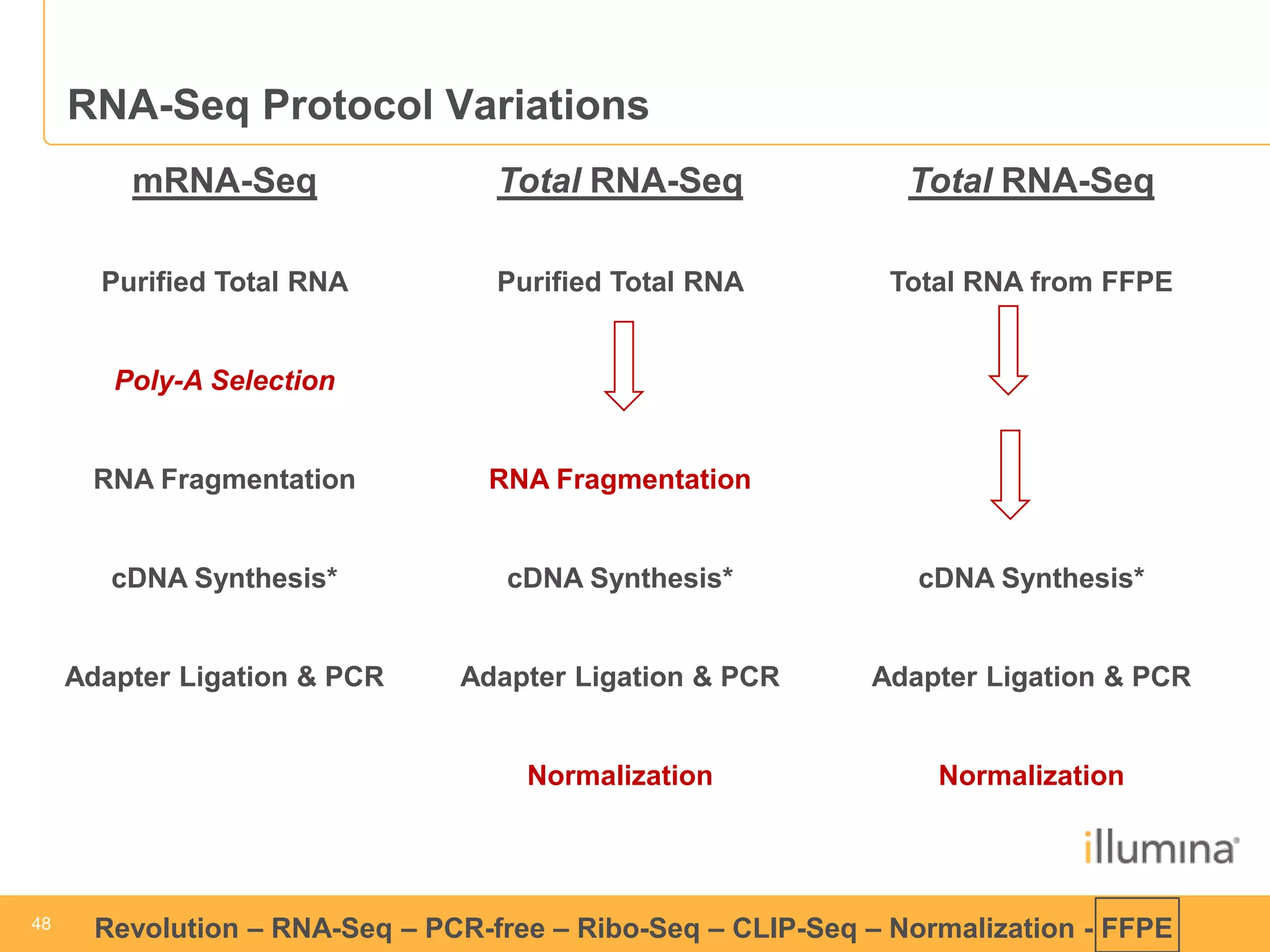
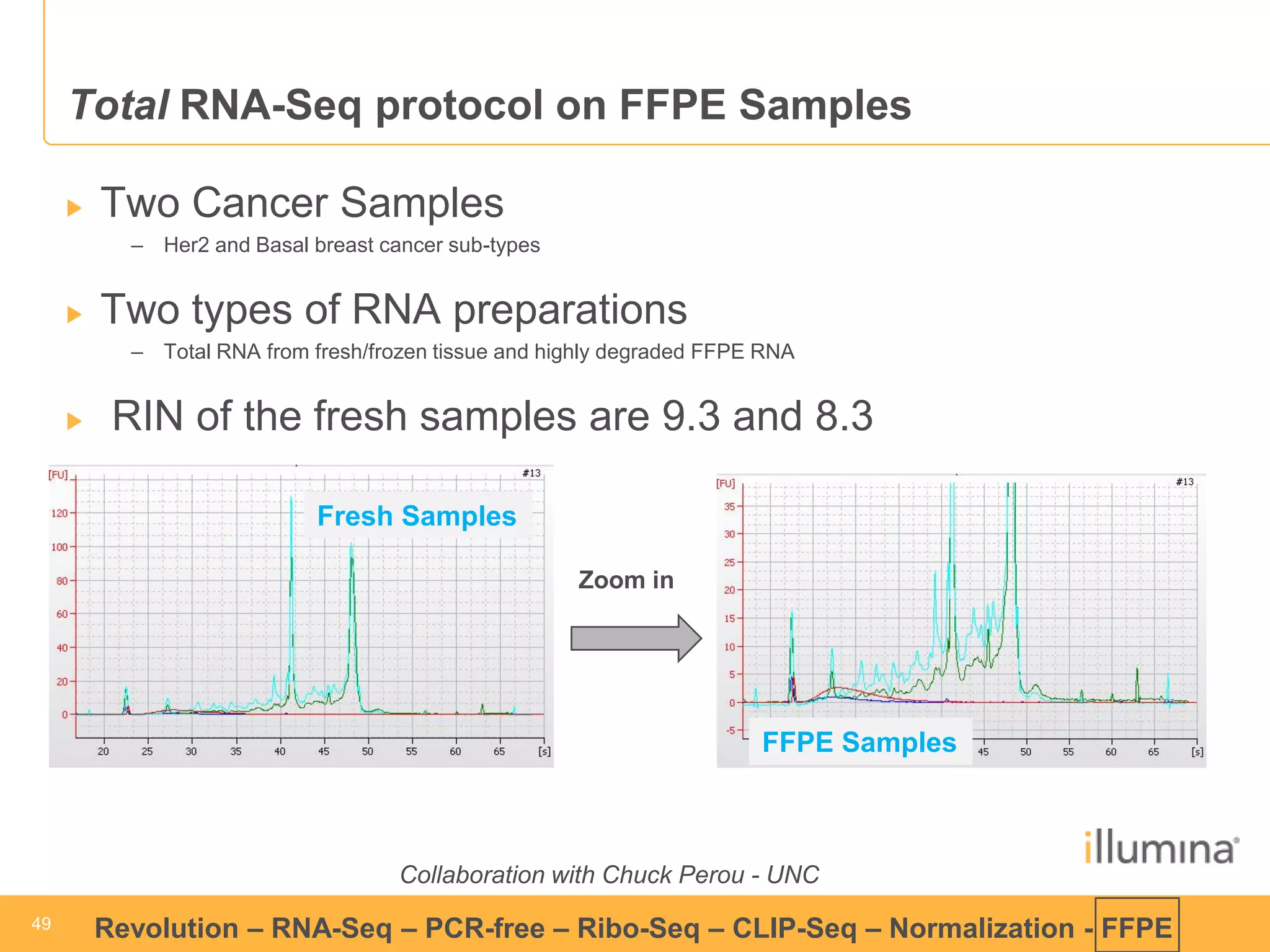
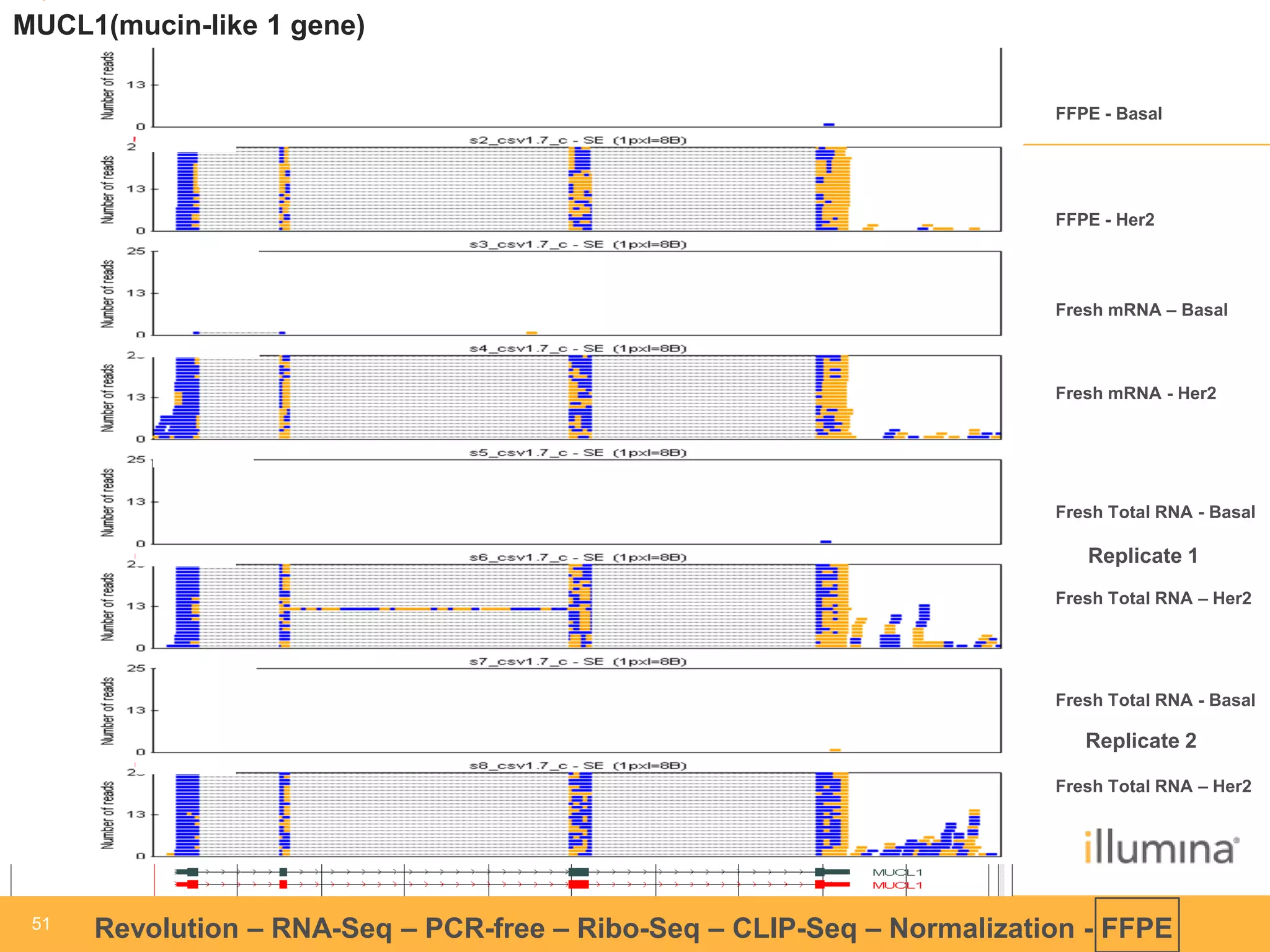
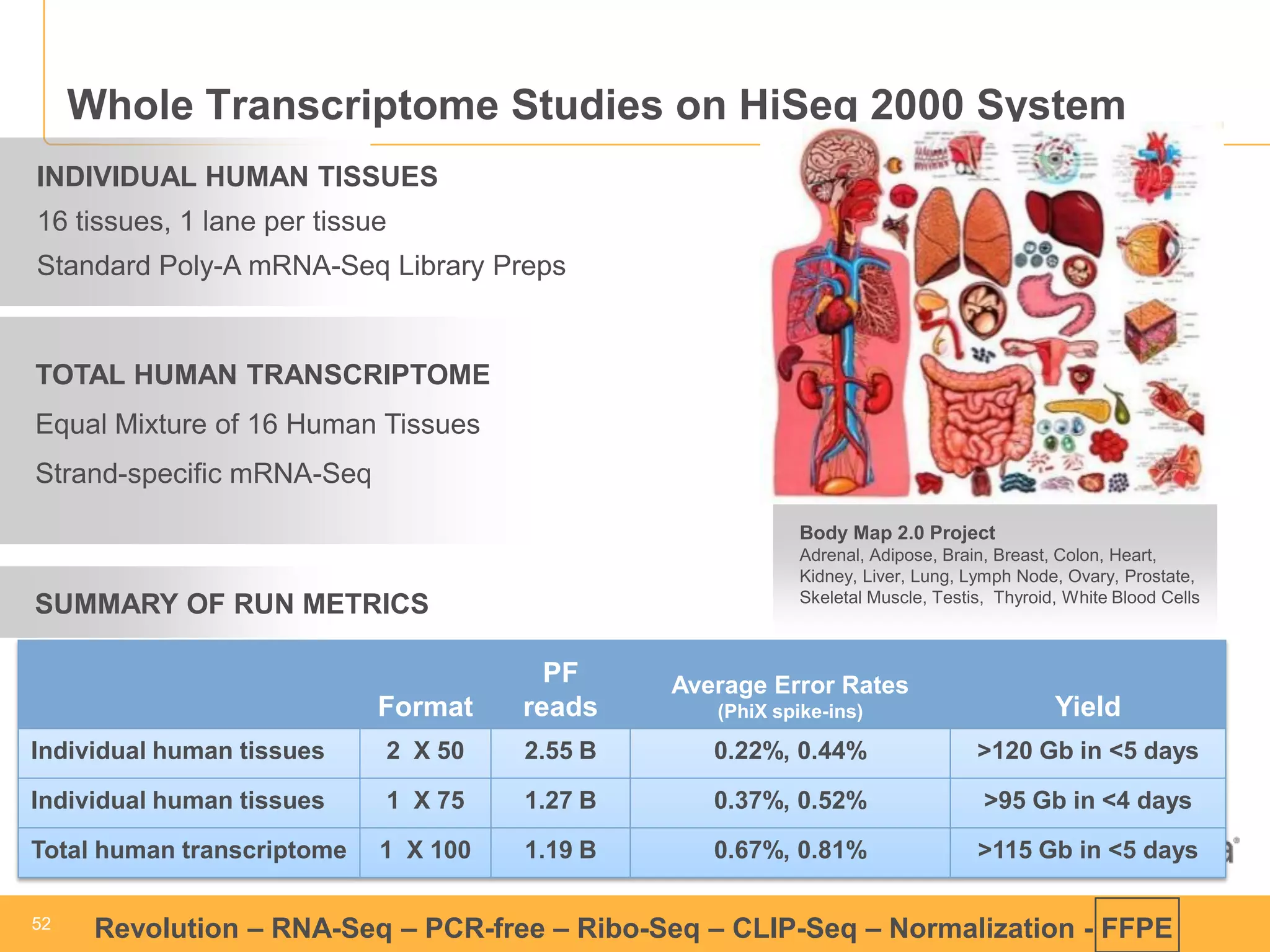
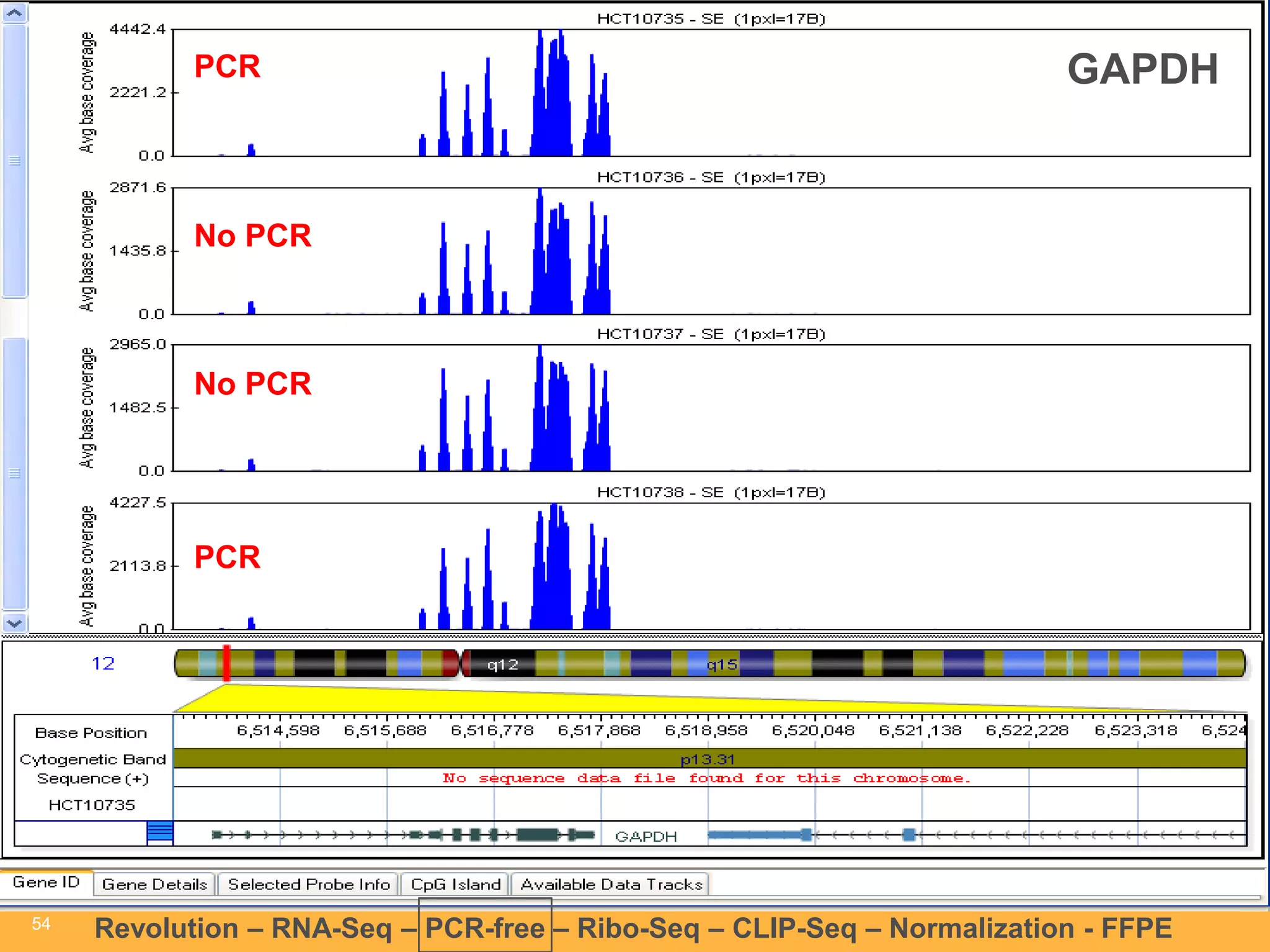
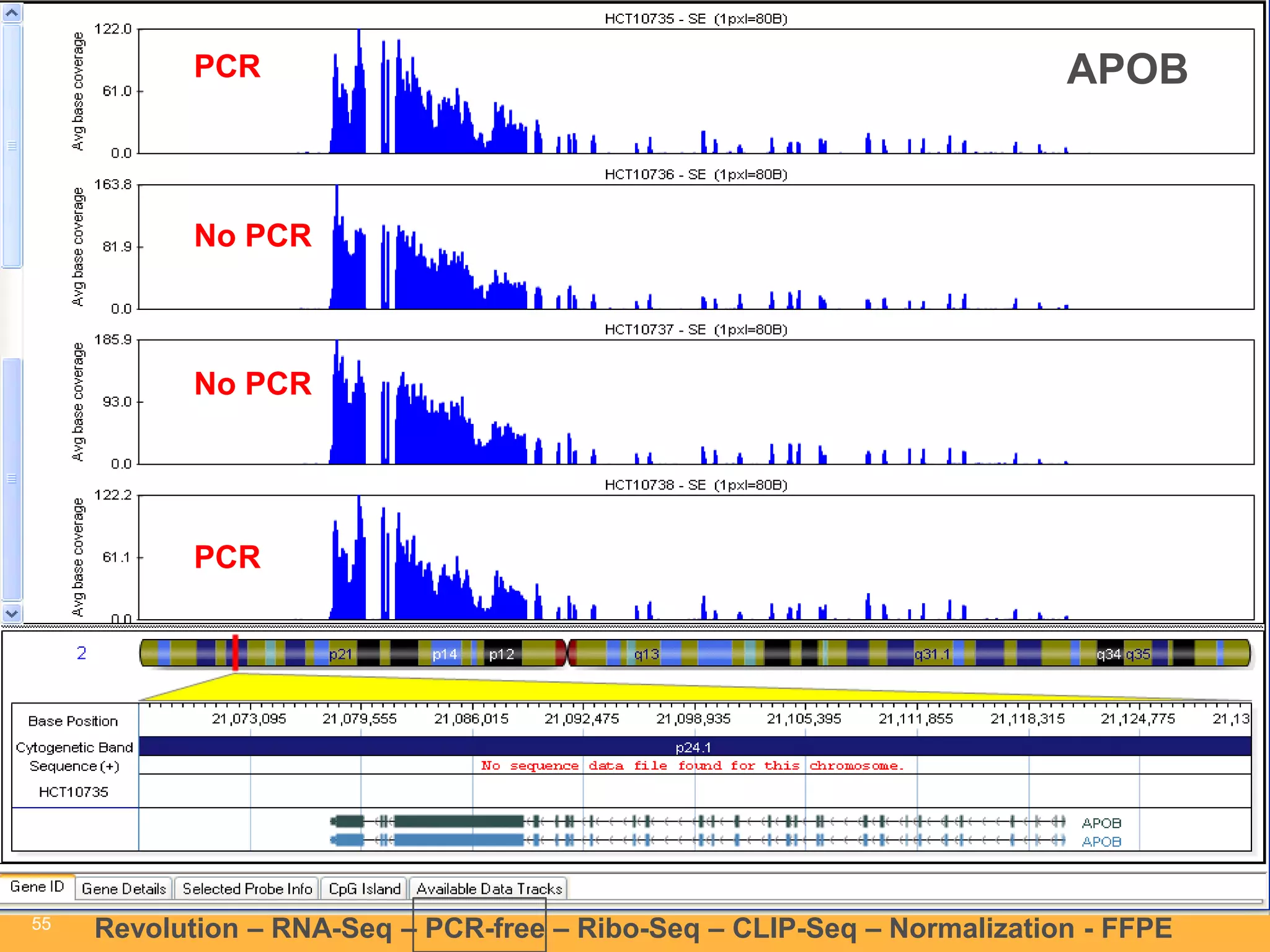
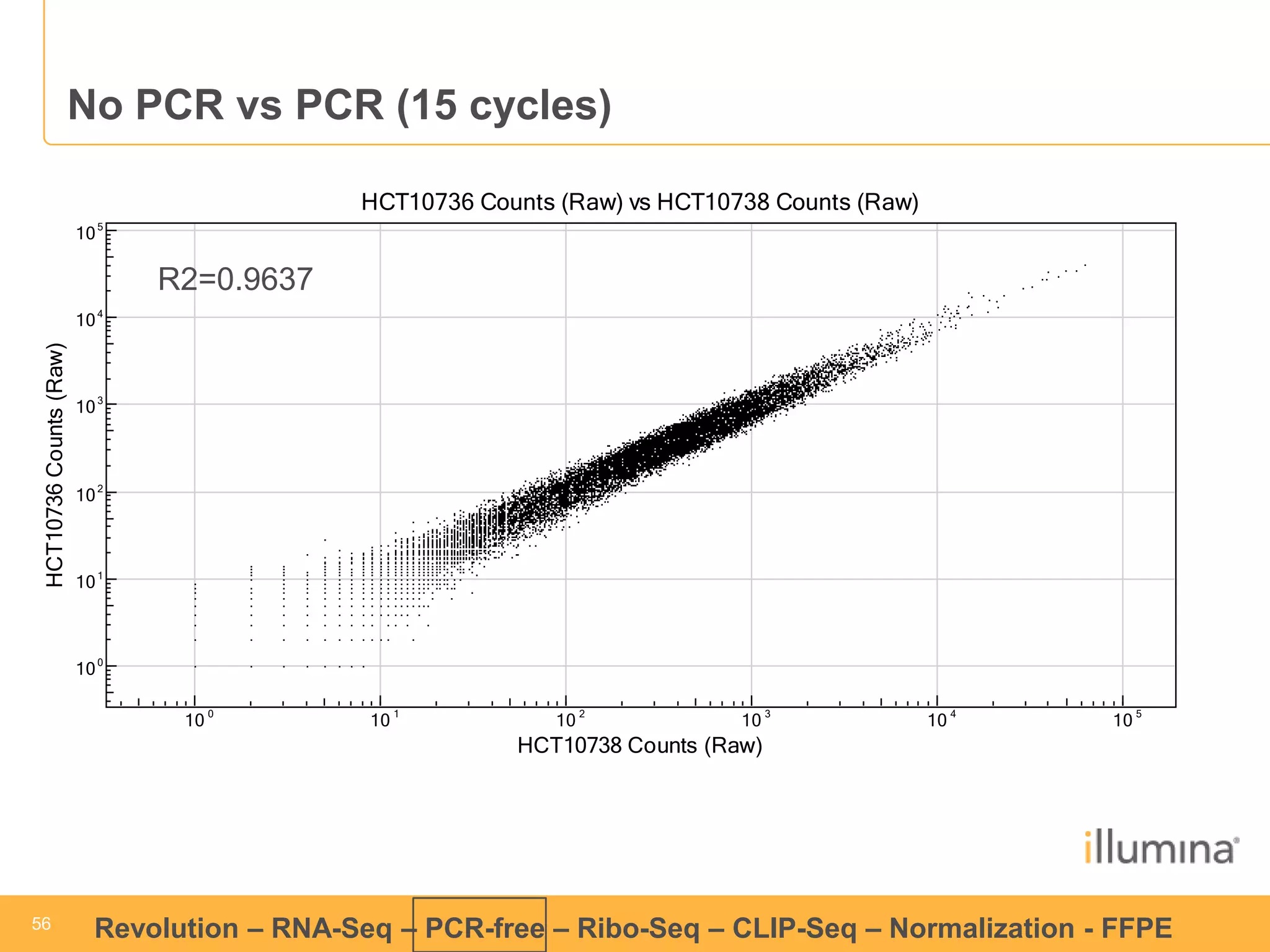
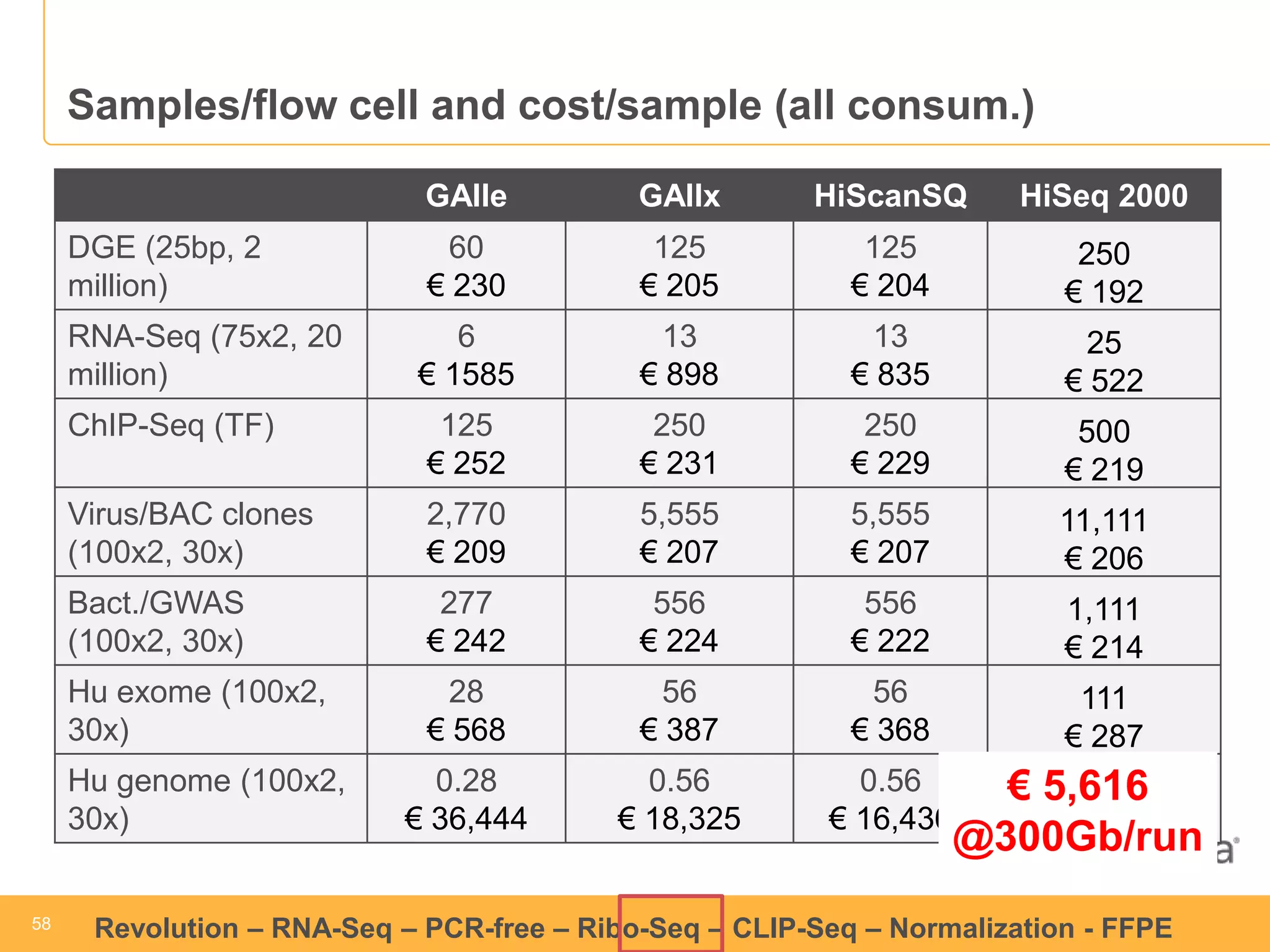
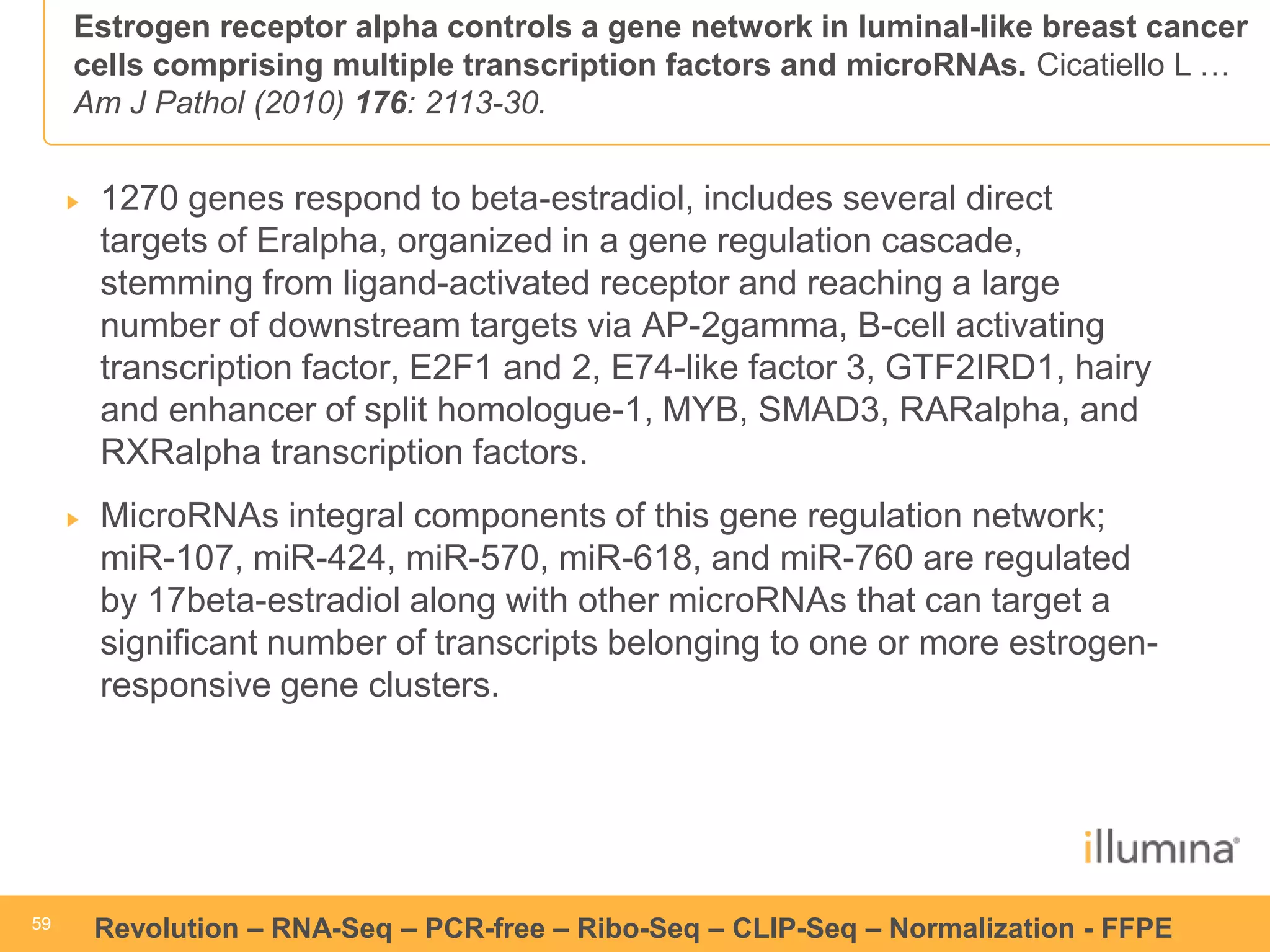
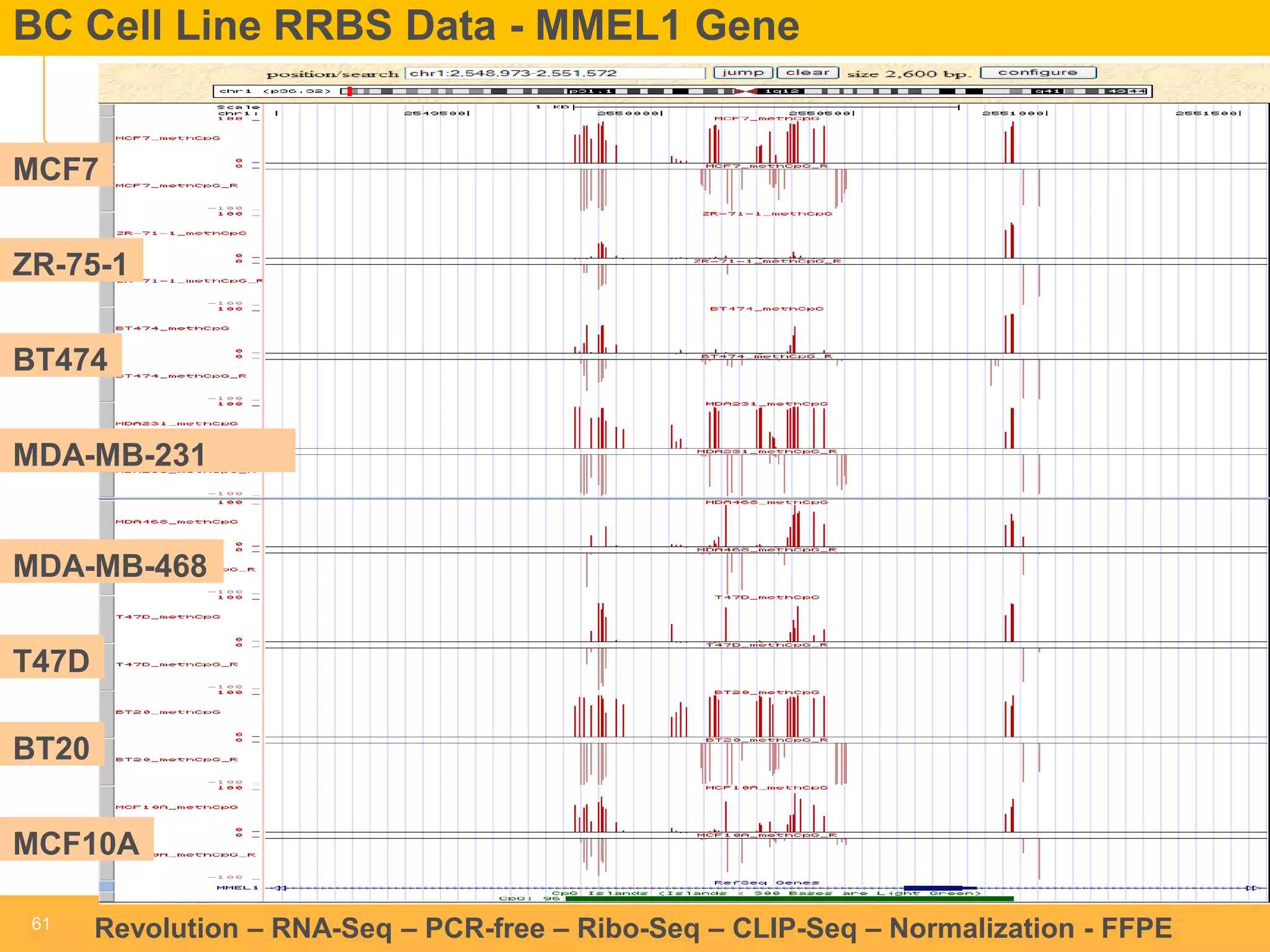
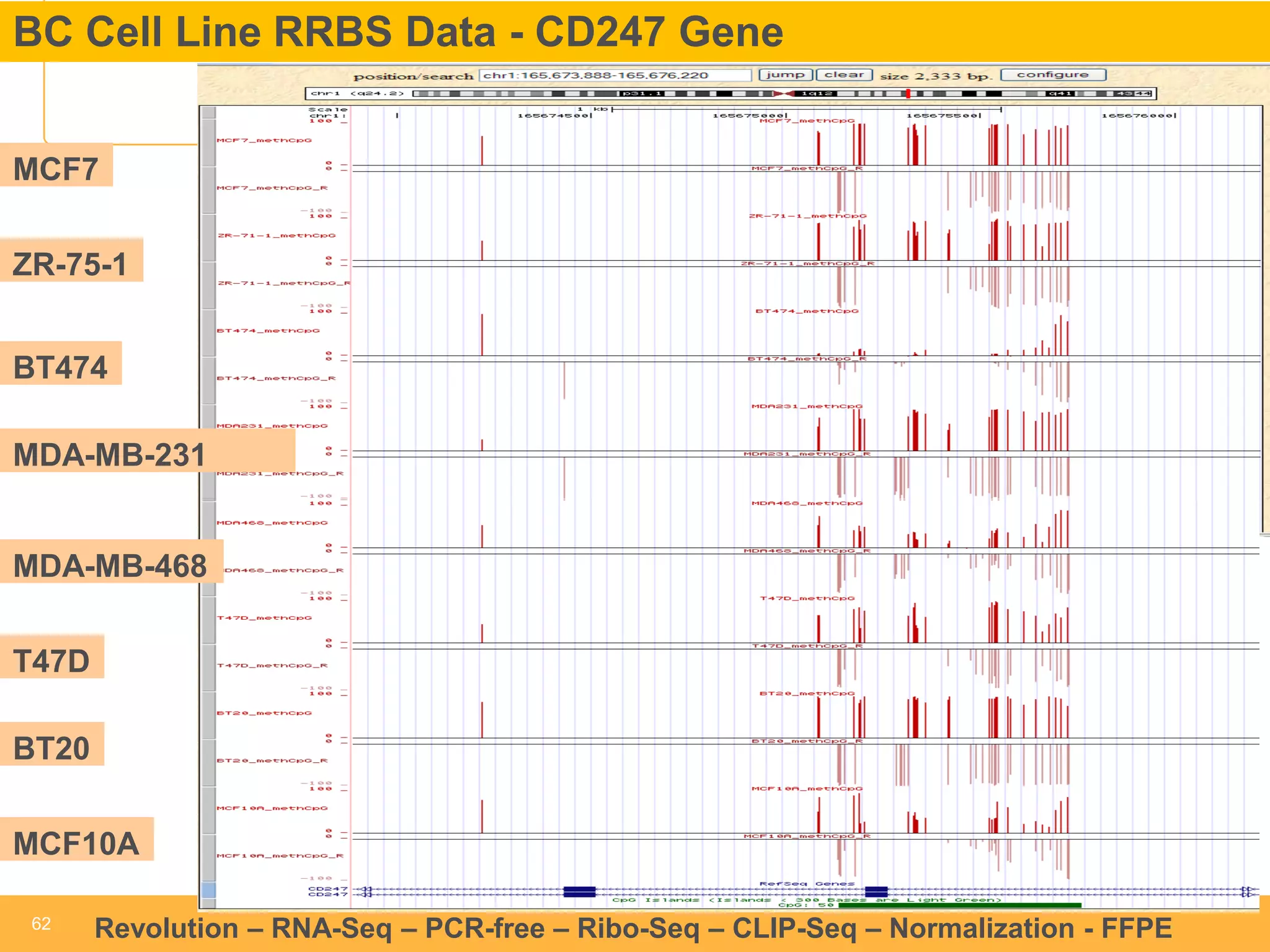
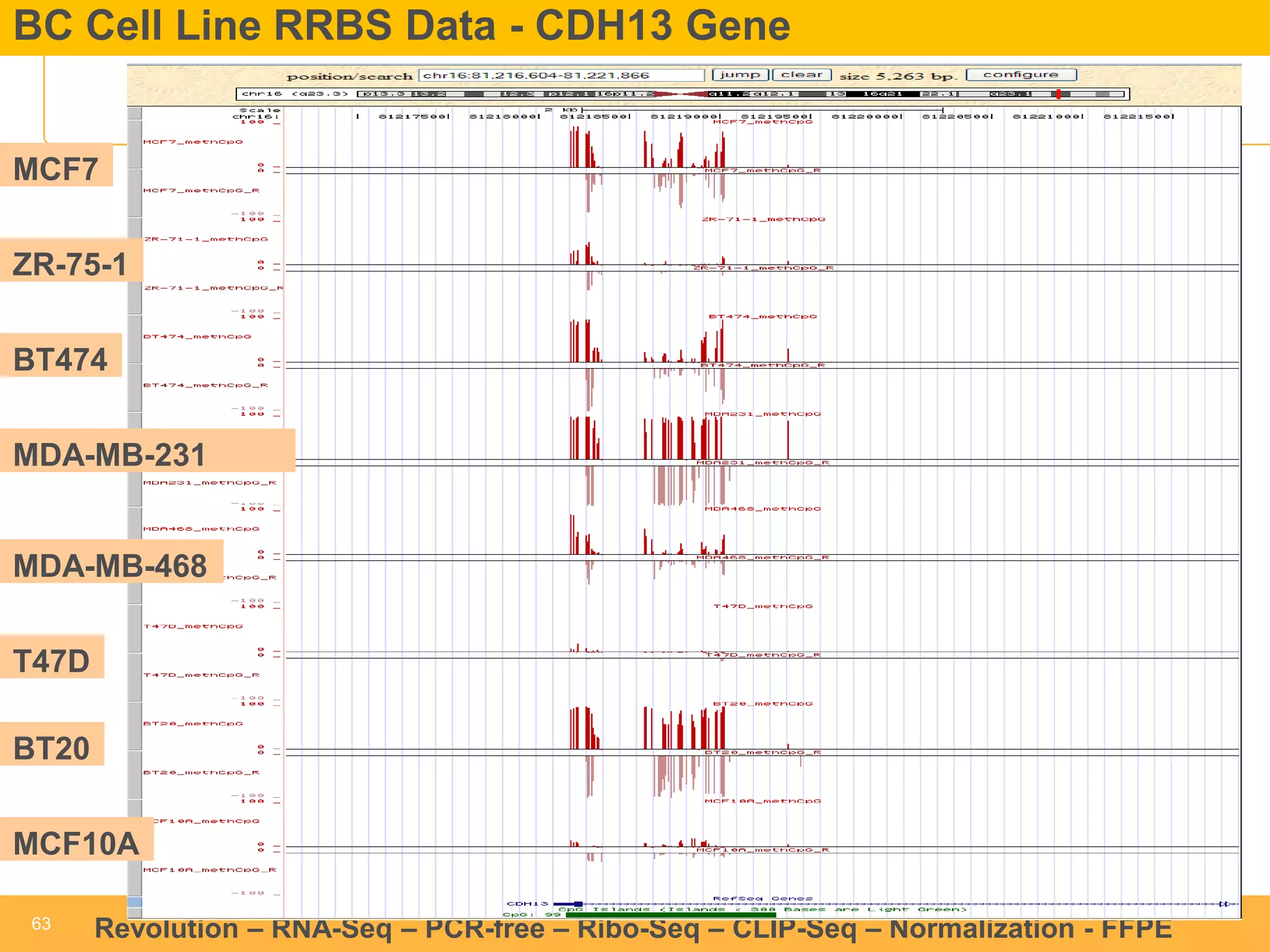
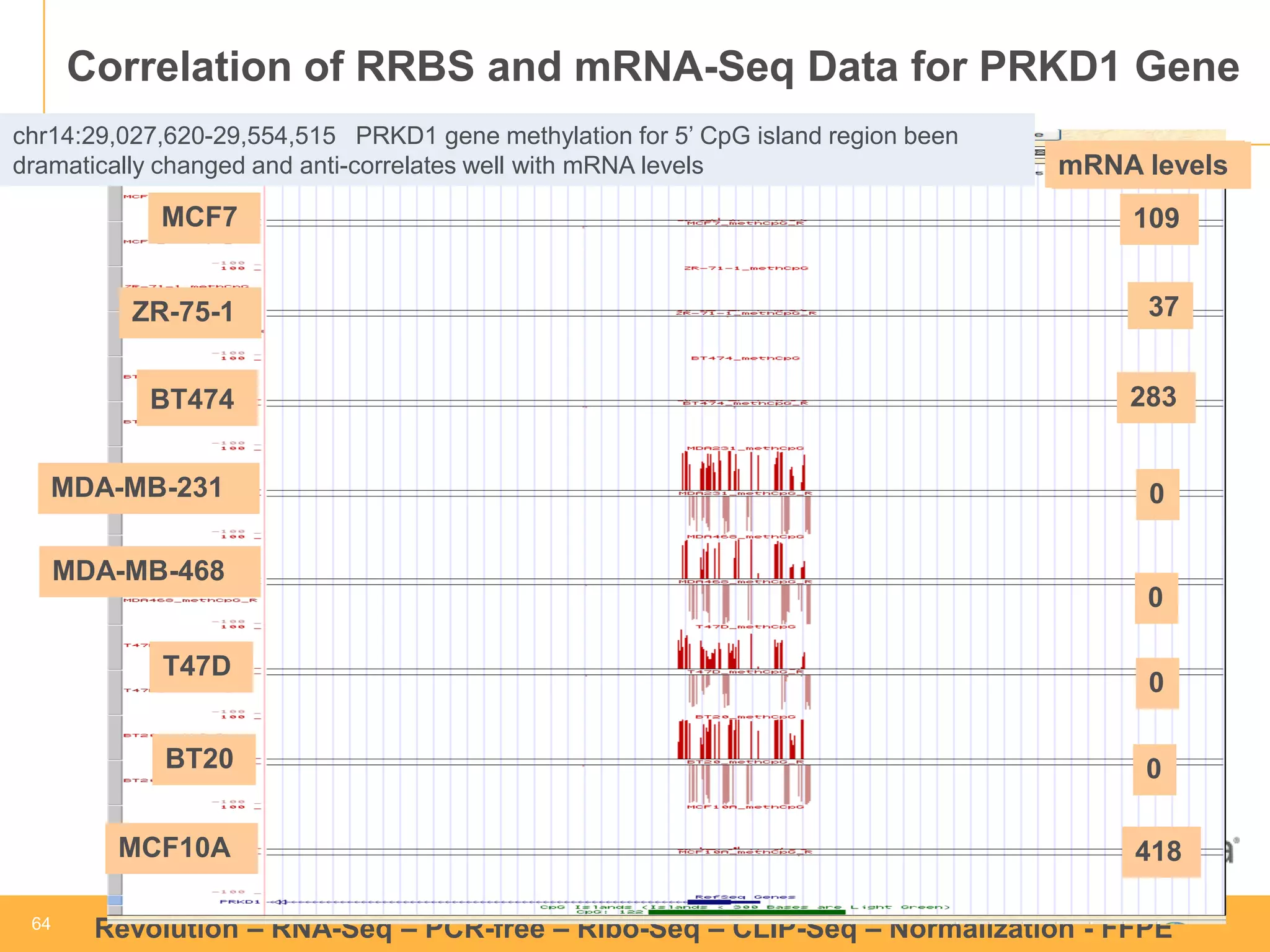
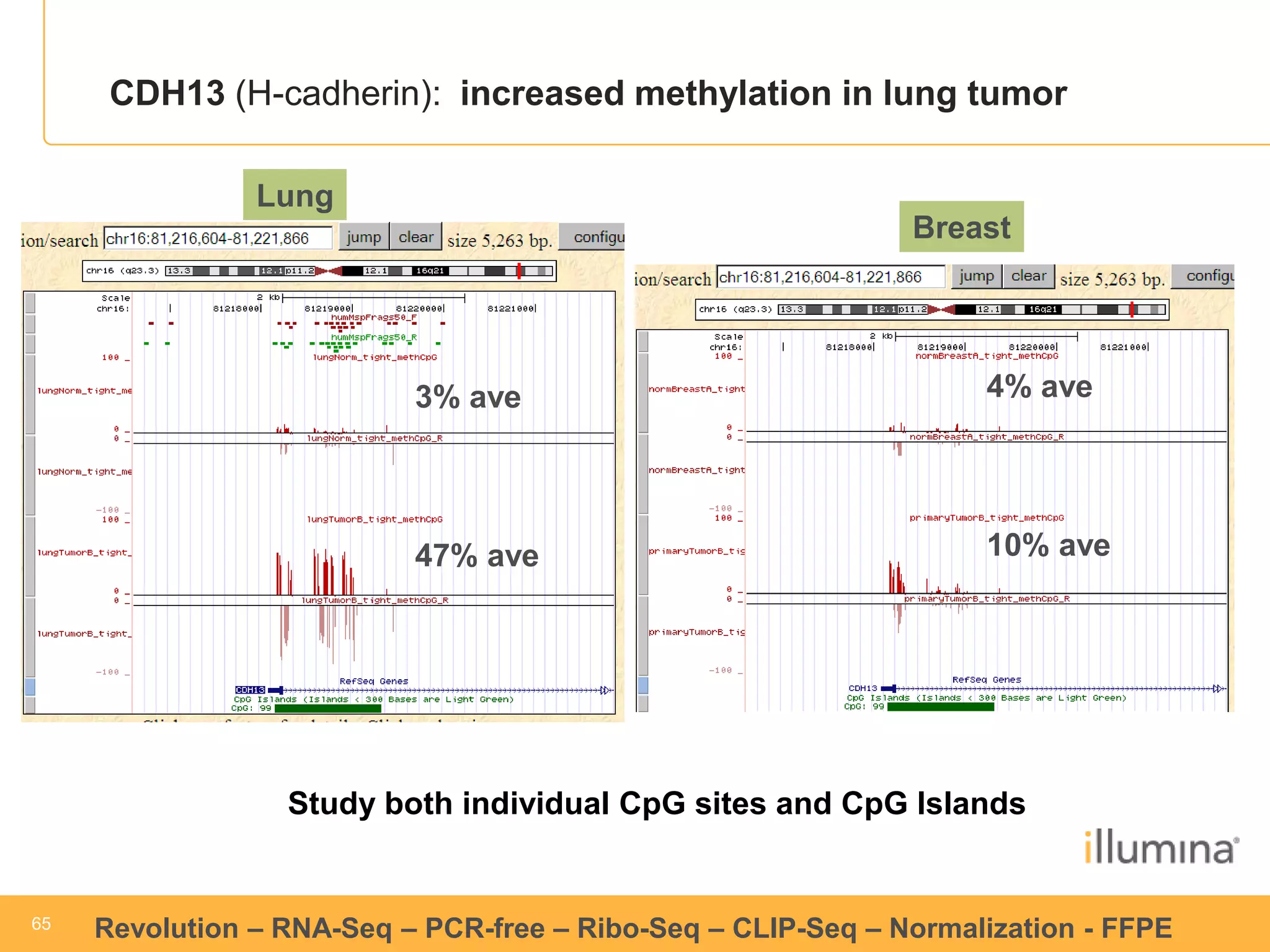
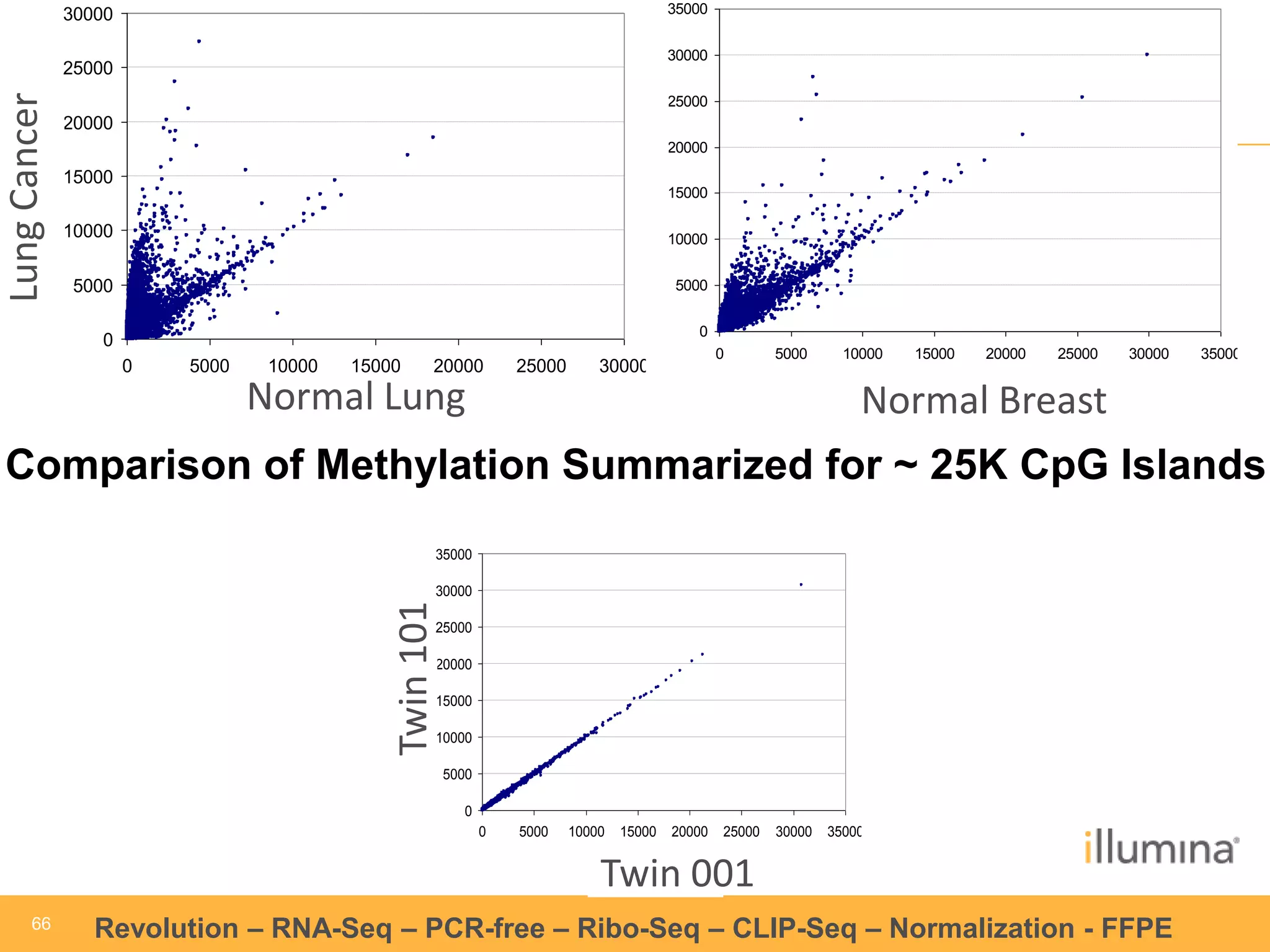
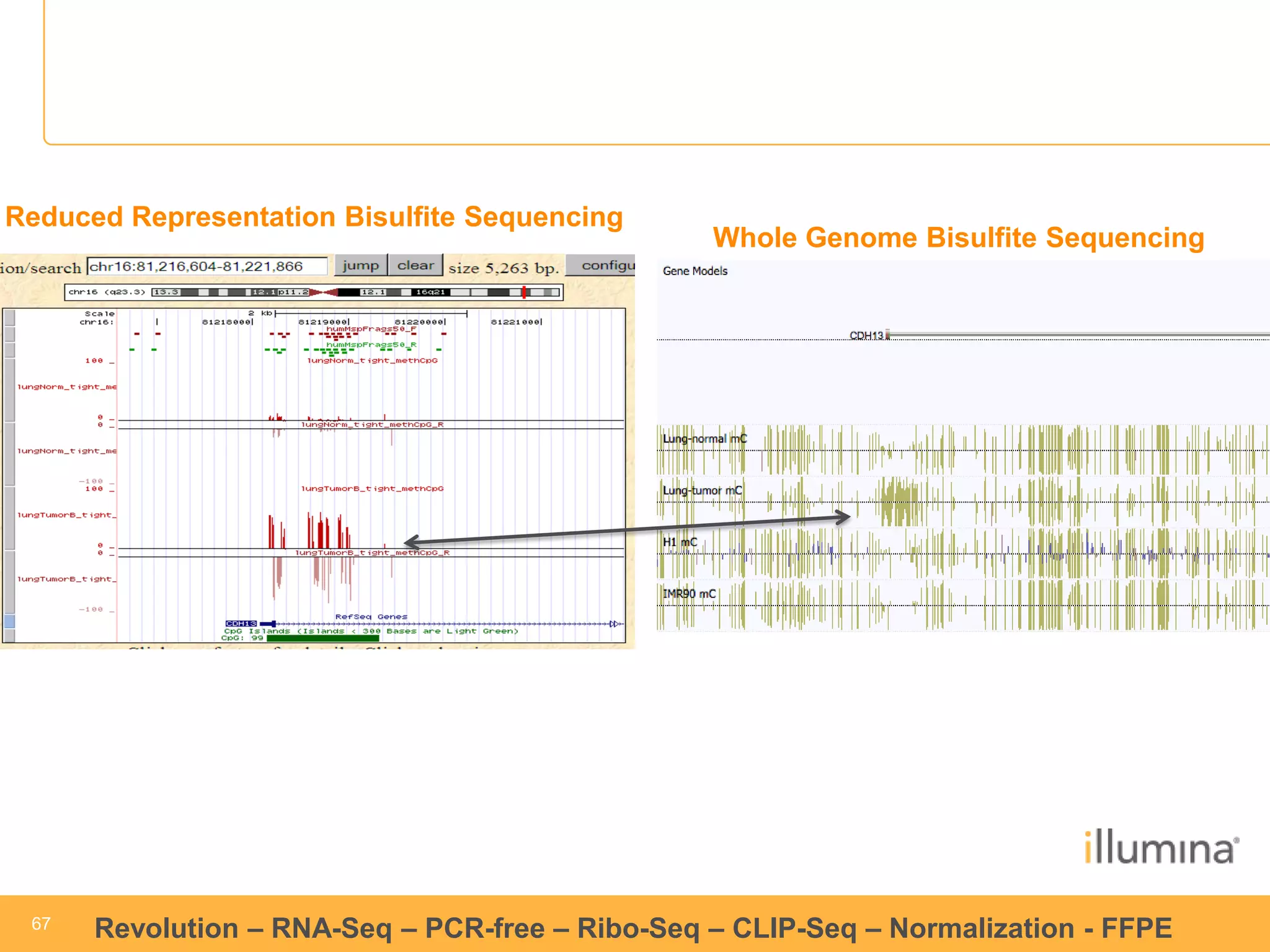
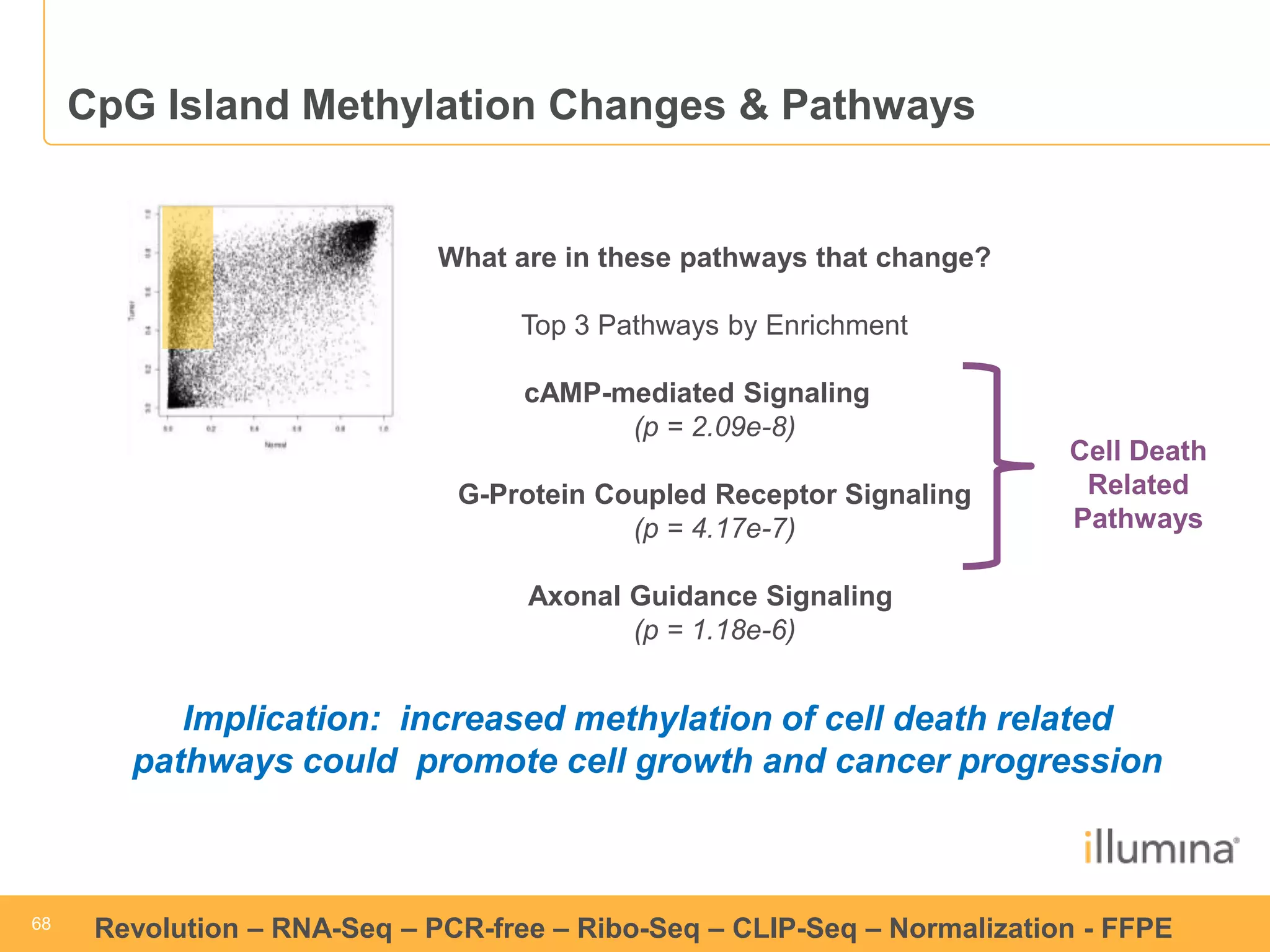
This document discusses RNA sequencing (RNA-Seq) and related techniques. It introduces total RNA sequencing which allows sequencing of all RNA molecules from a sample, including non-coding RNA, using less than 100ng of input RNA. Double-strand specific nuclease (DSN) normalization is described as a method to normalize total RNA sequencing libraries. The document also discusses applying RNA sequencing to analyze formalin-fixed paraffin-embedded (FFPE) samples.