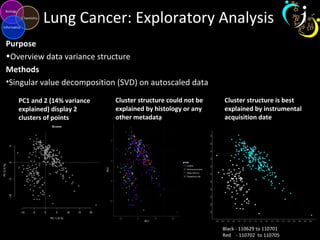
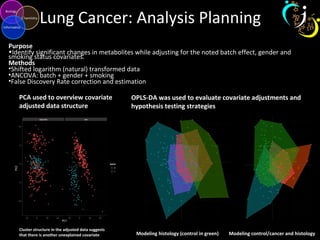
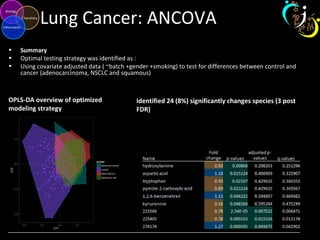
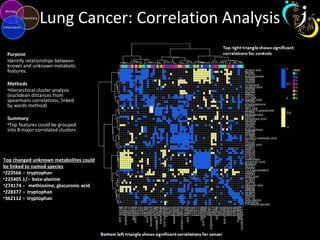
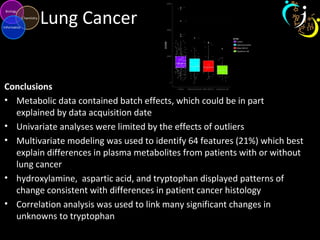
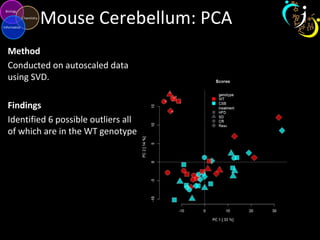
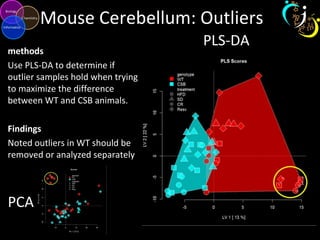
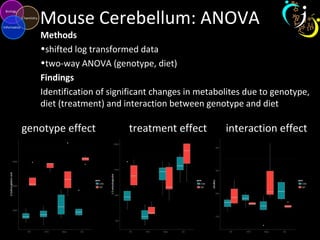
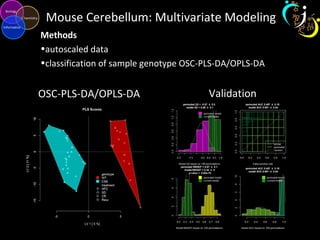
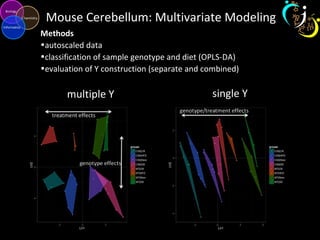
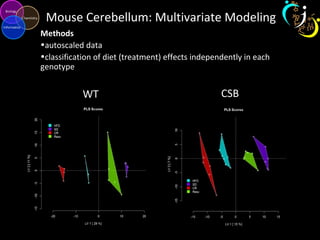
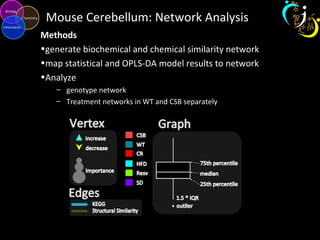
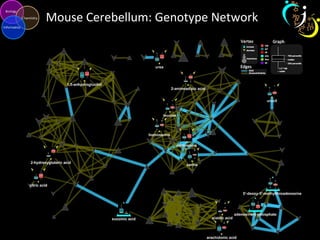
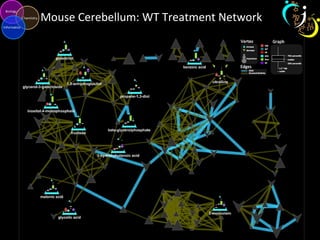
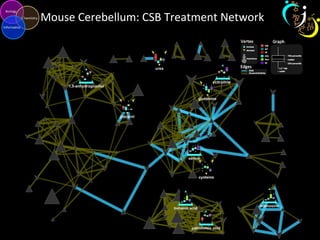
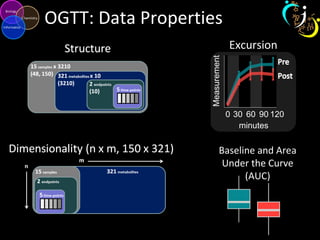
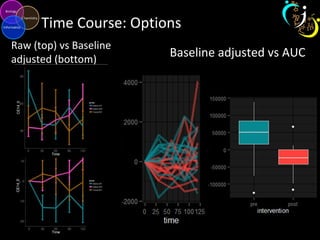
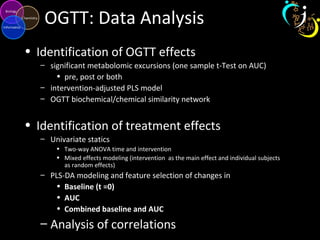
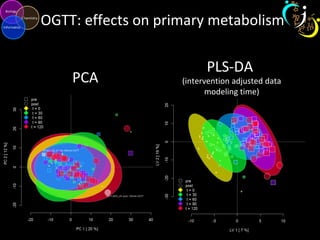
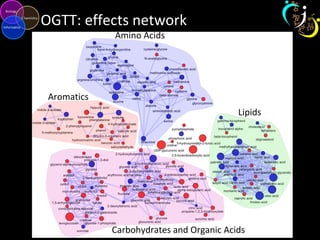
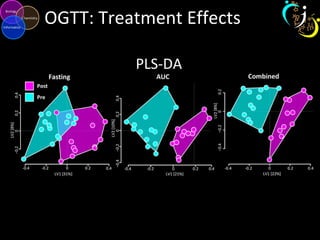
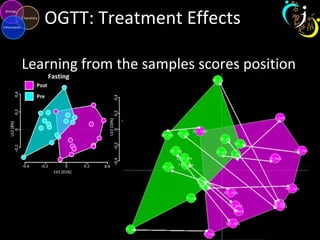
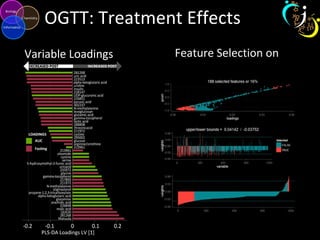
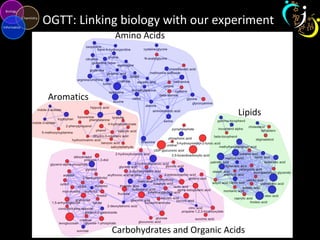
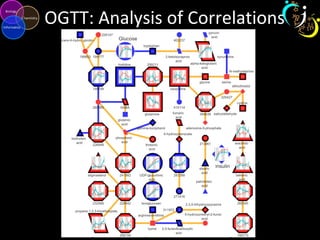
The document outlines various case studies on metabolomic data analysis focusing on lung cancer, mouse cerebellum, and an oral glucose tolerance test (OGTT). It discusses methodologies such as principal components analysis (PCA), analysis of covariance (ANCOVA), and orthogonal partial least squares discriminant analysis (OPLS-DA). The findings reveal significant metabolic changes linked with disease conditions and the influence of genotype and diet on metabolic alterations.