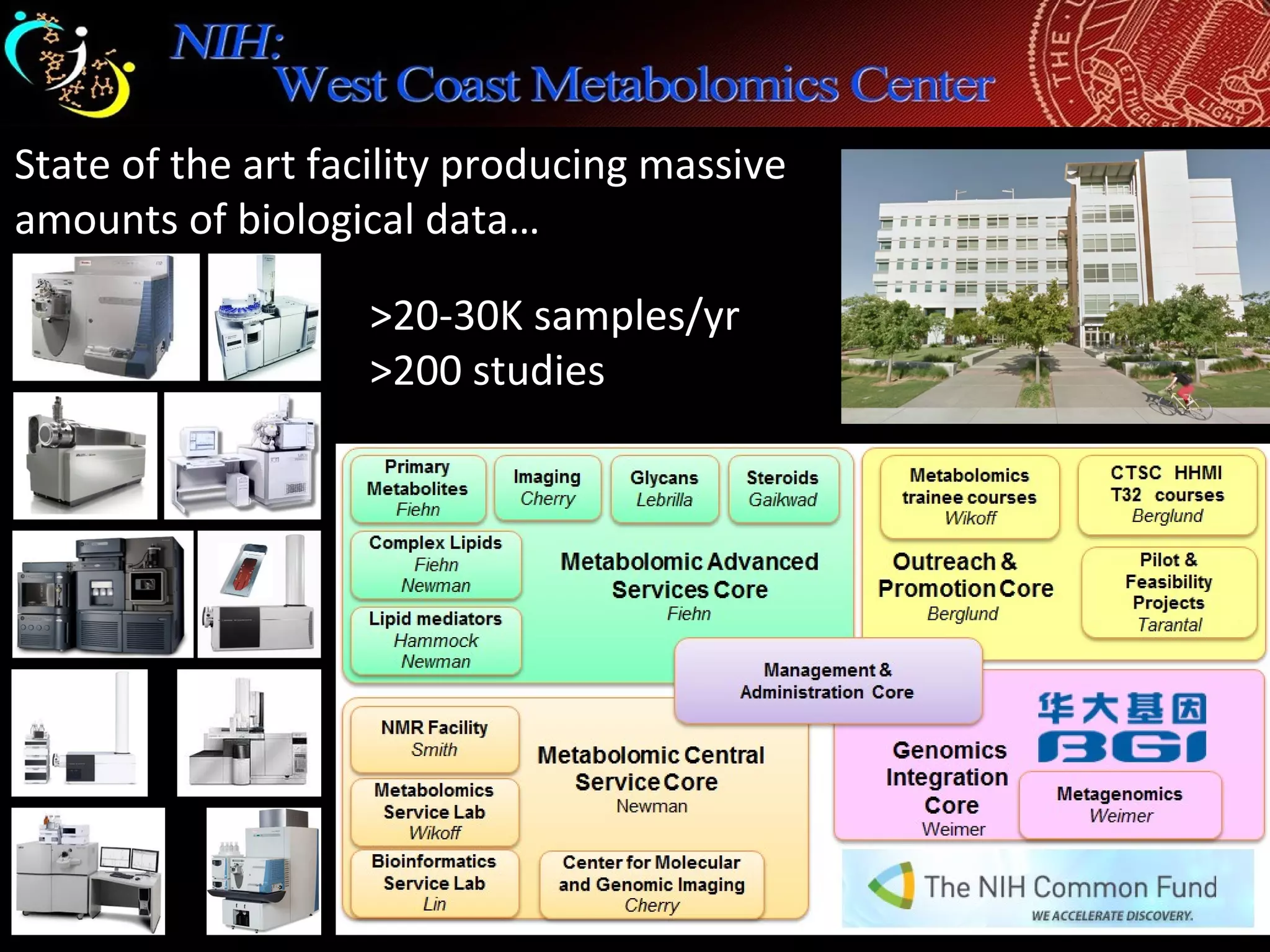
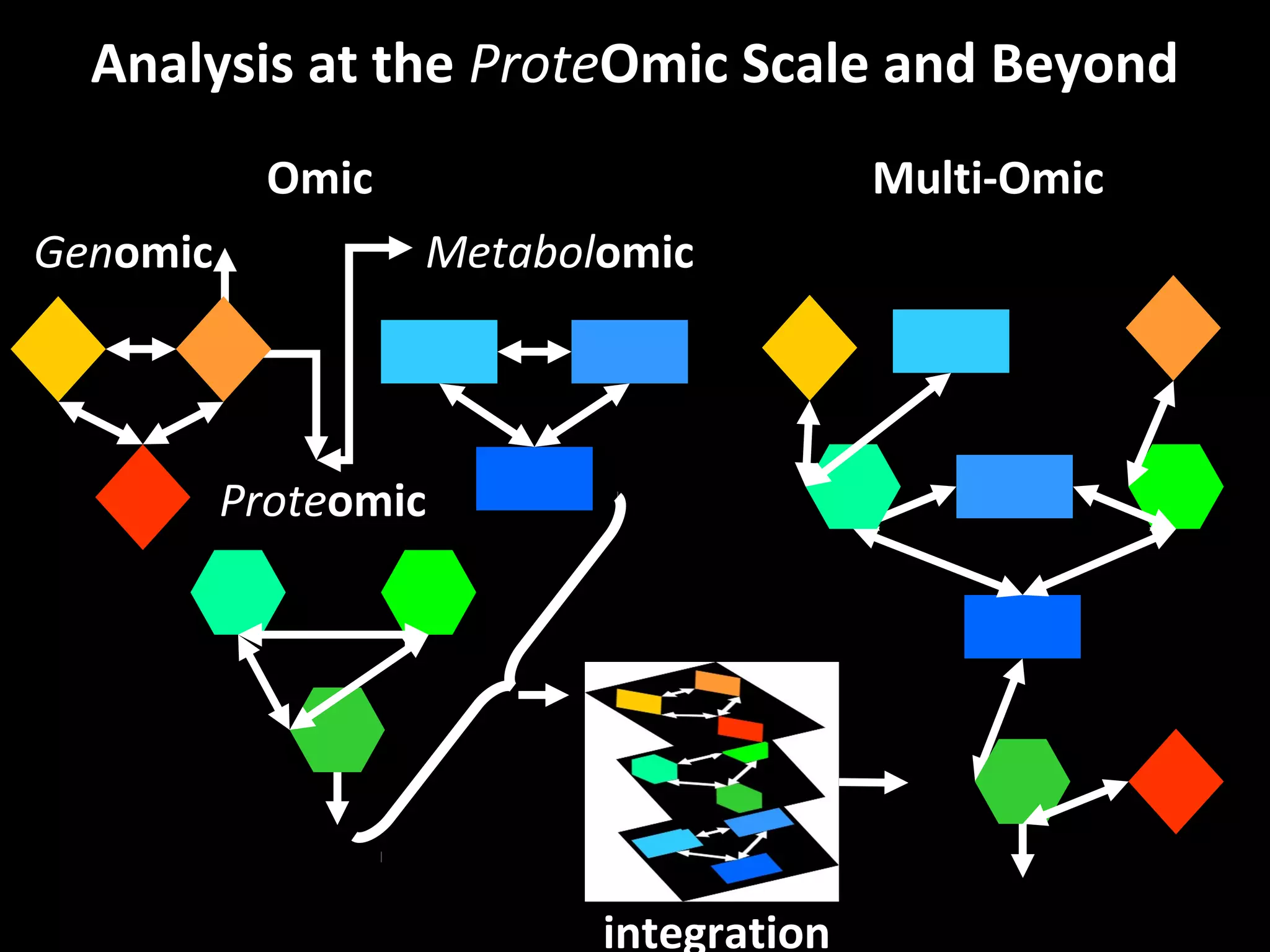
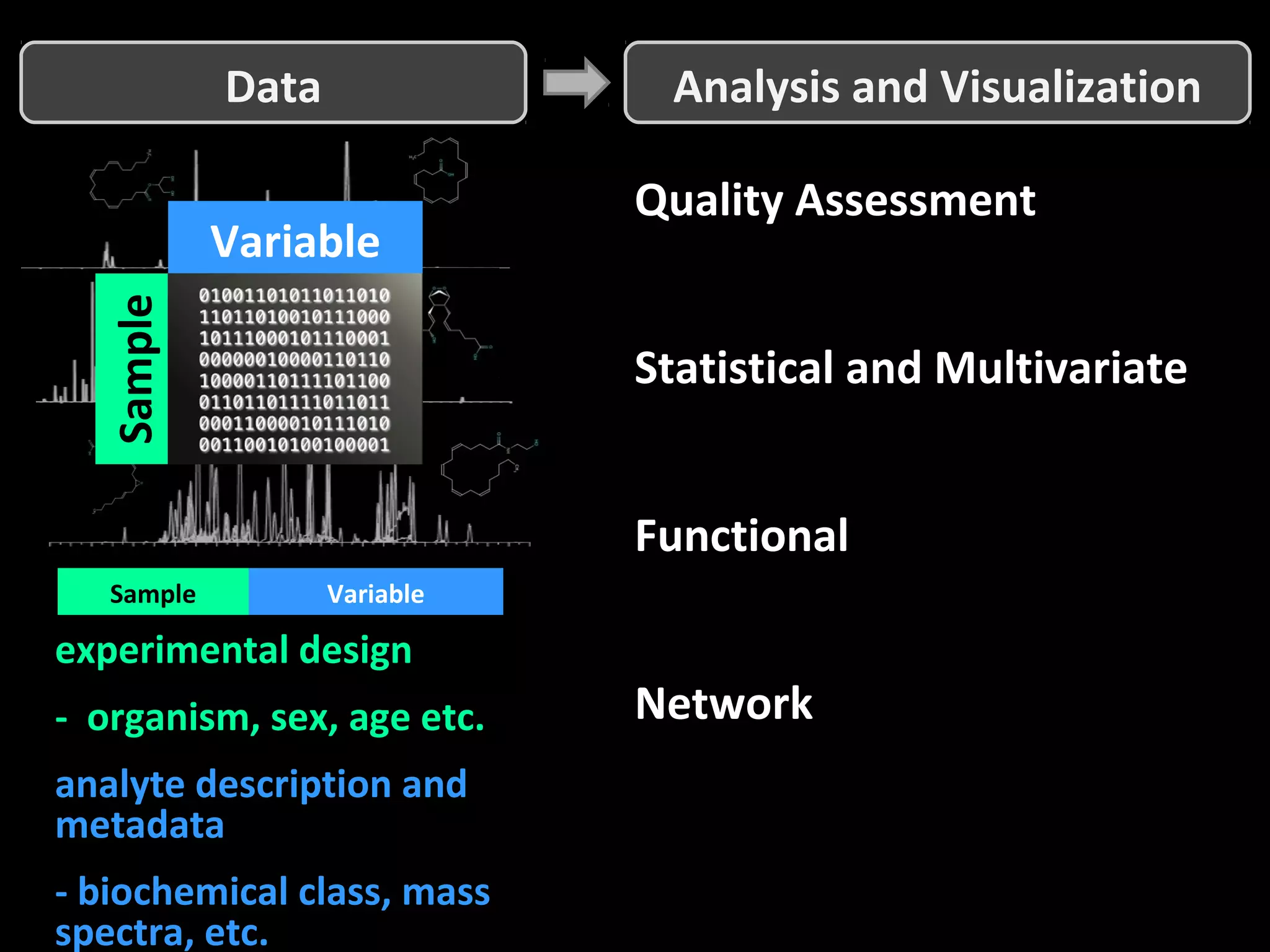
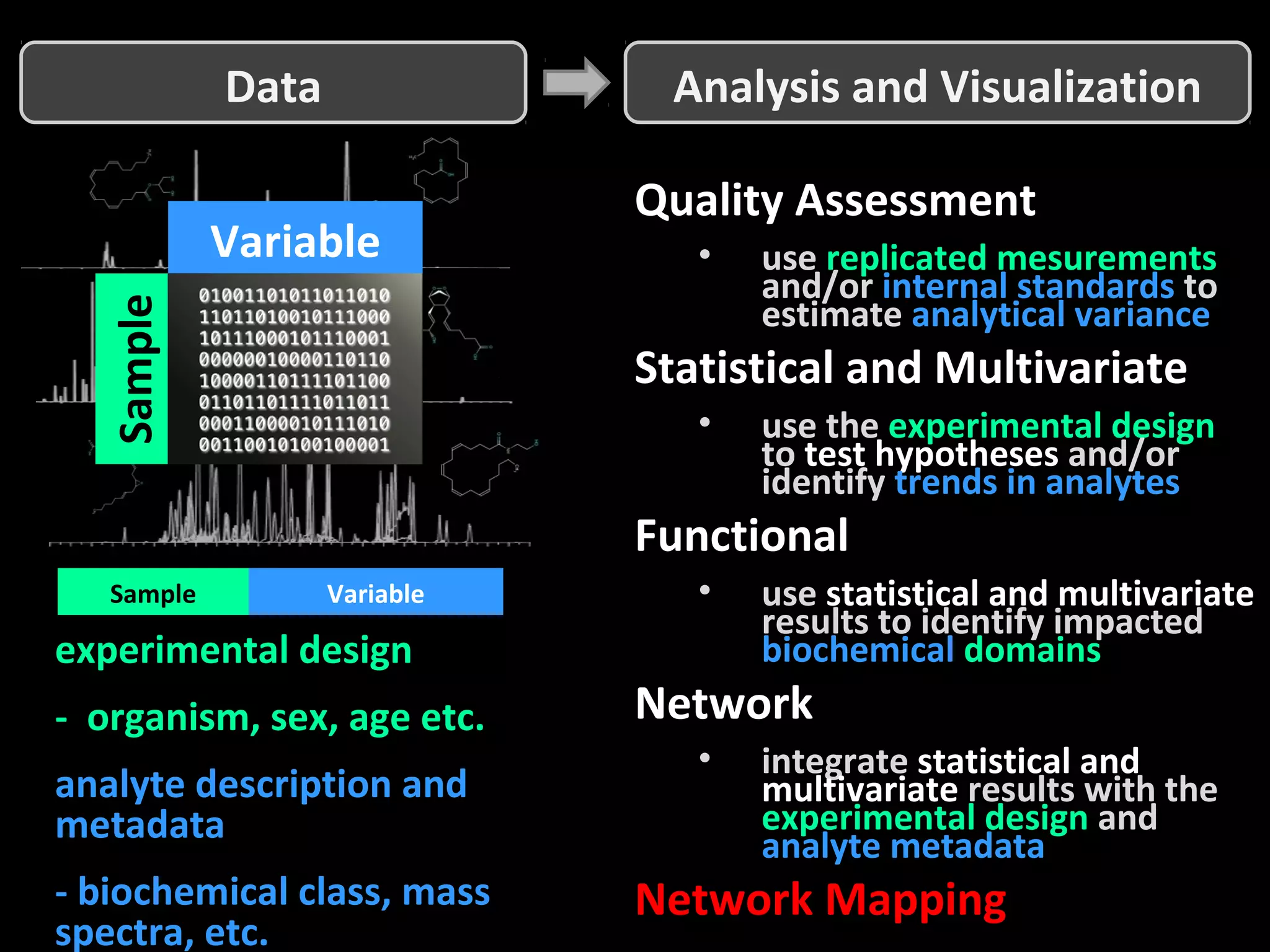
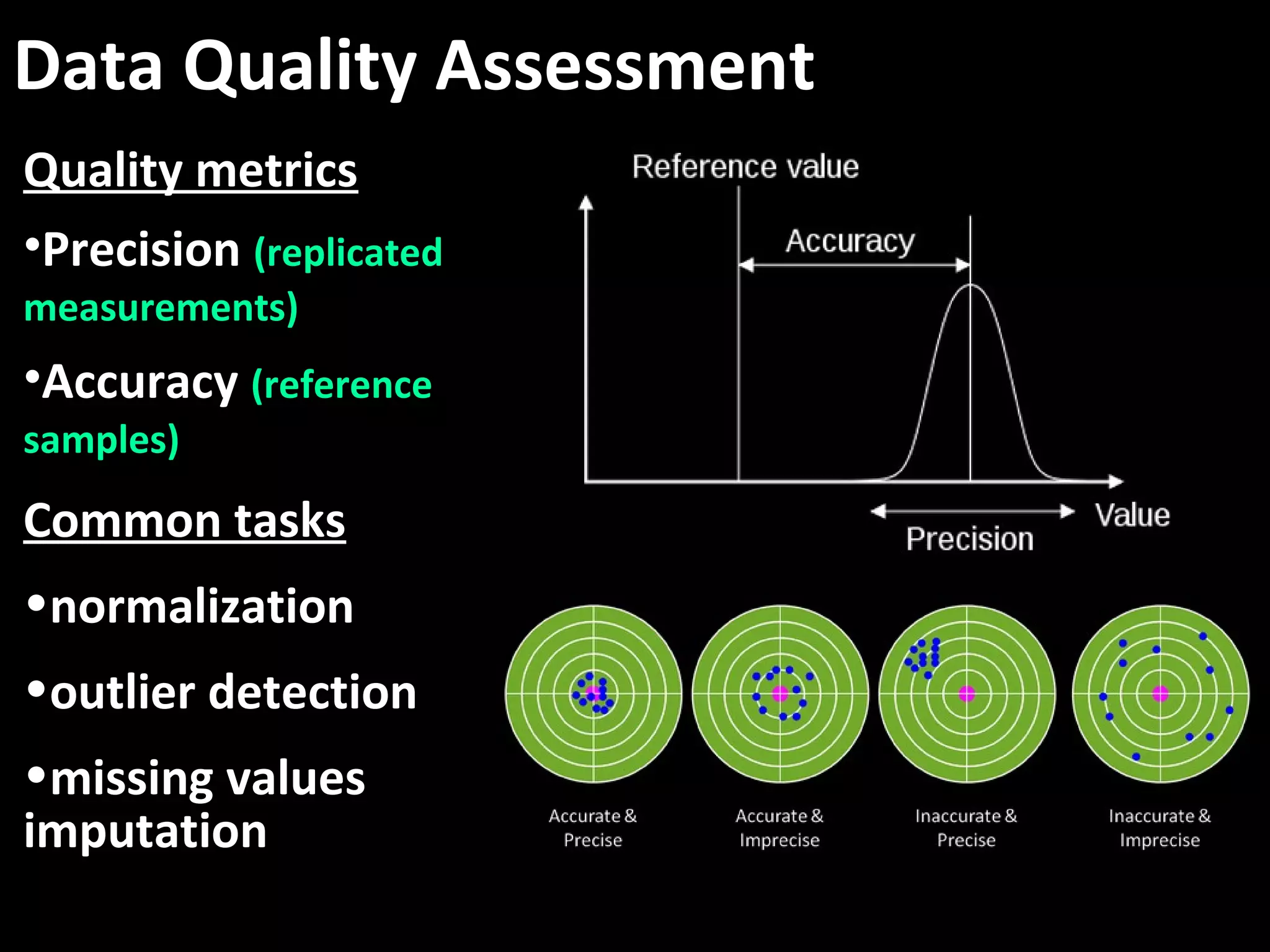
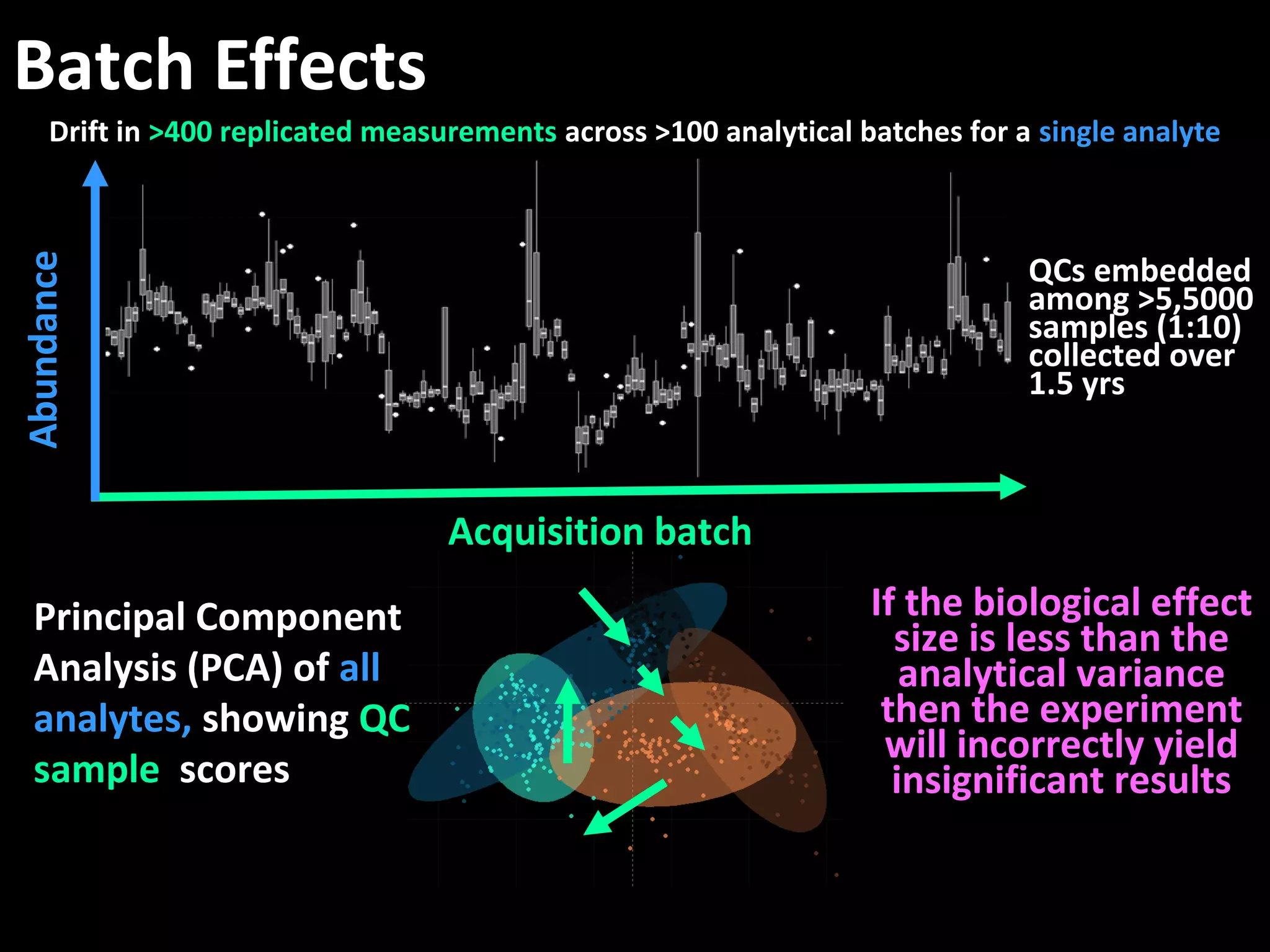
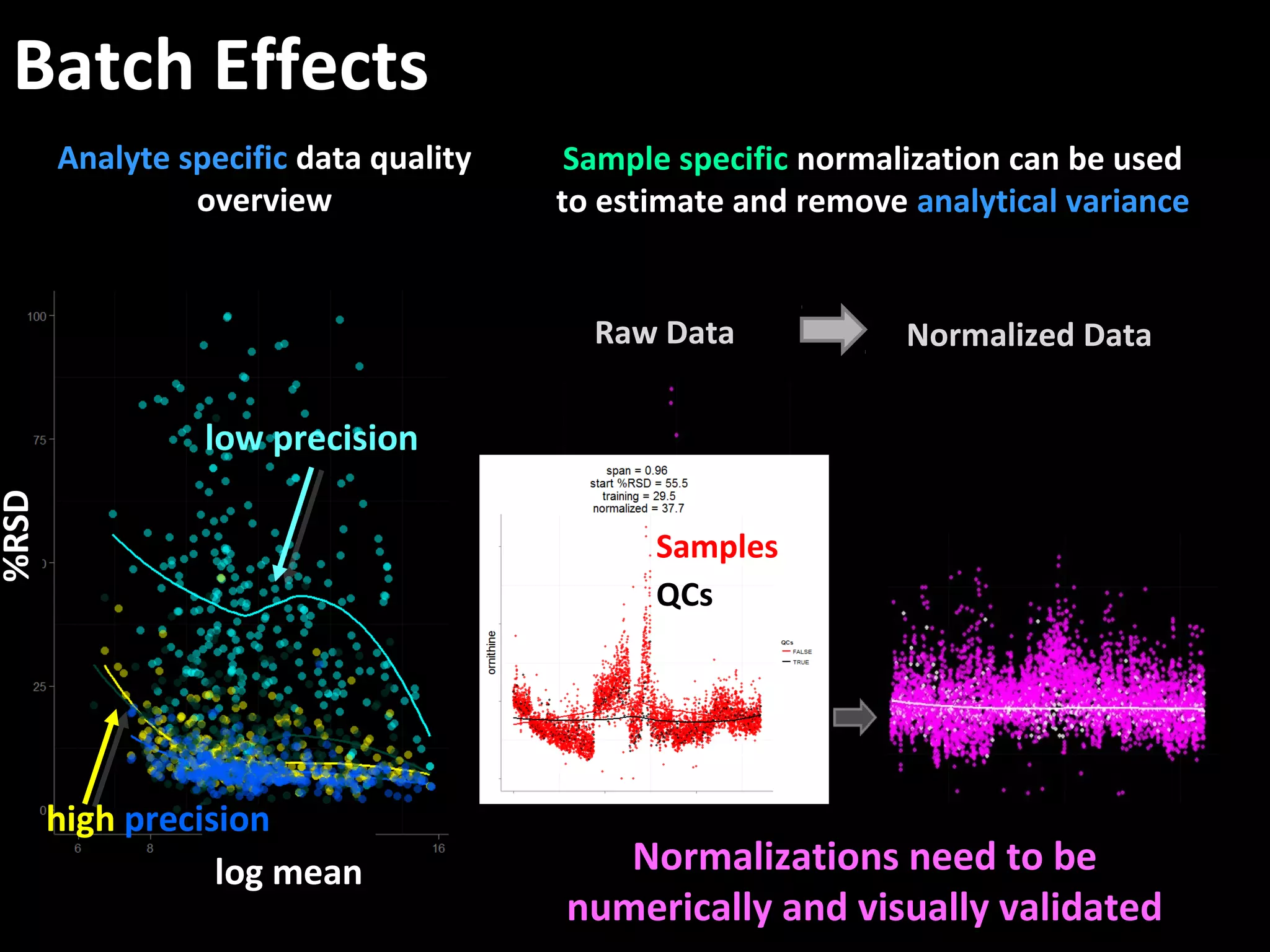
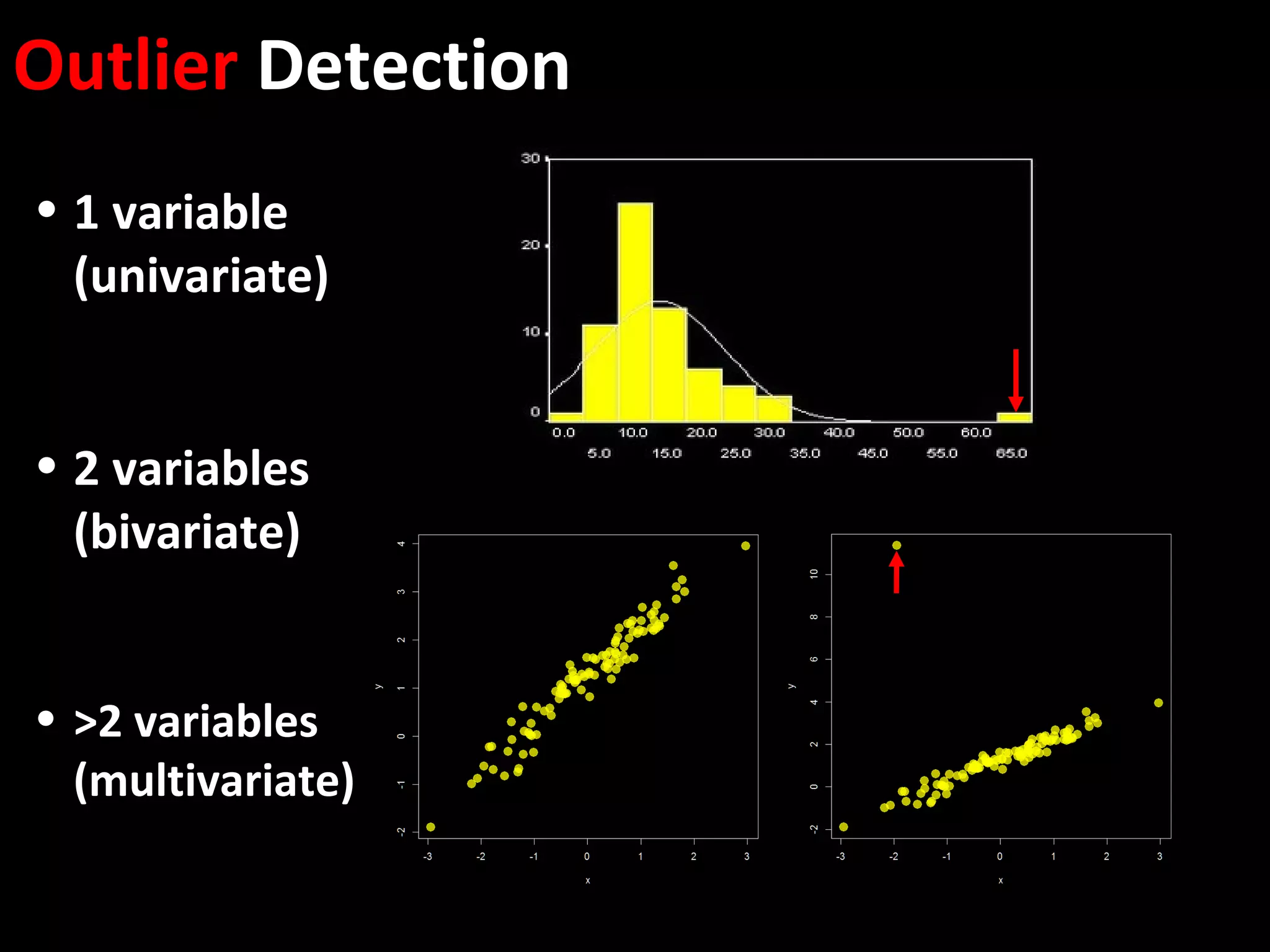
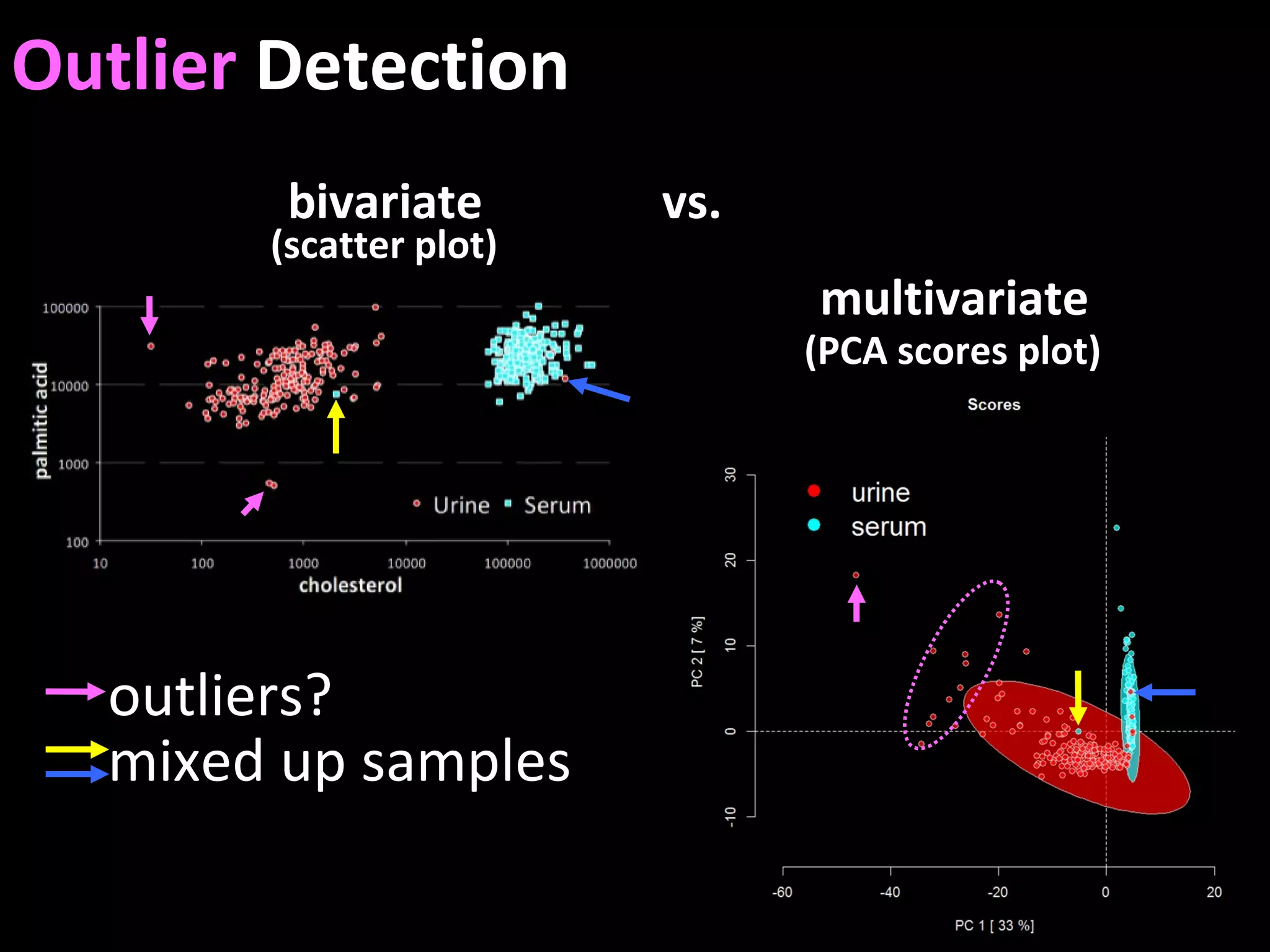
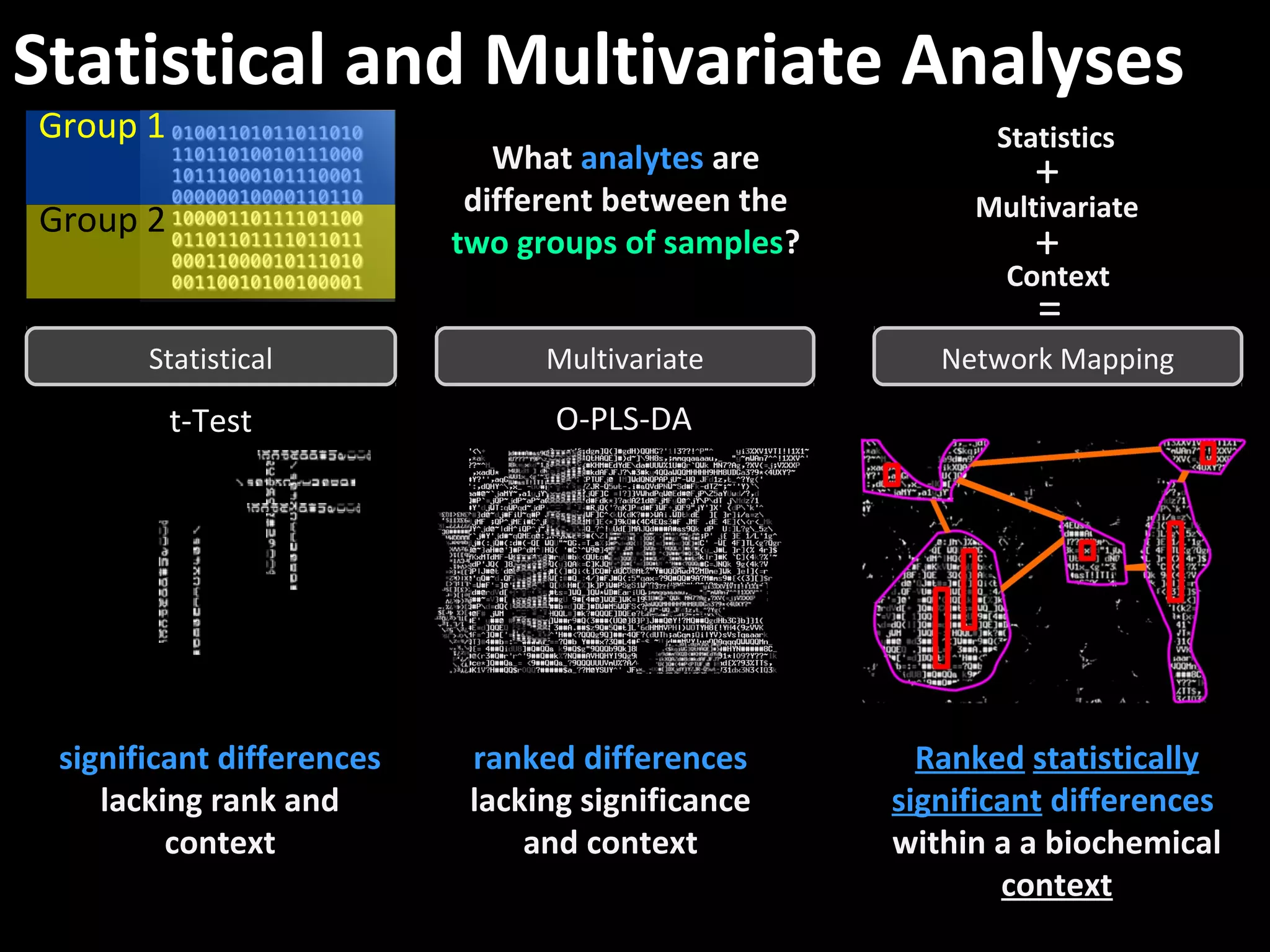
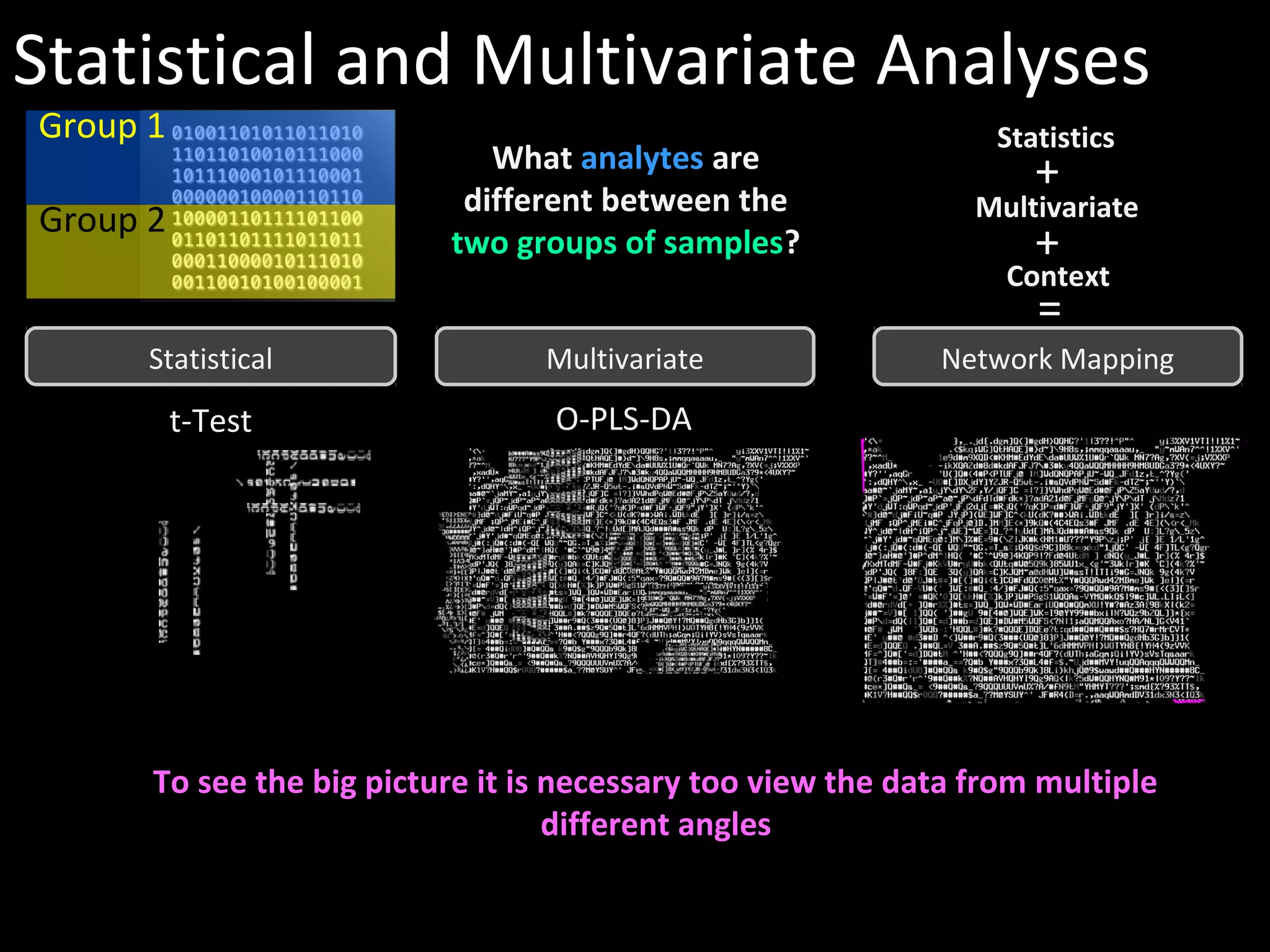
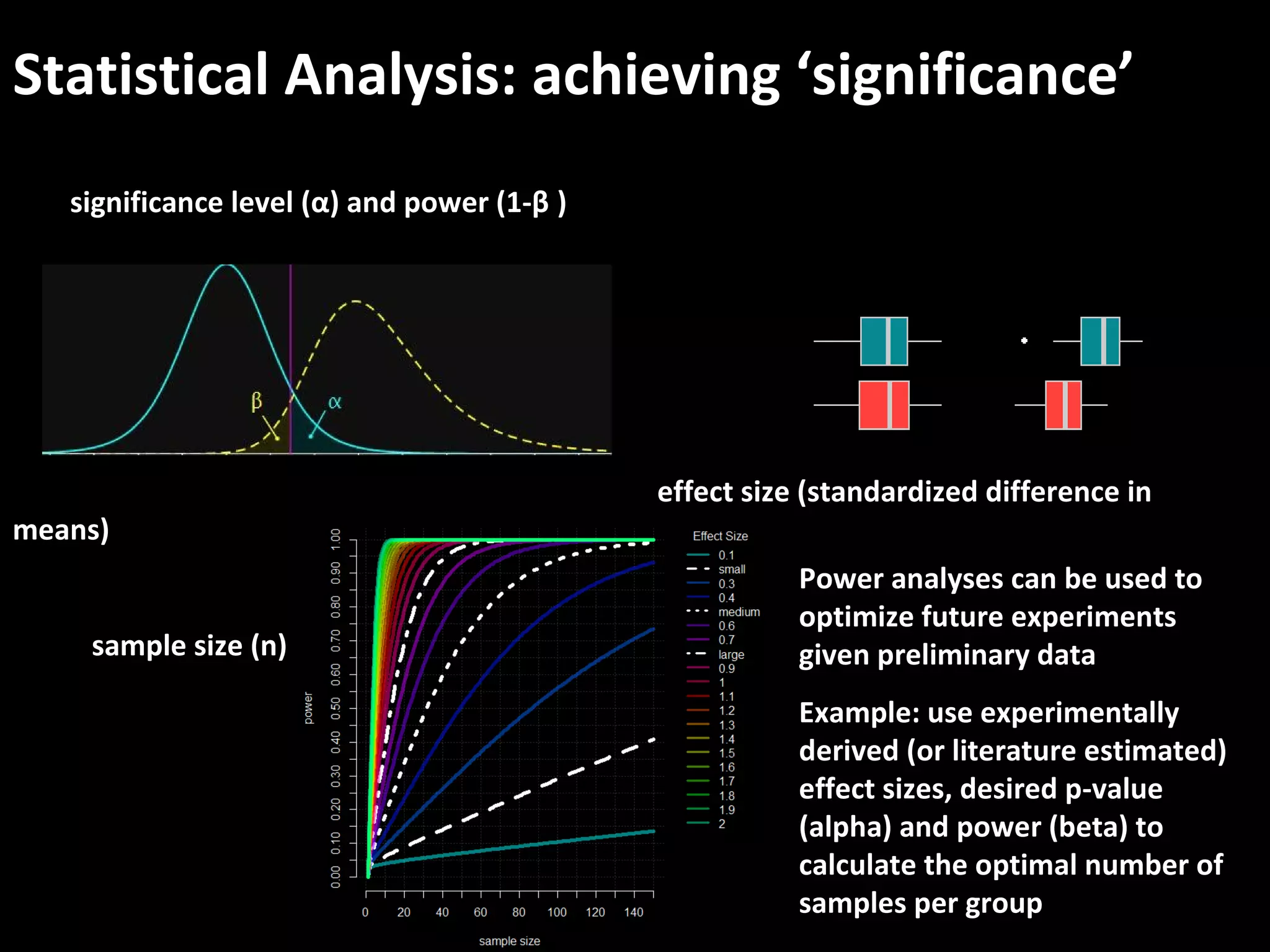
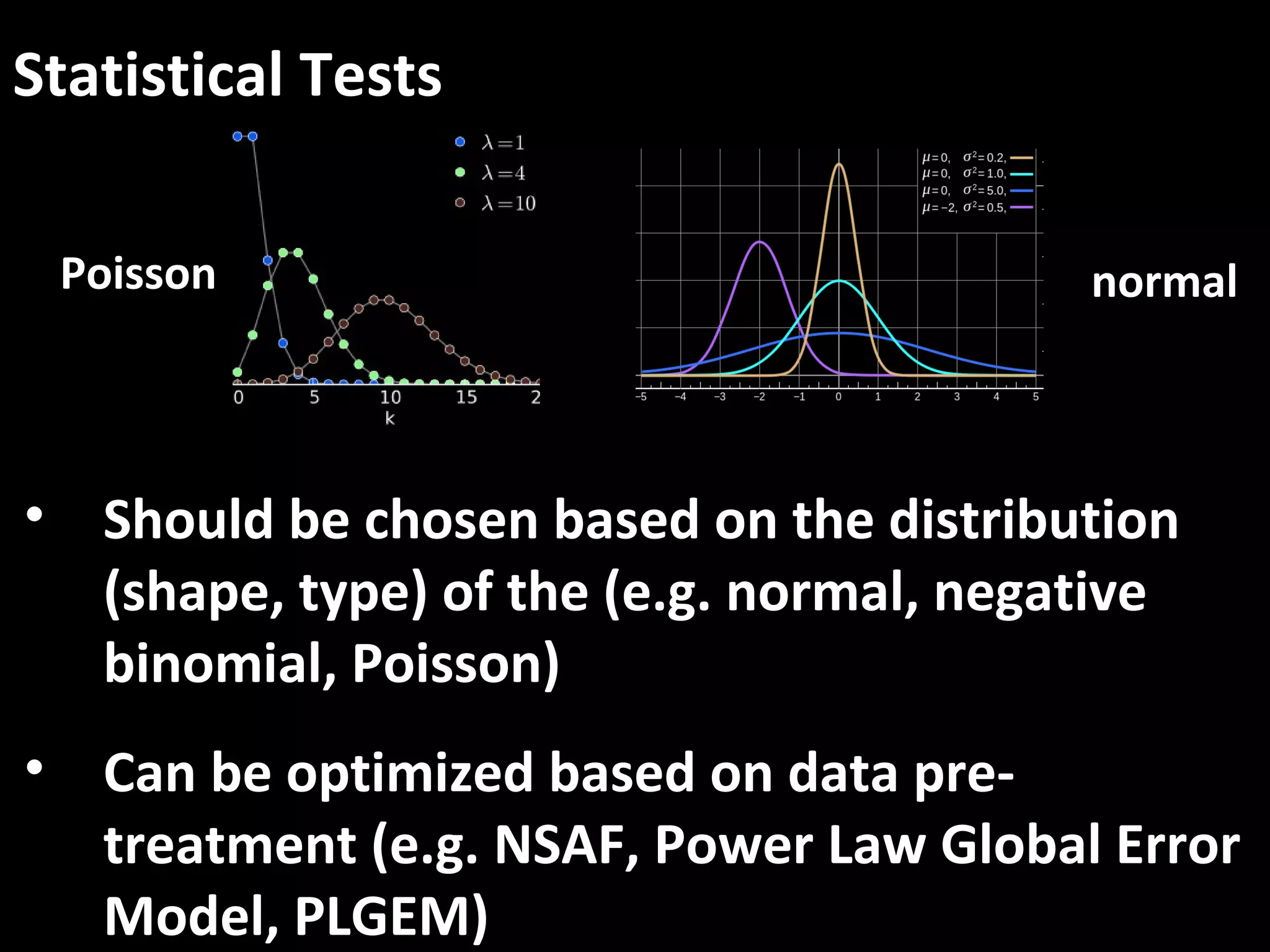
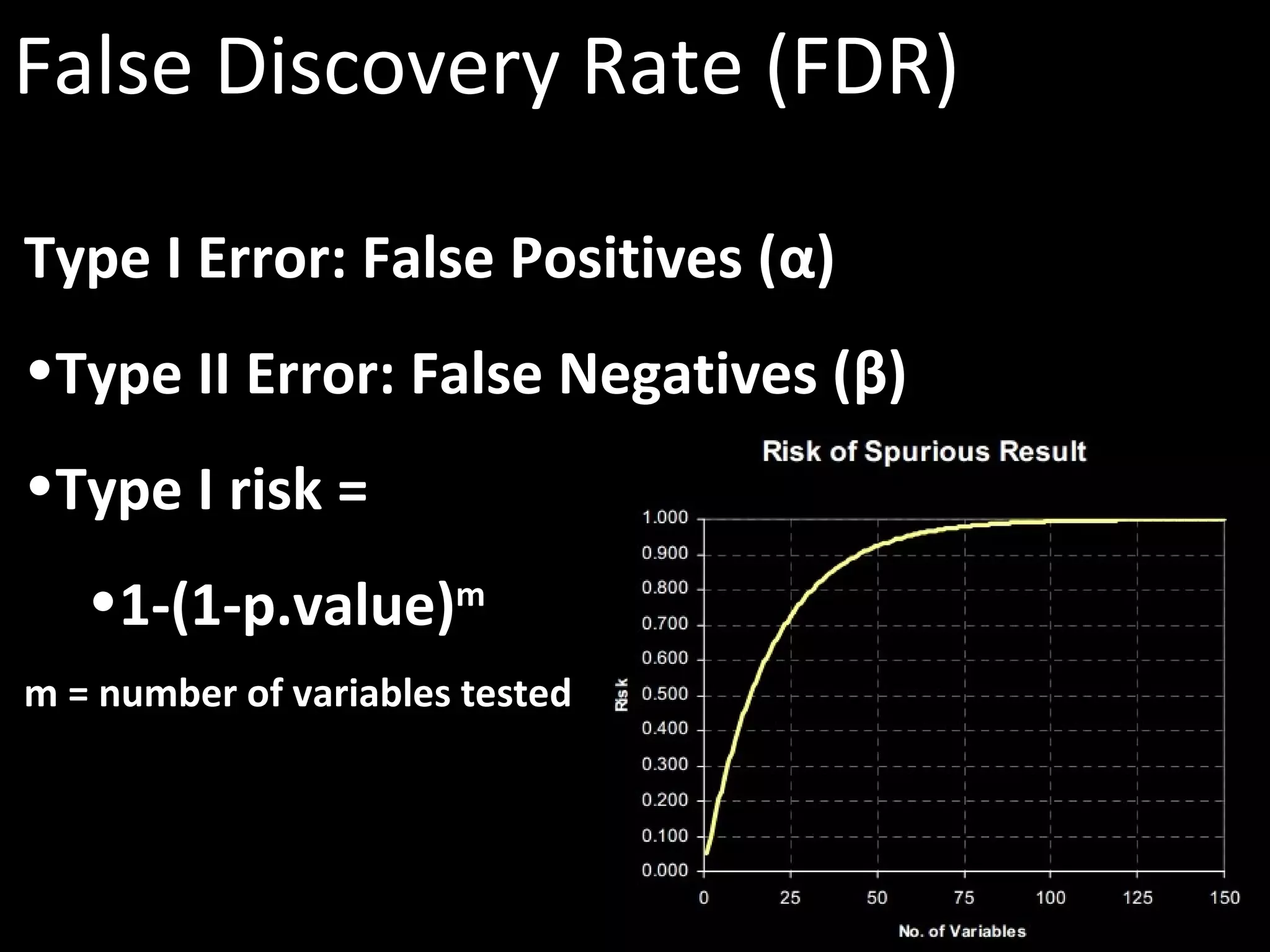
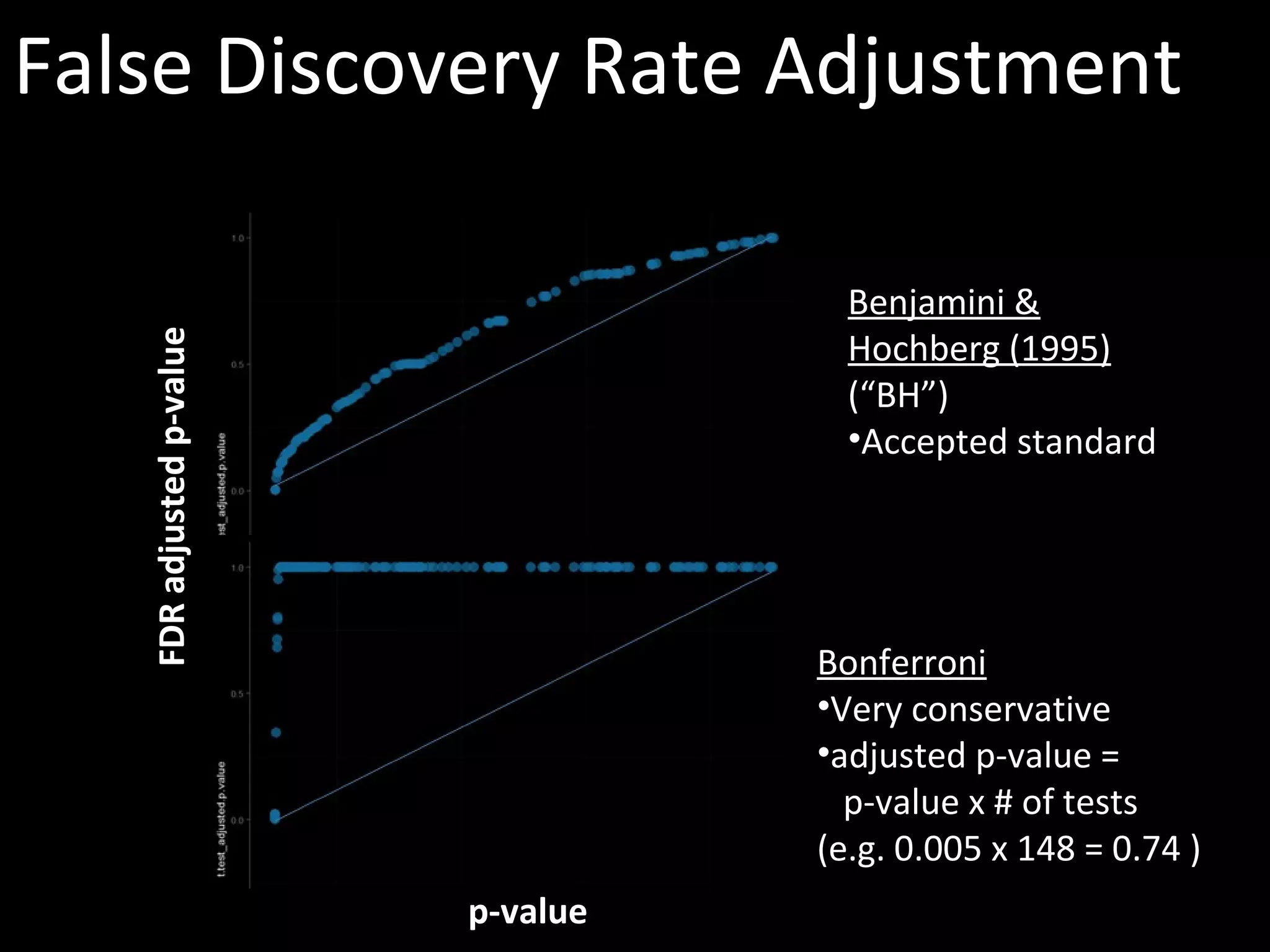
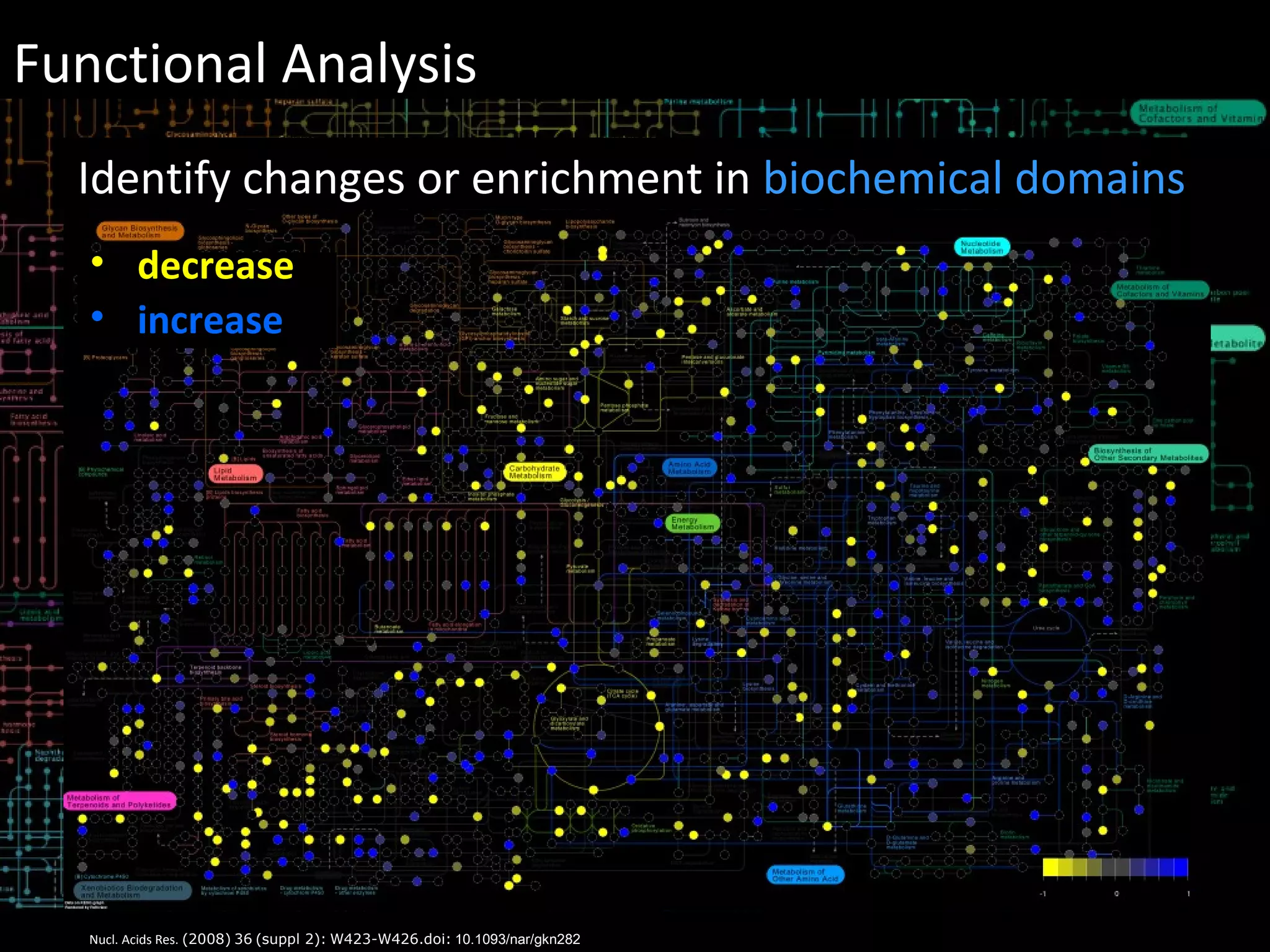
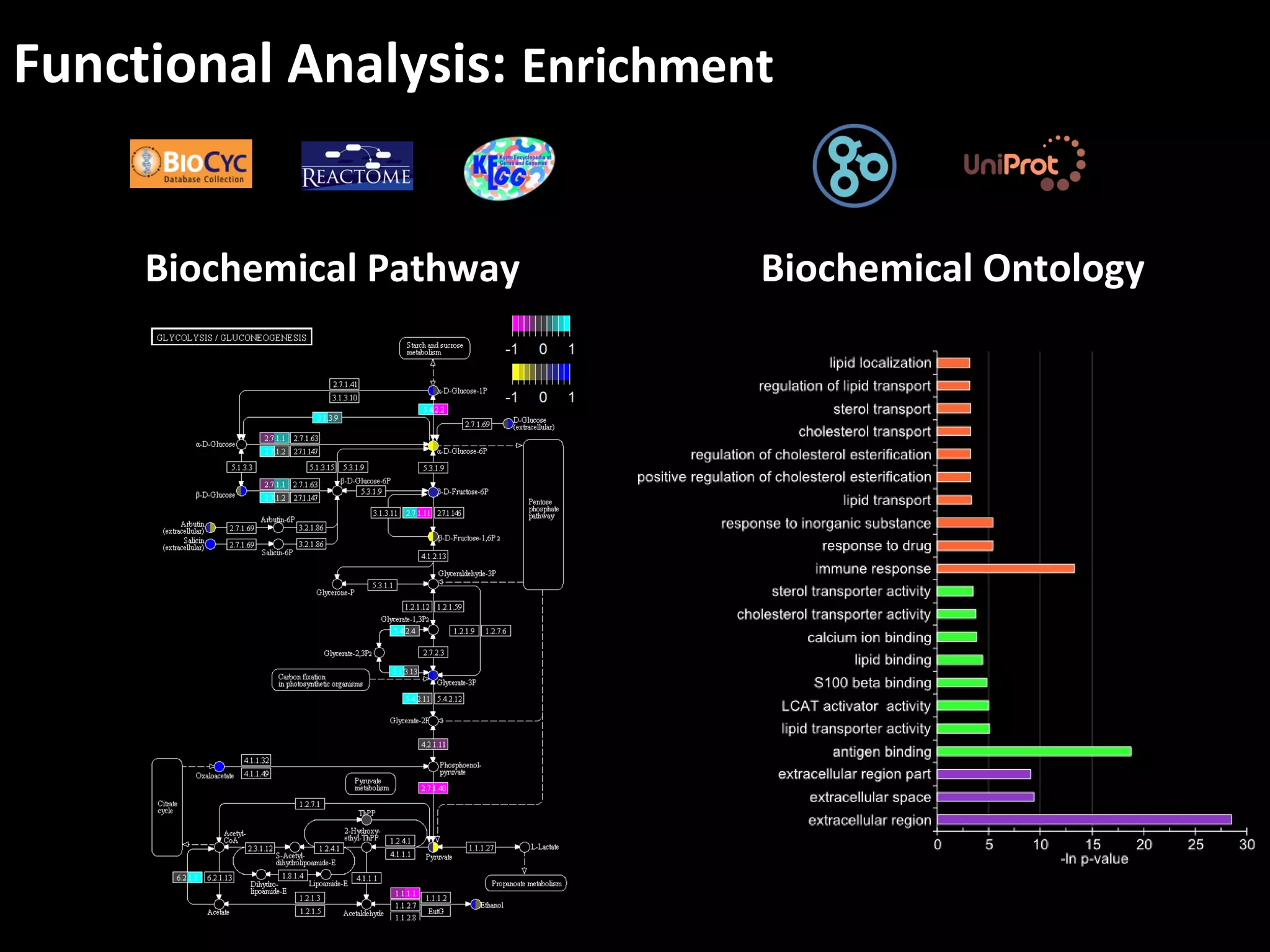
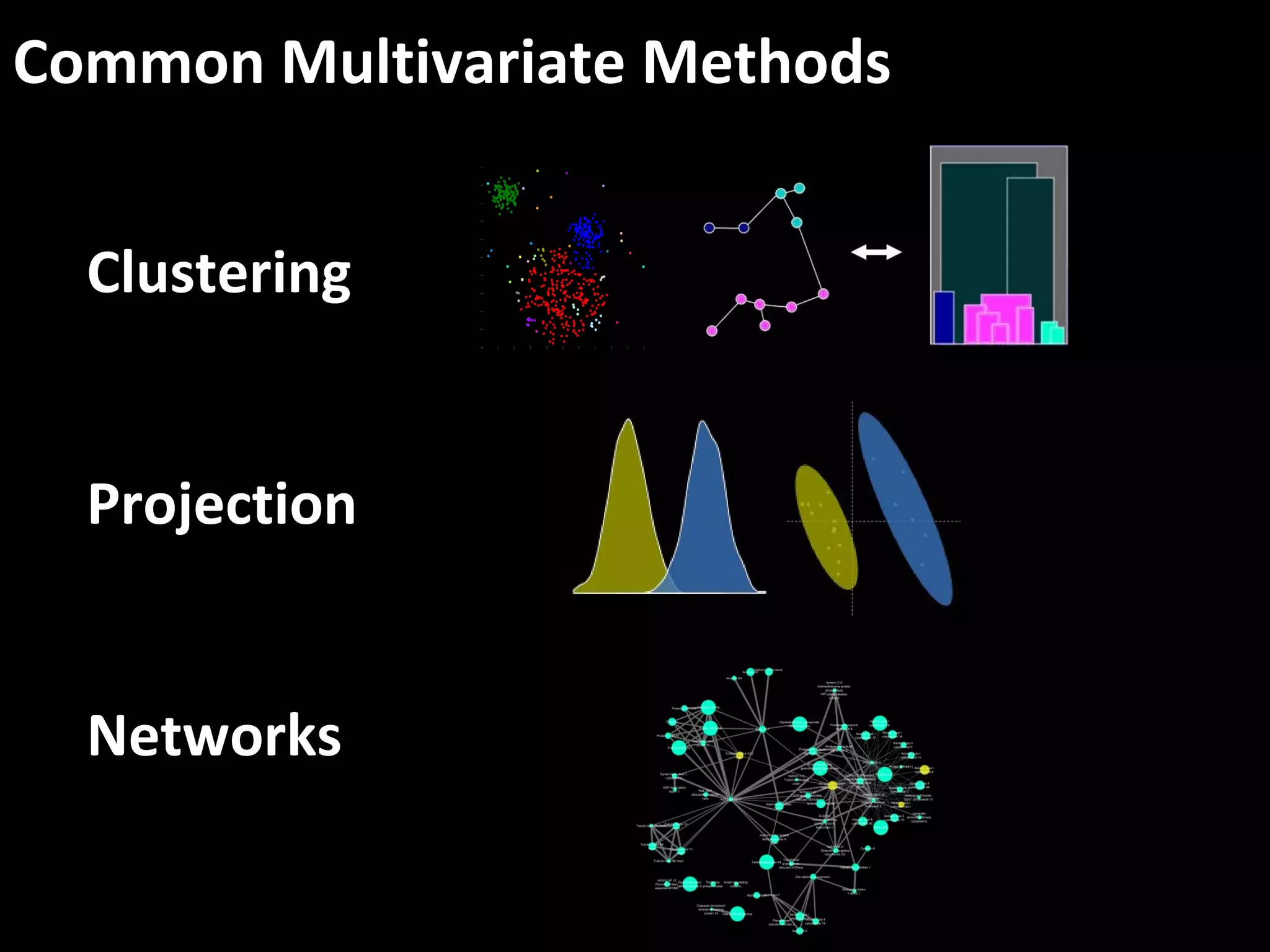
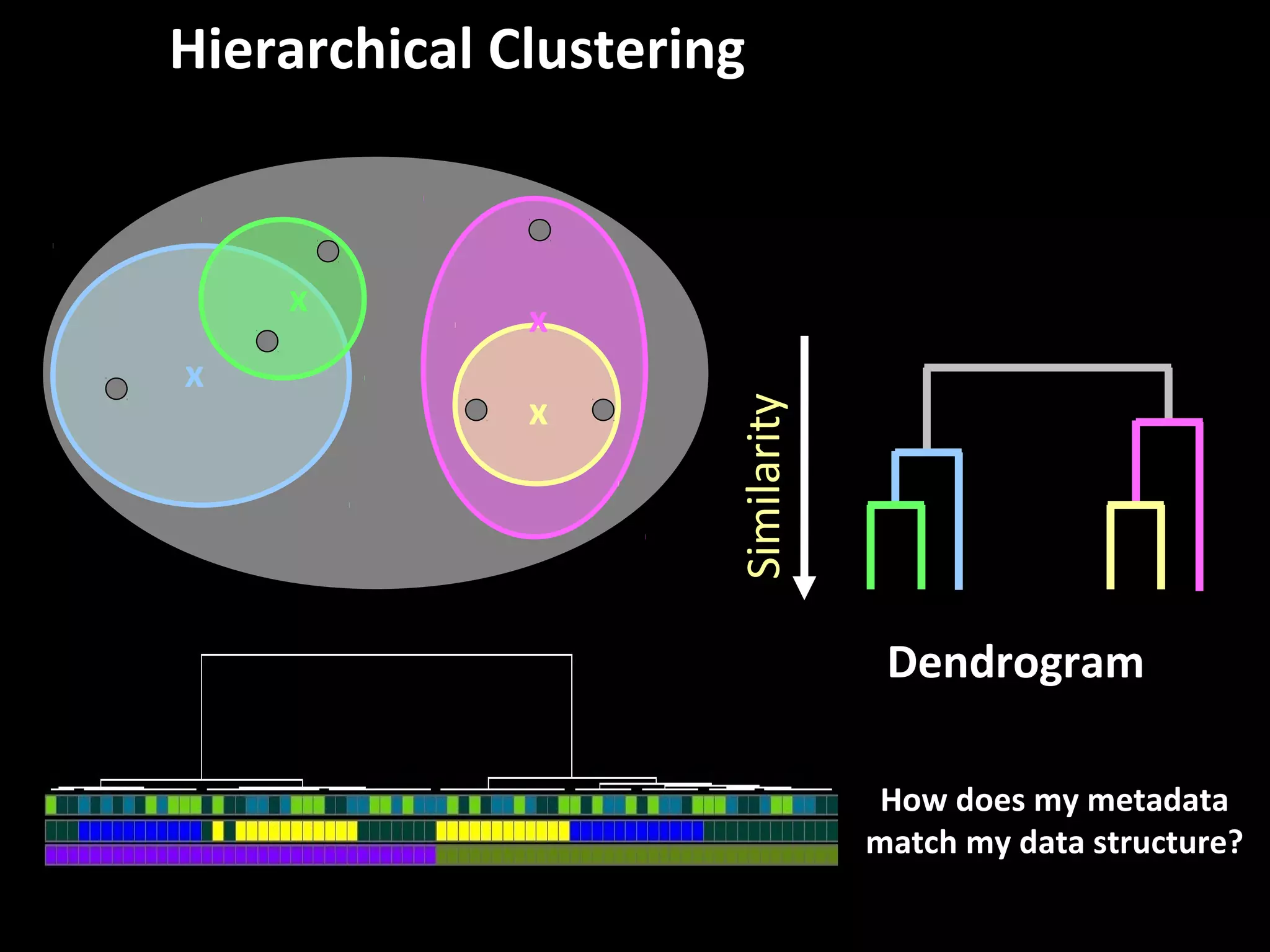
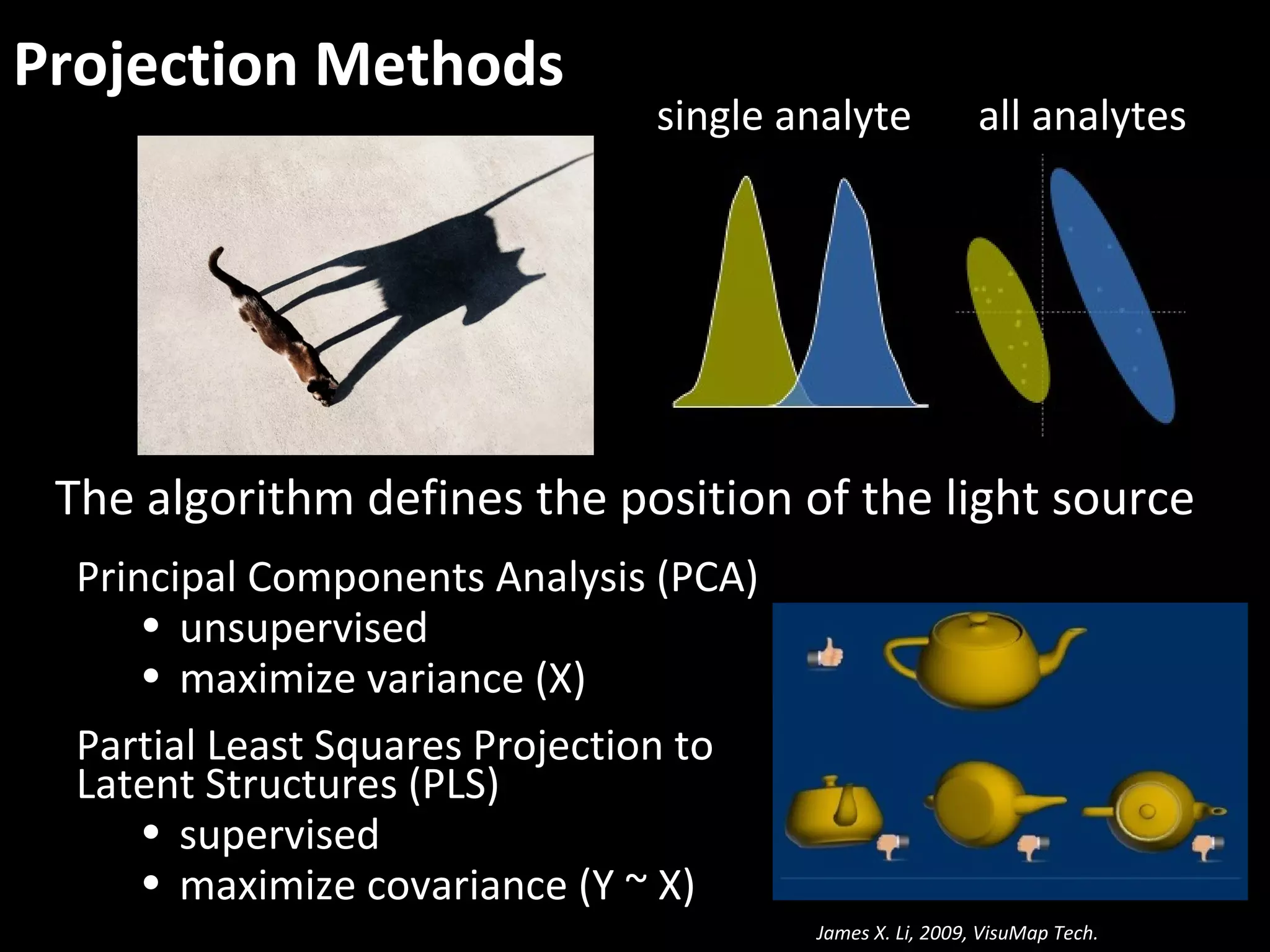
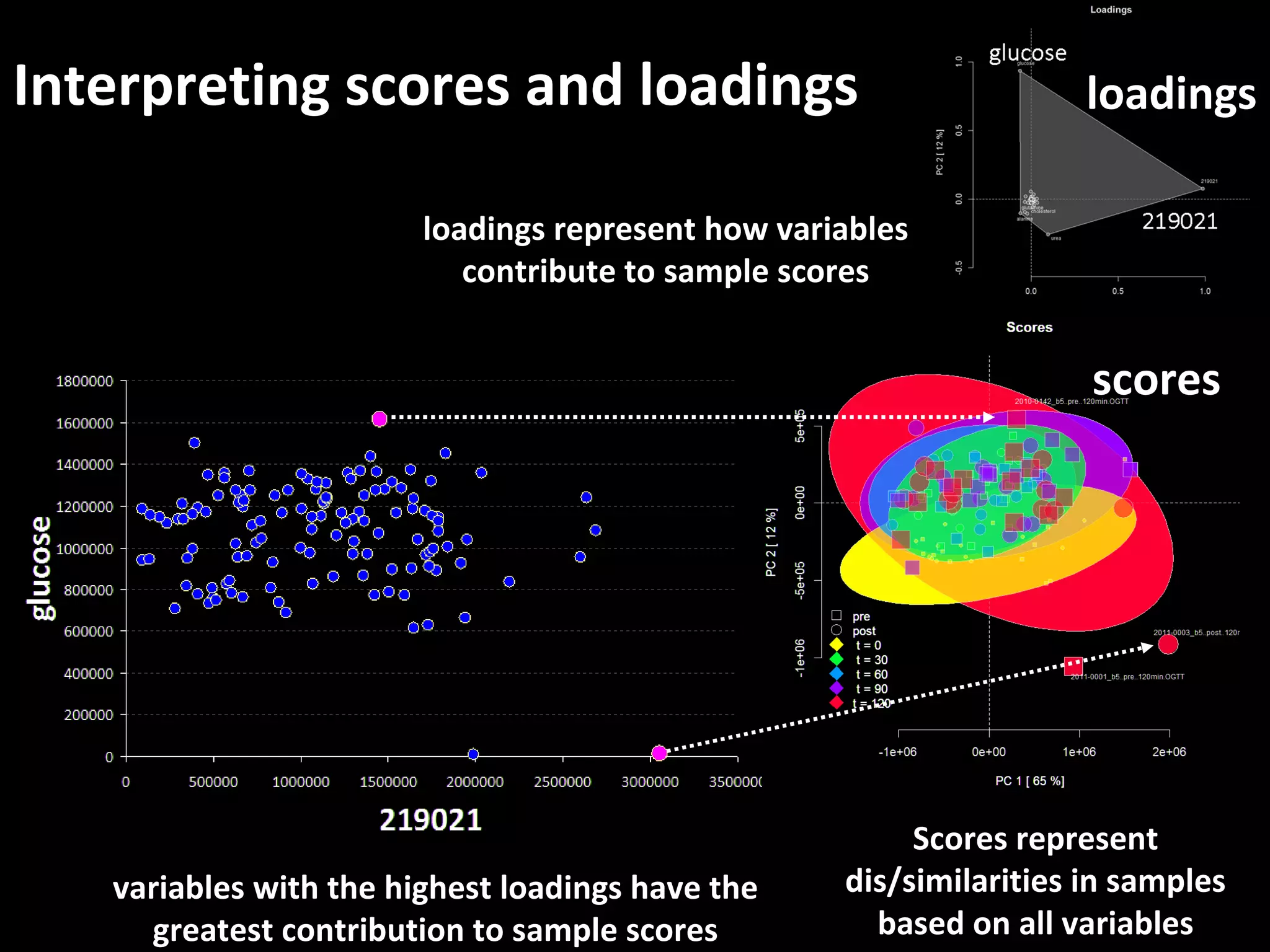
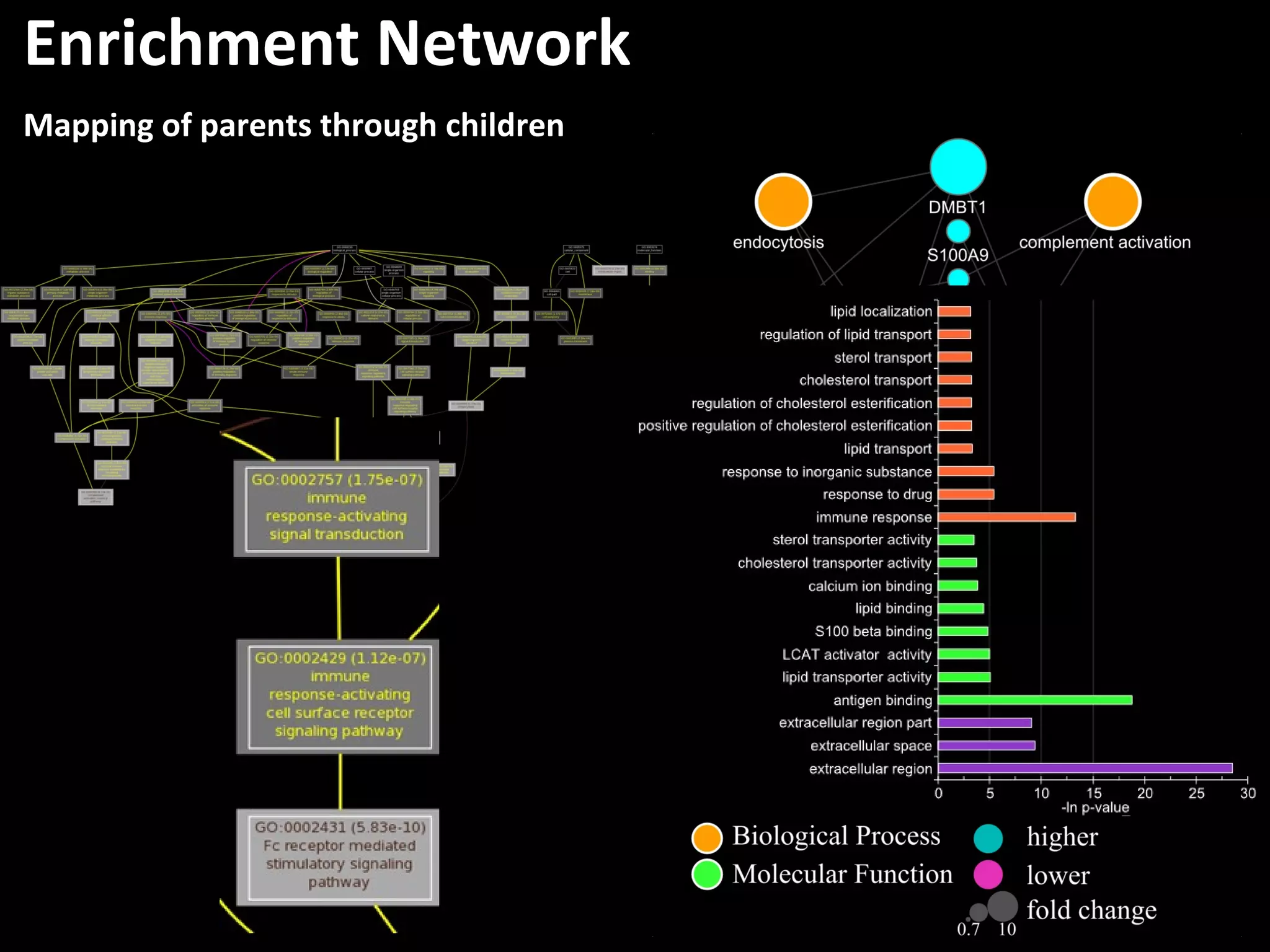
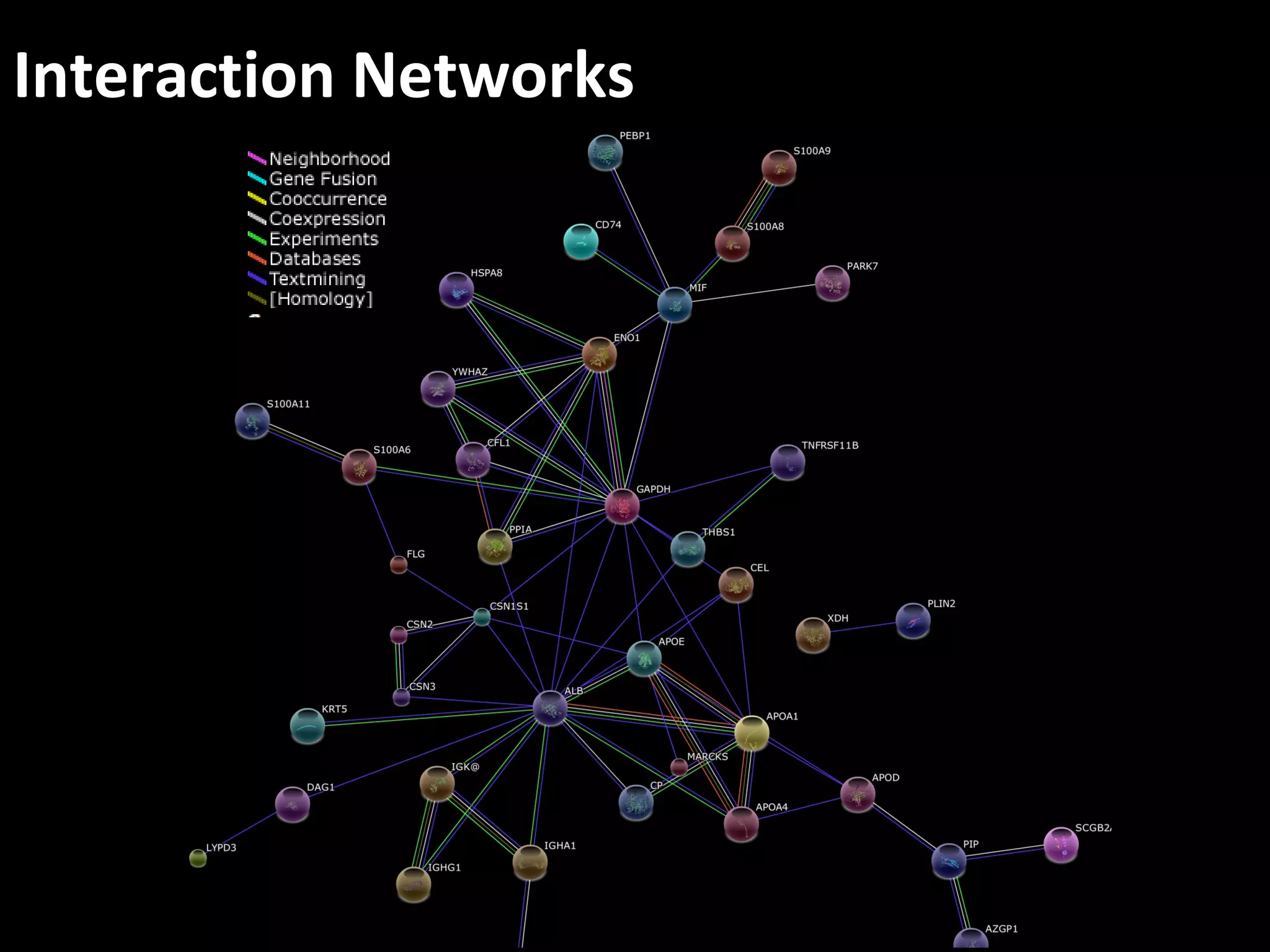
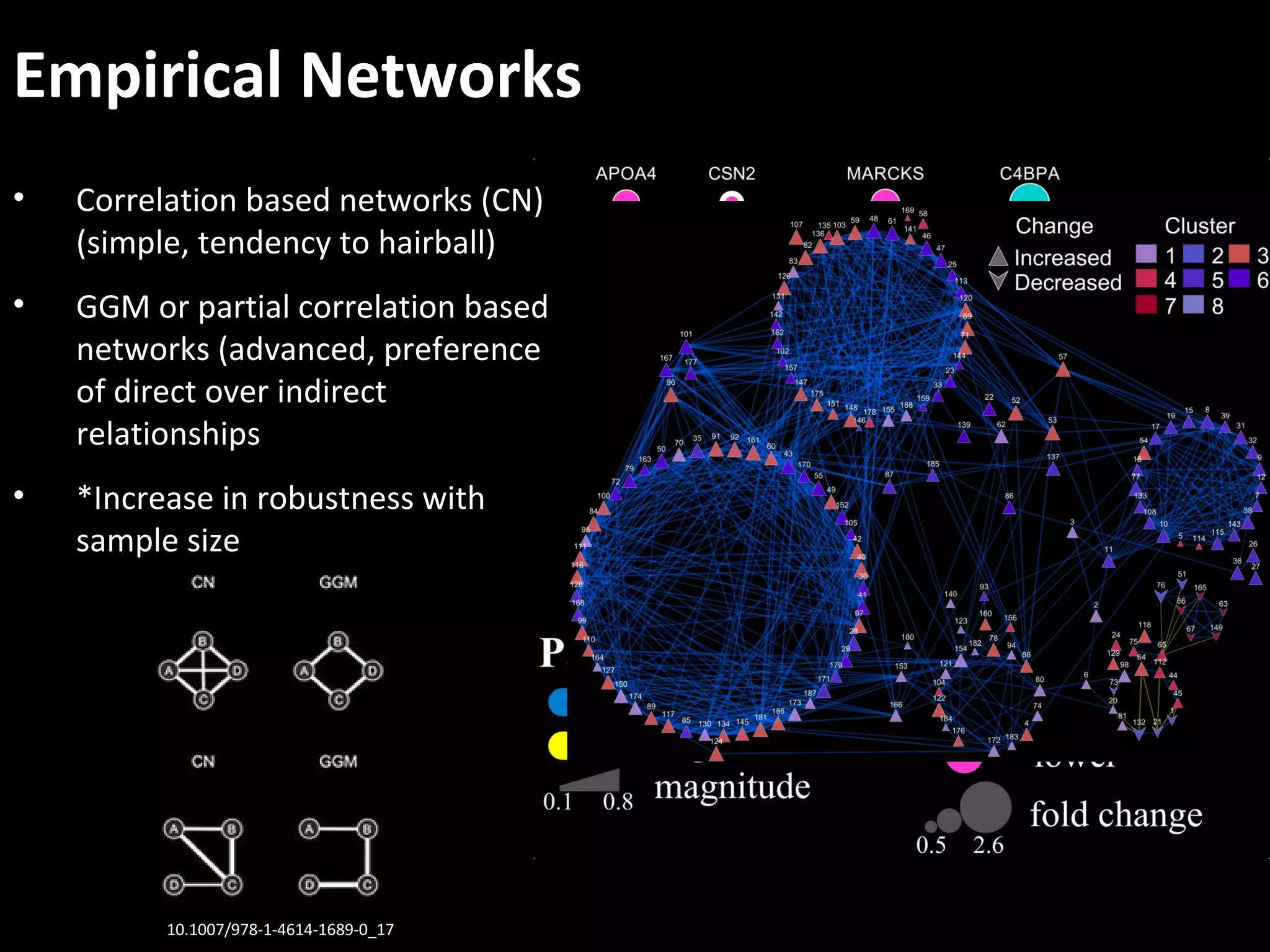
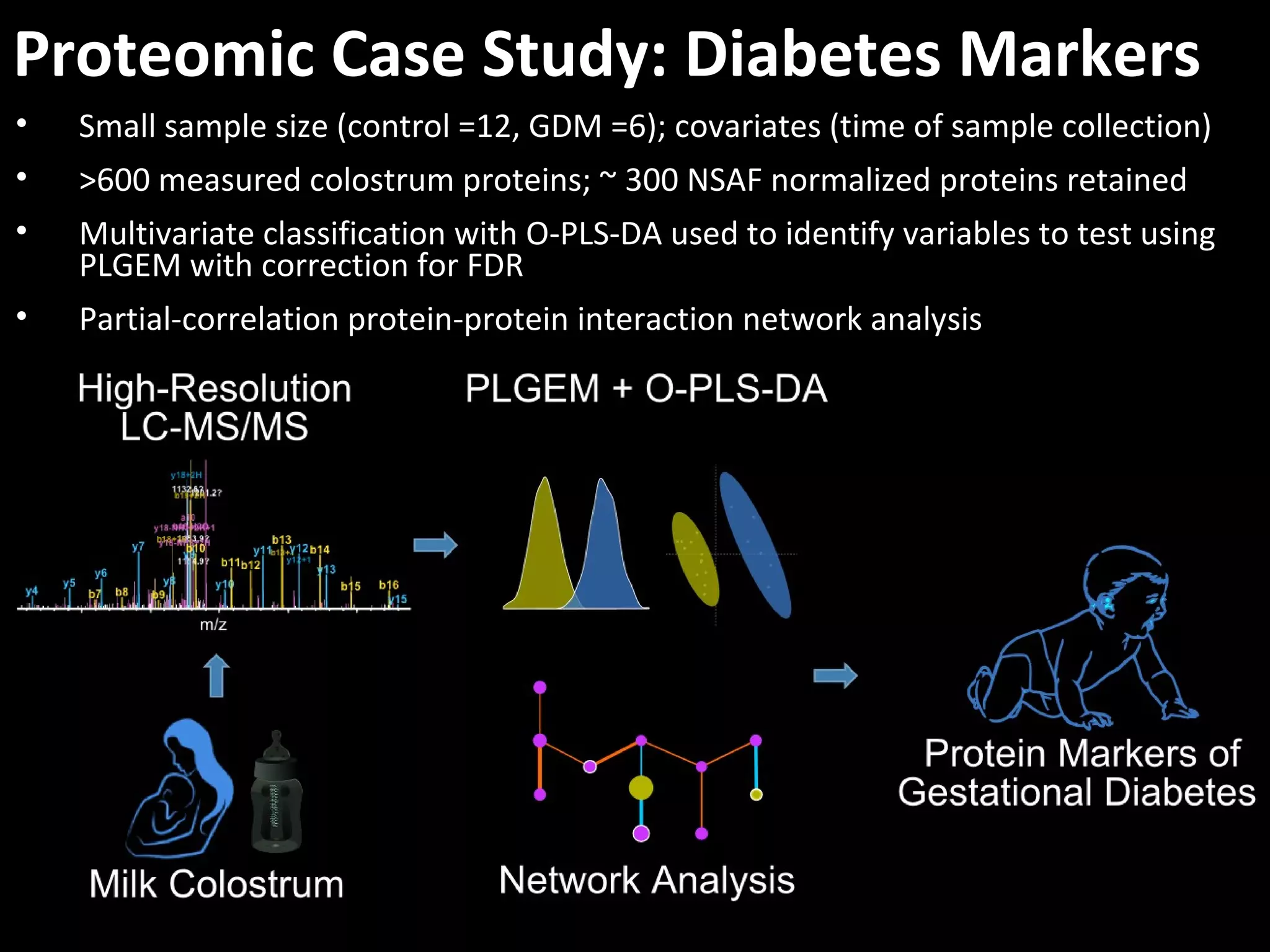
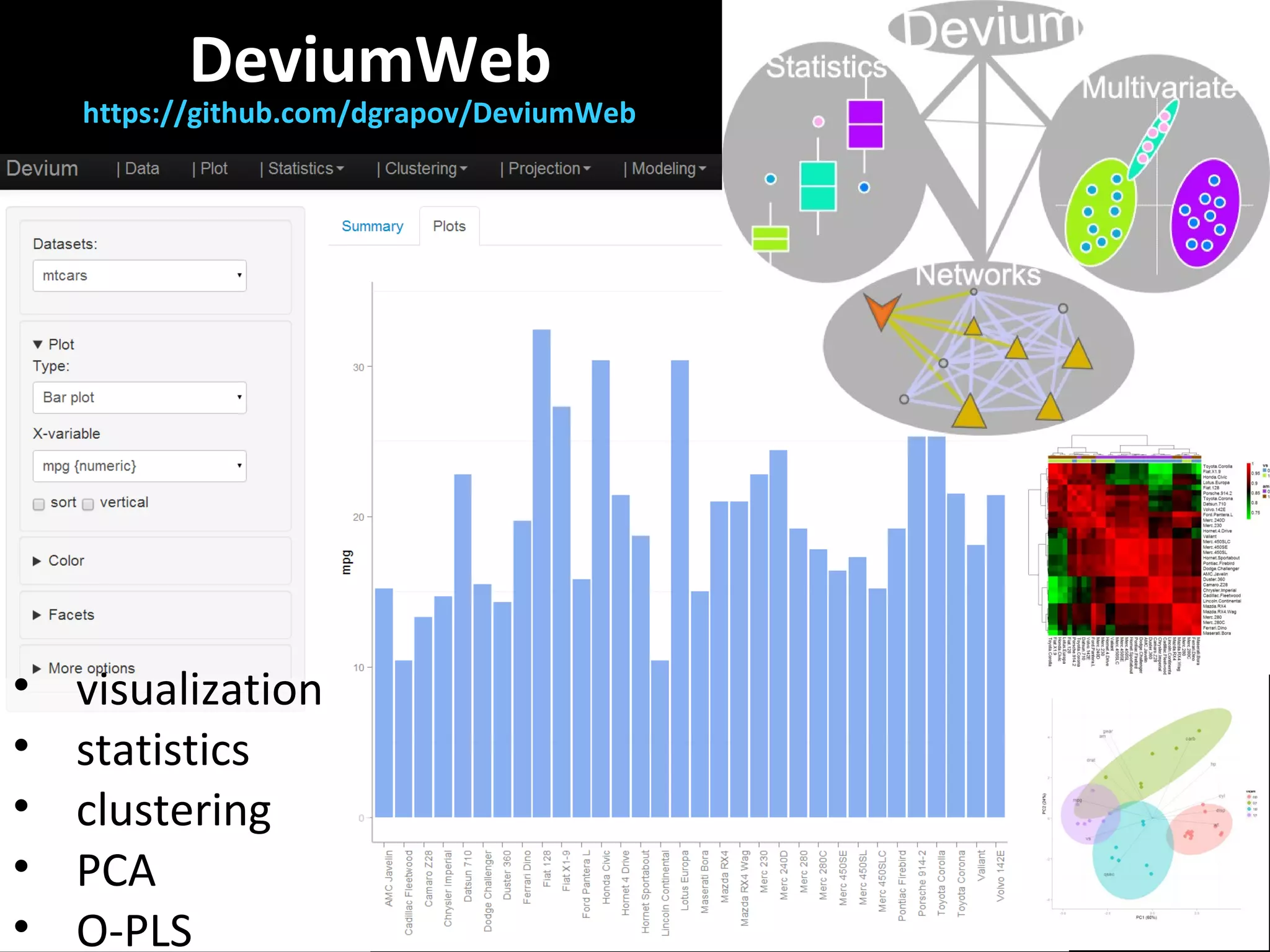
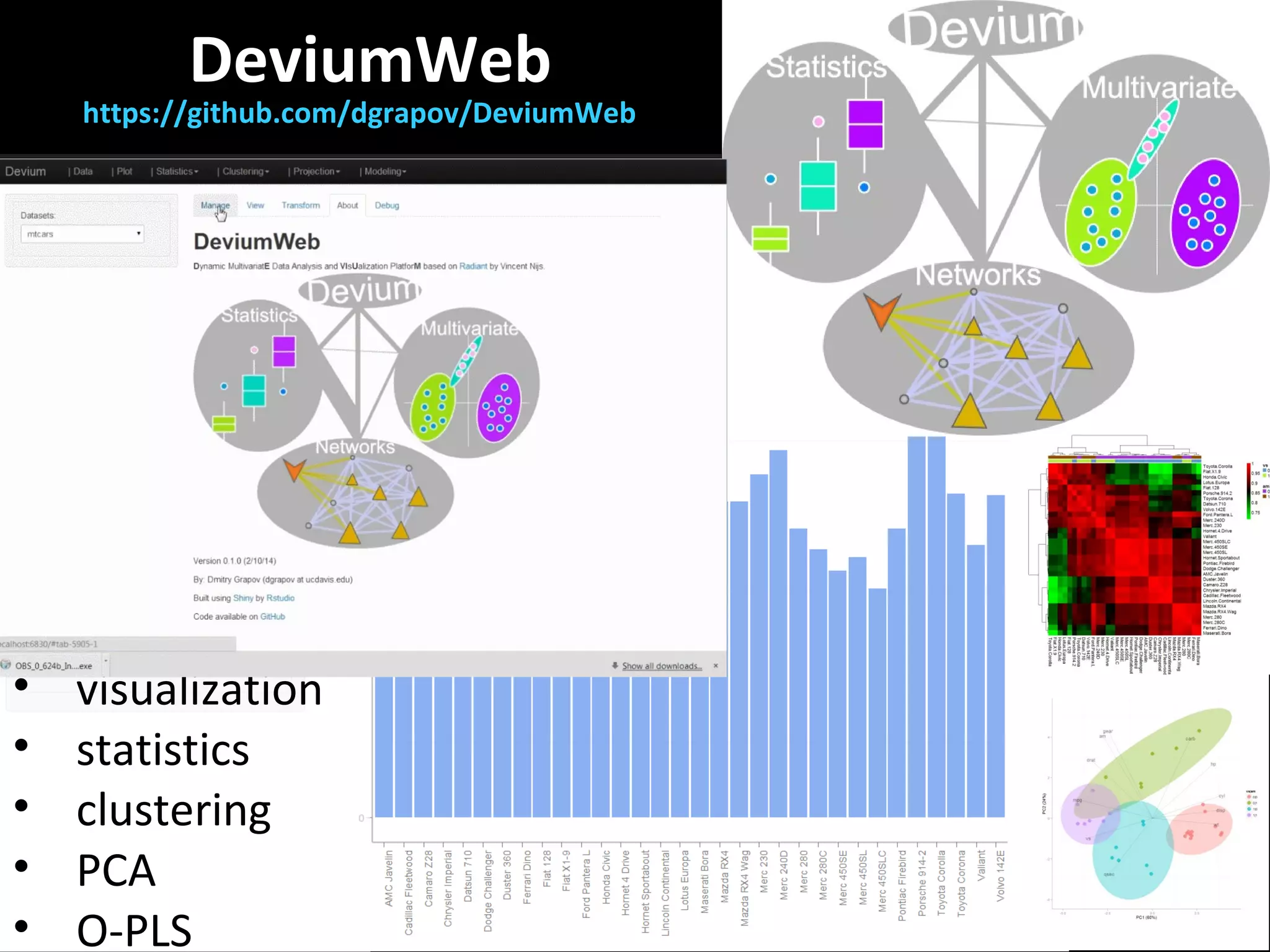
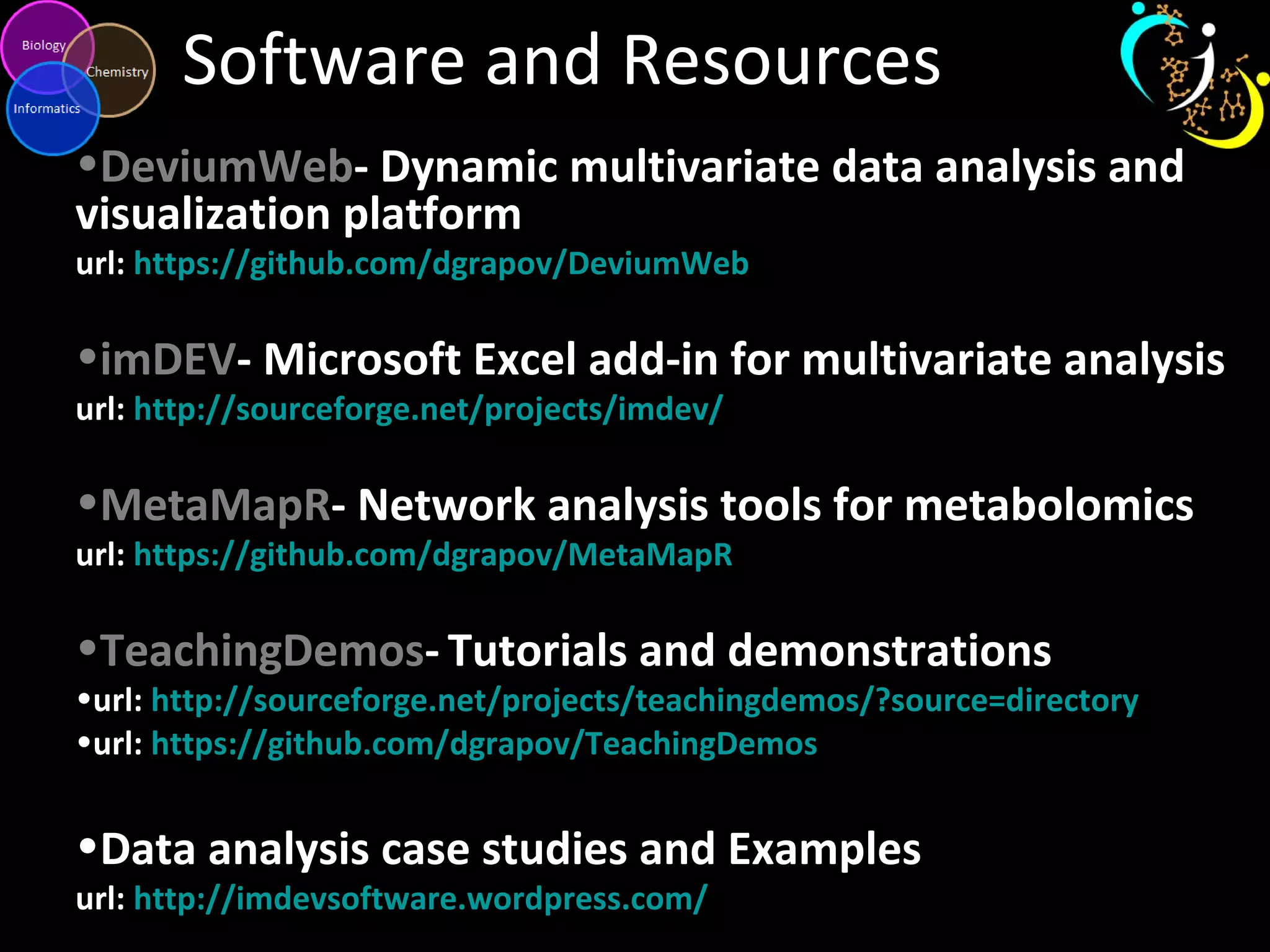
The document discusses advanced methodologies for multivariate analysis and visualization of proteomic data in a state-of-the-art facility, managing significant biological data outputs. It covers various statistical techniques, data quality assessments, experimental designs, and provides insights into analytical variability and significance testing for effective data interpretation. Additionally, it introduces software tools for dynamic data analysis and visualization, aiding researchers in extracting meaningful biological insights from complex datasets.