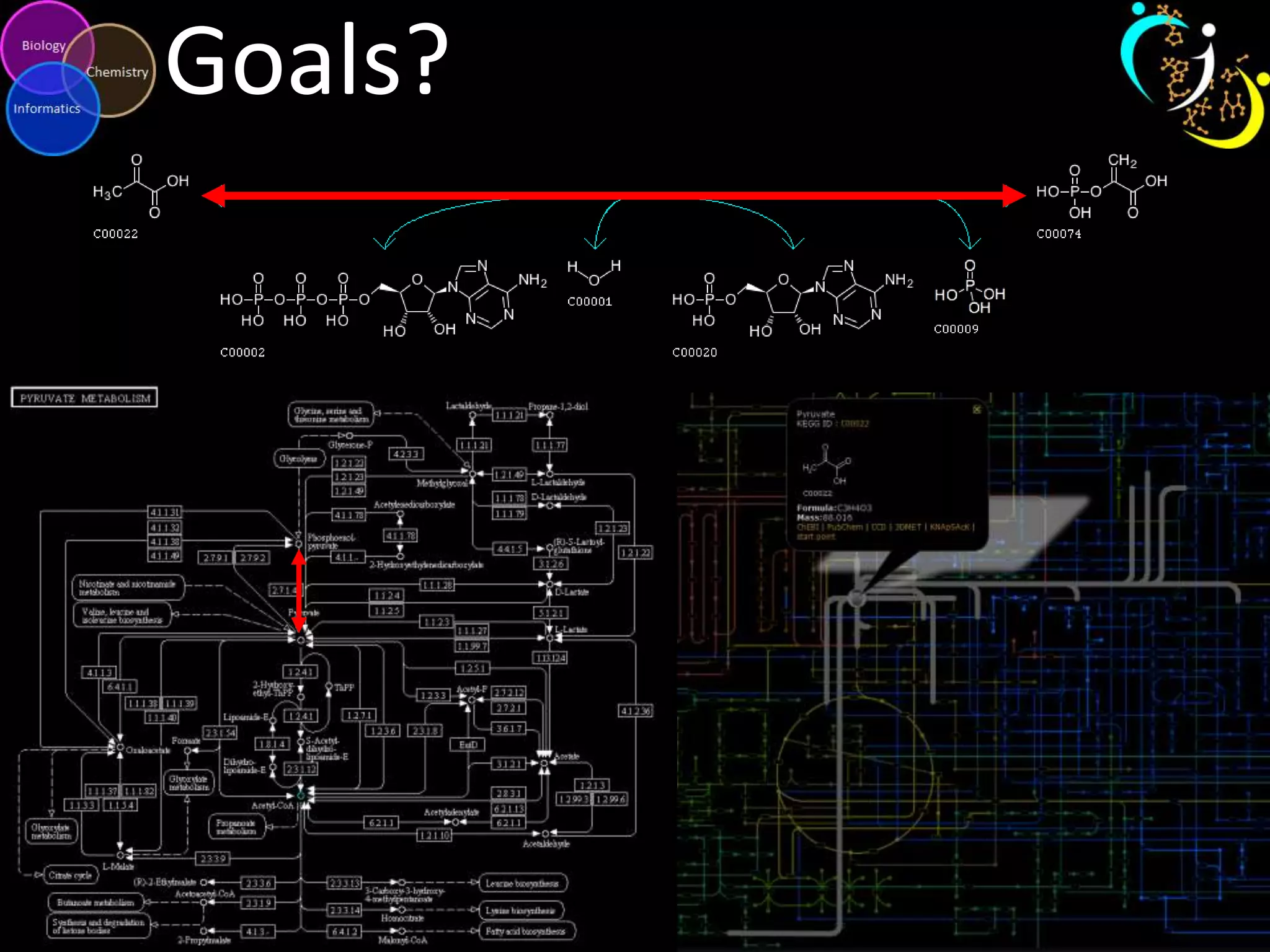
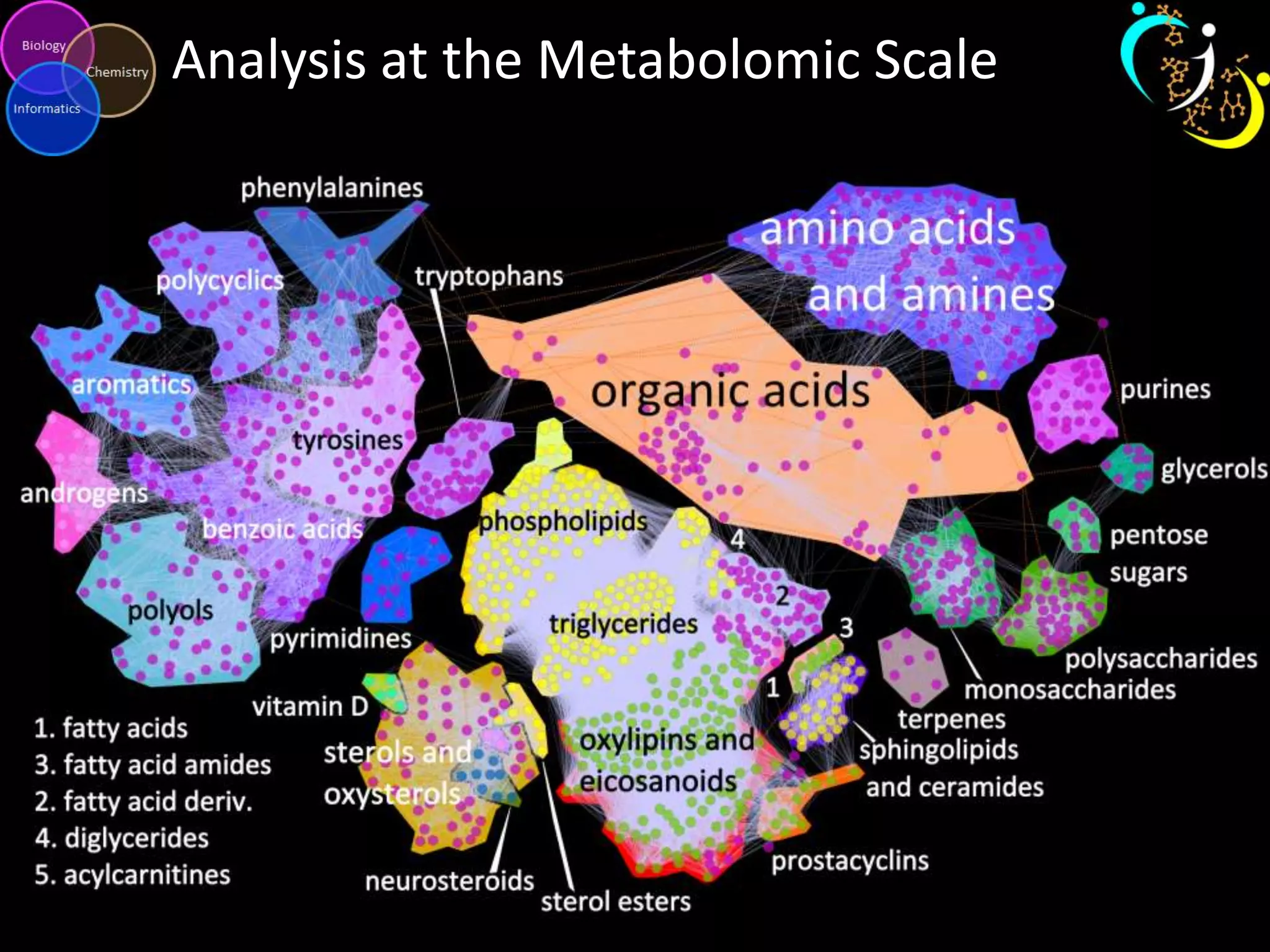
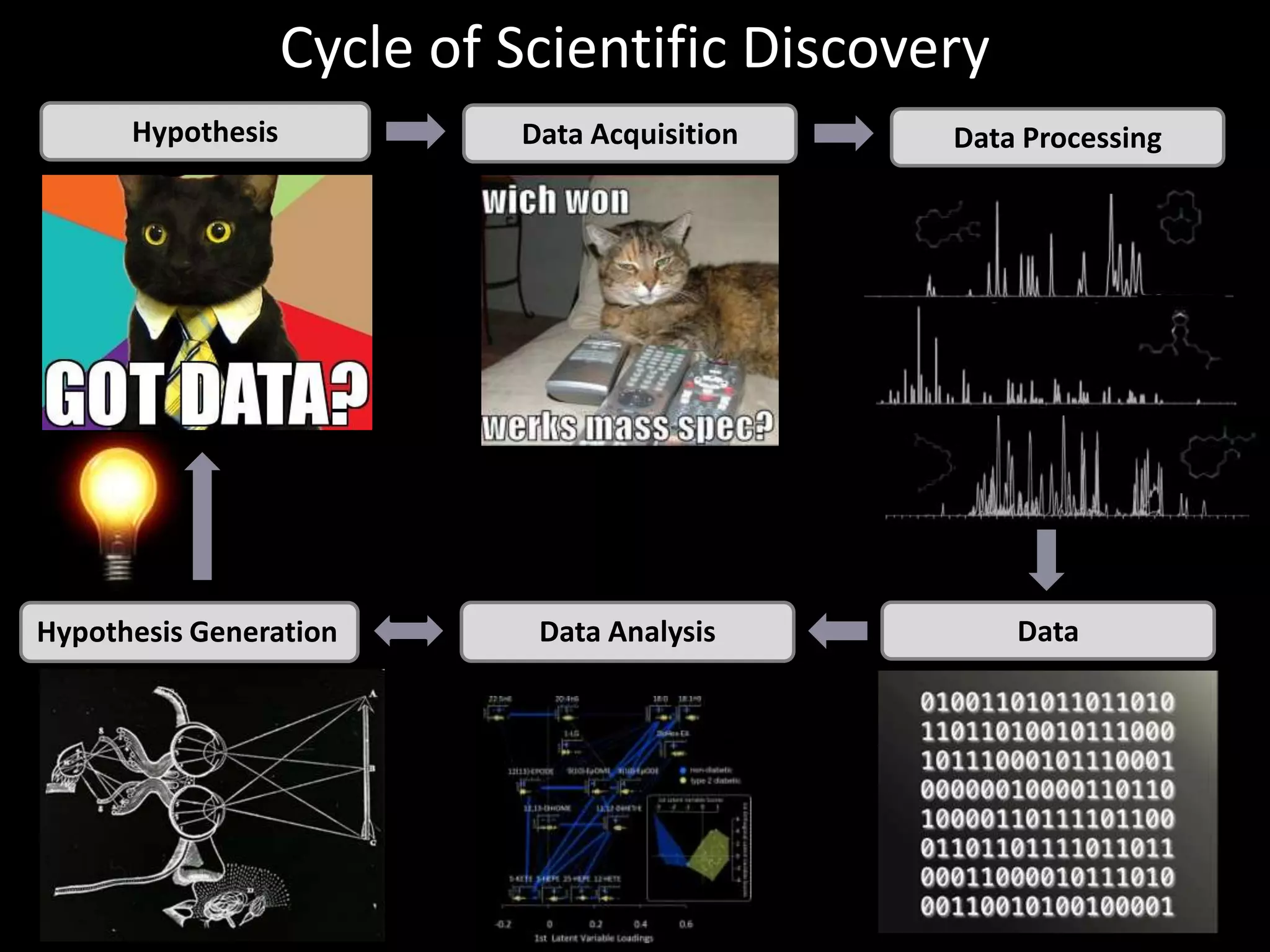
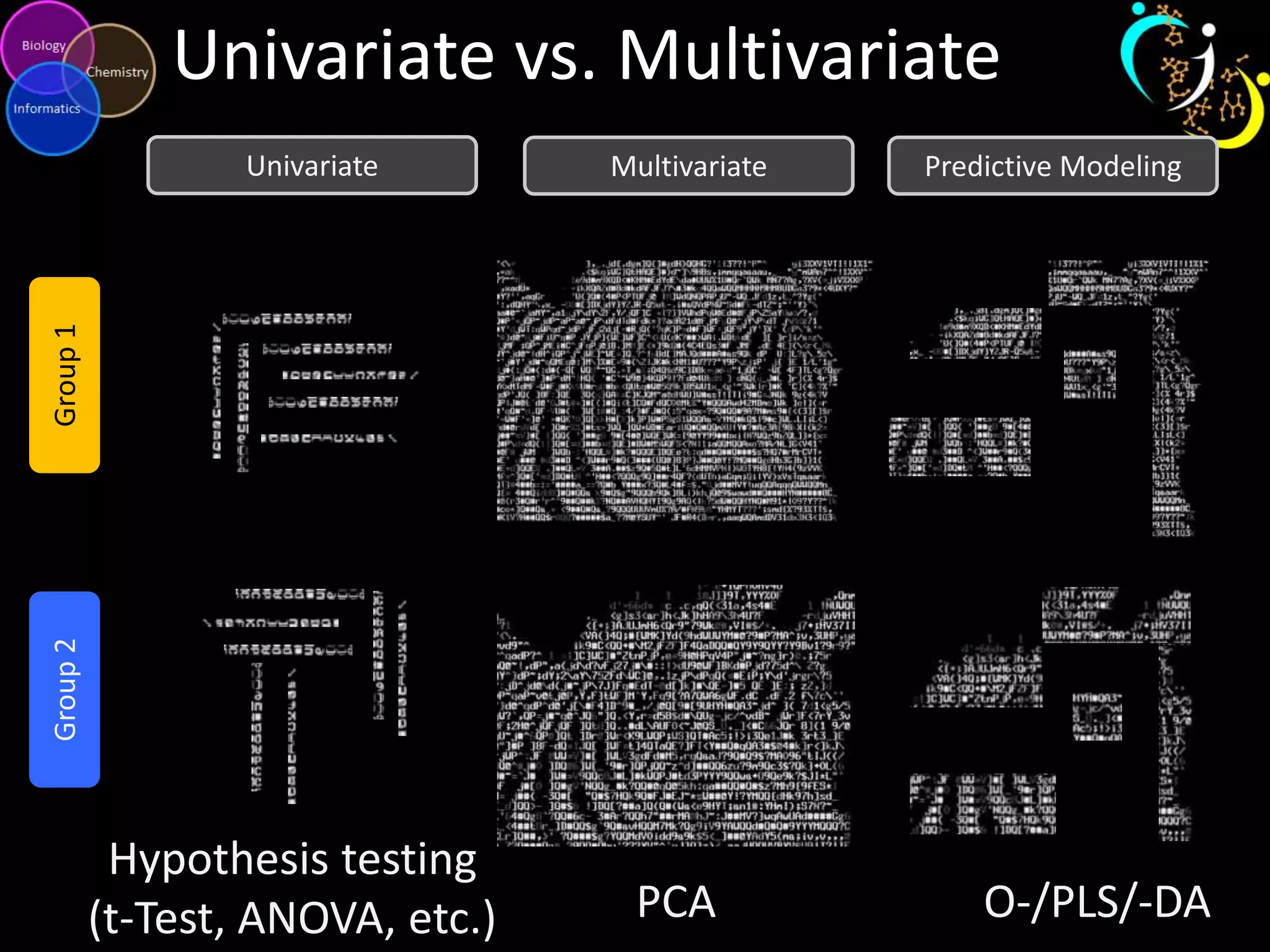
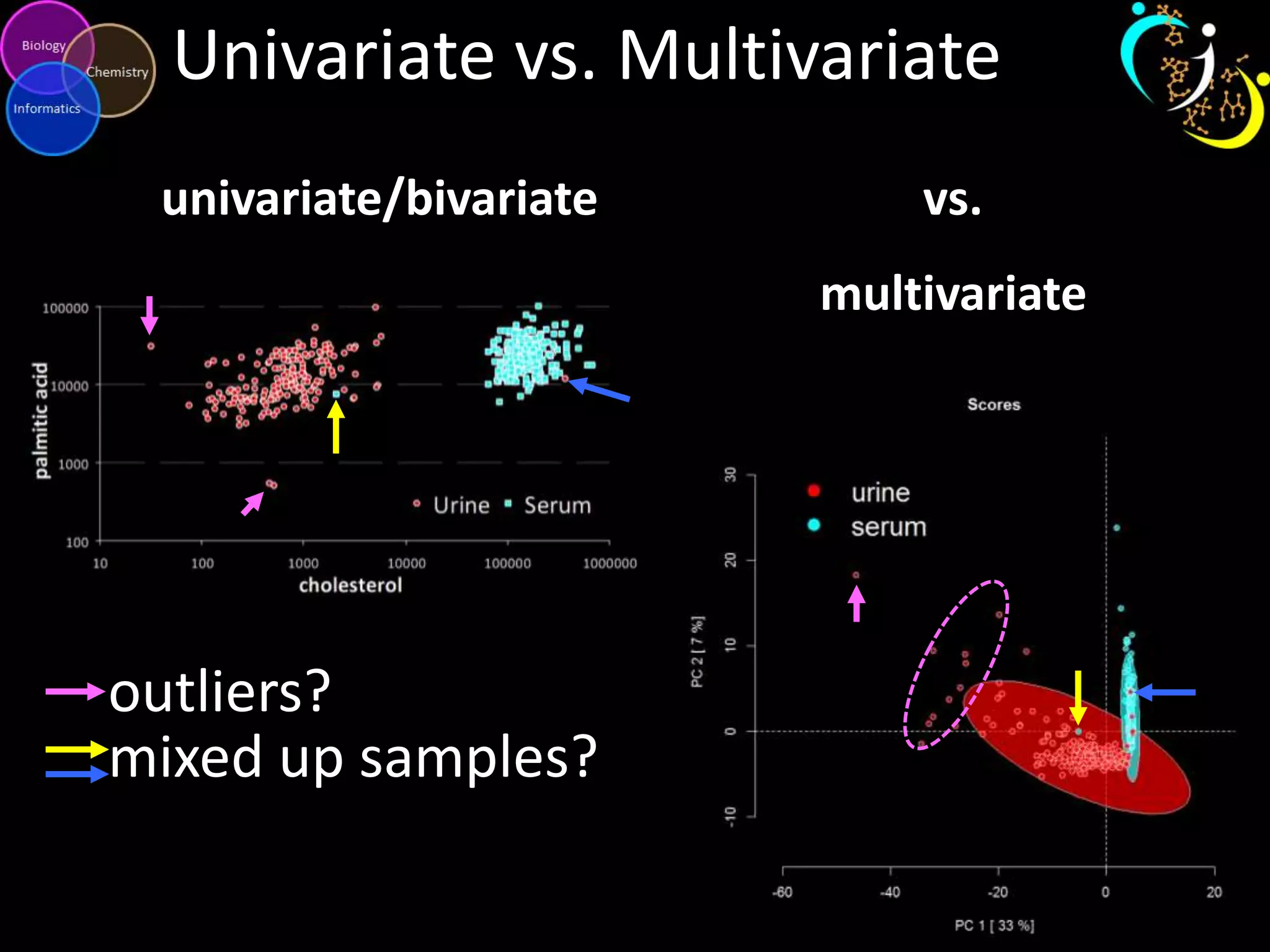
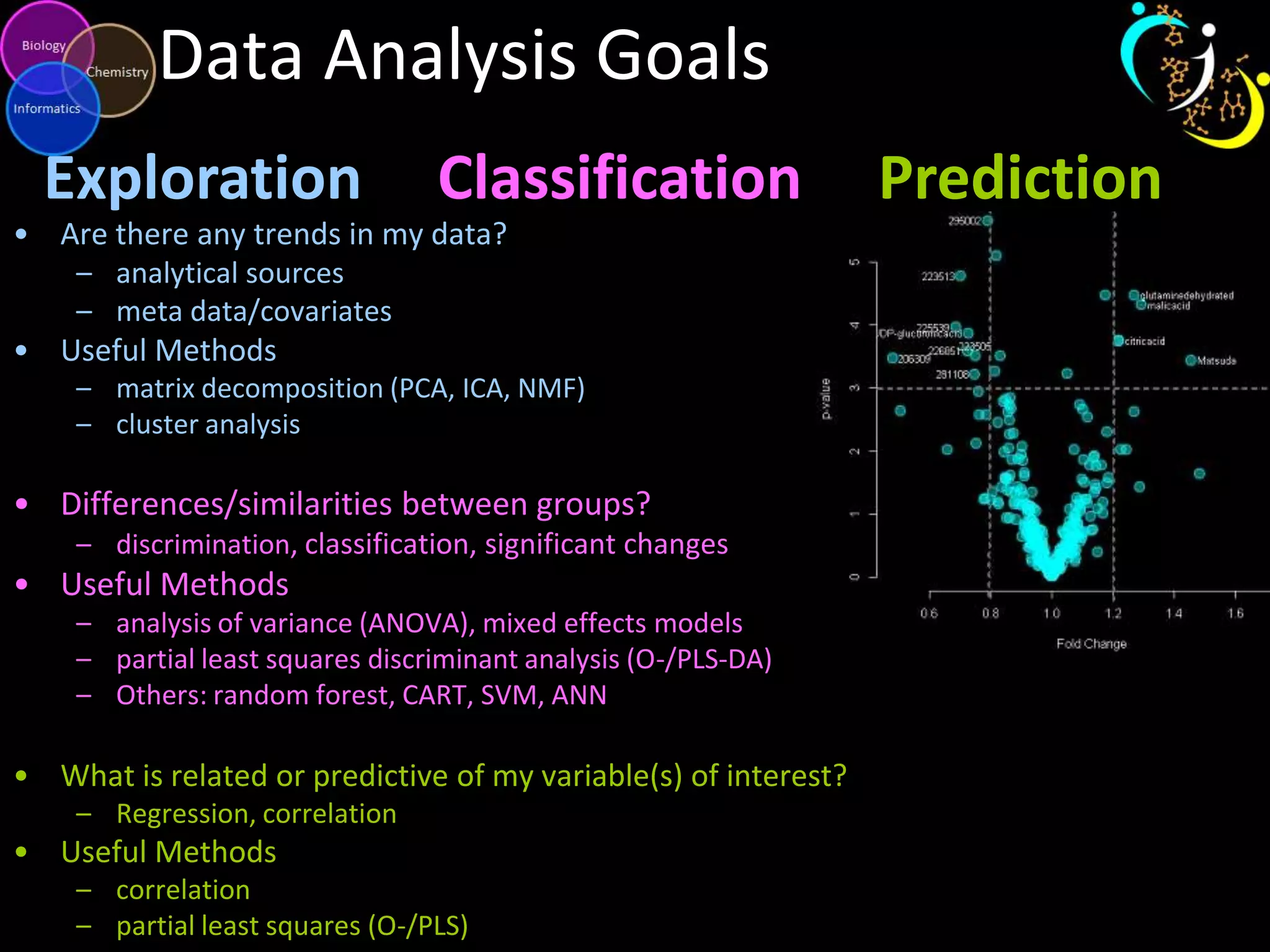
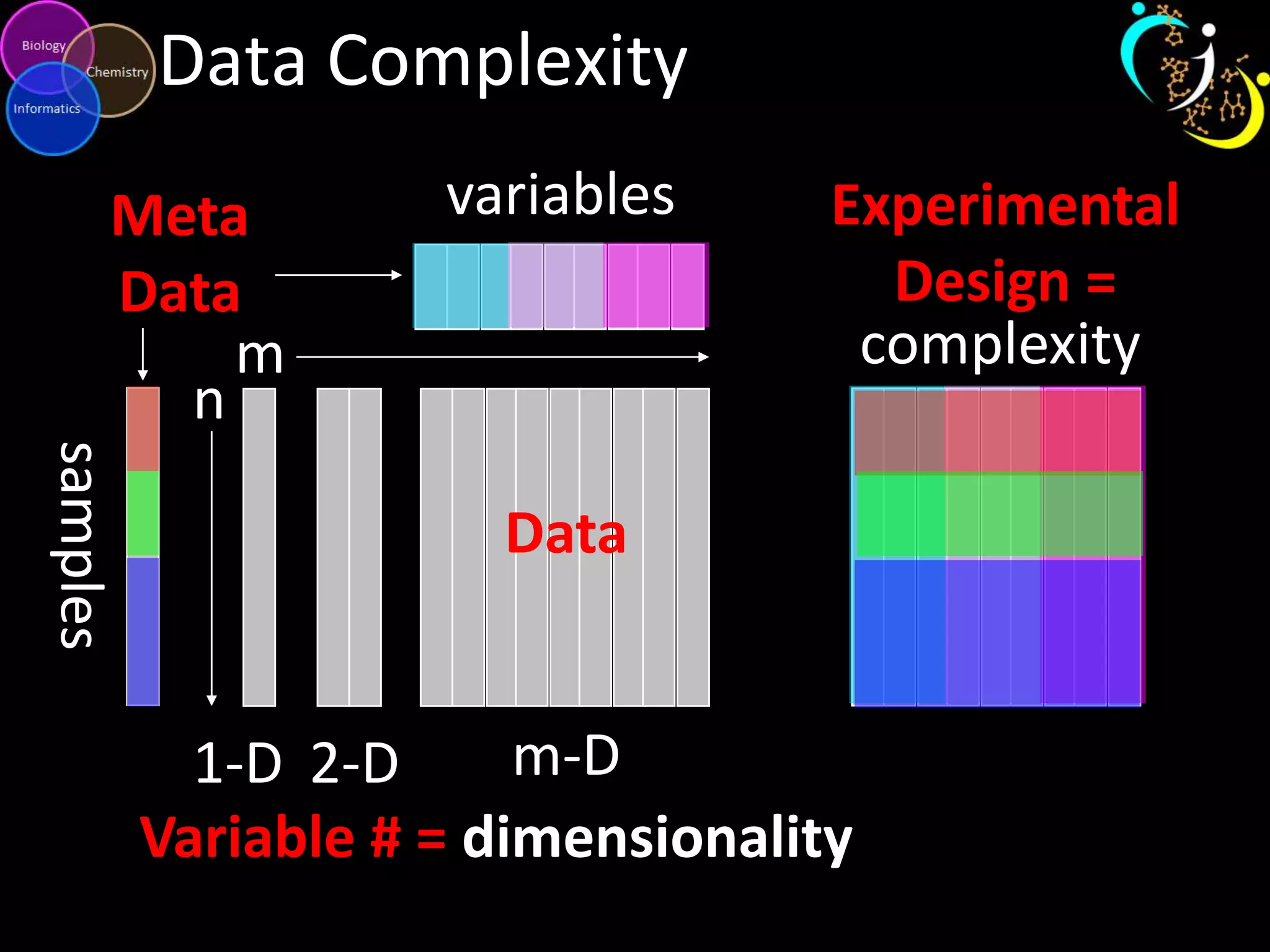
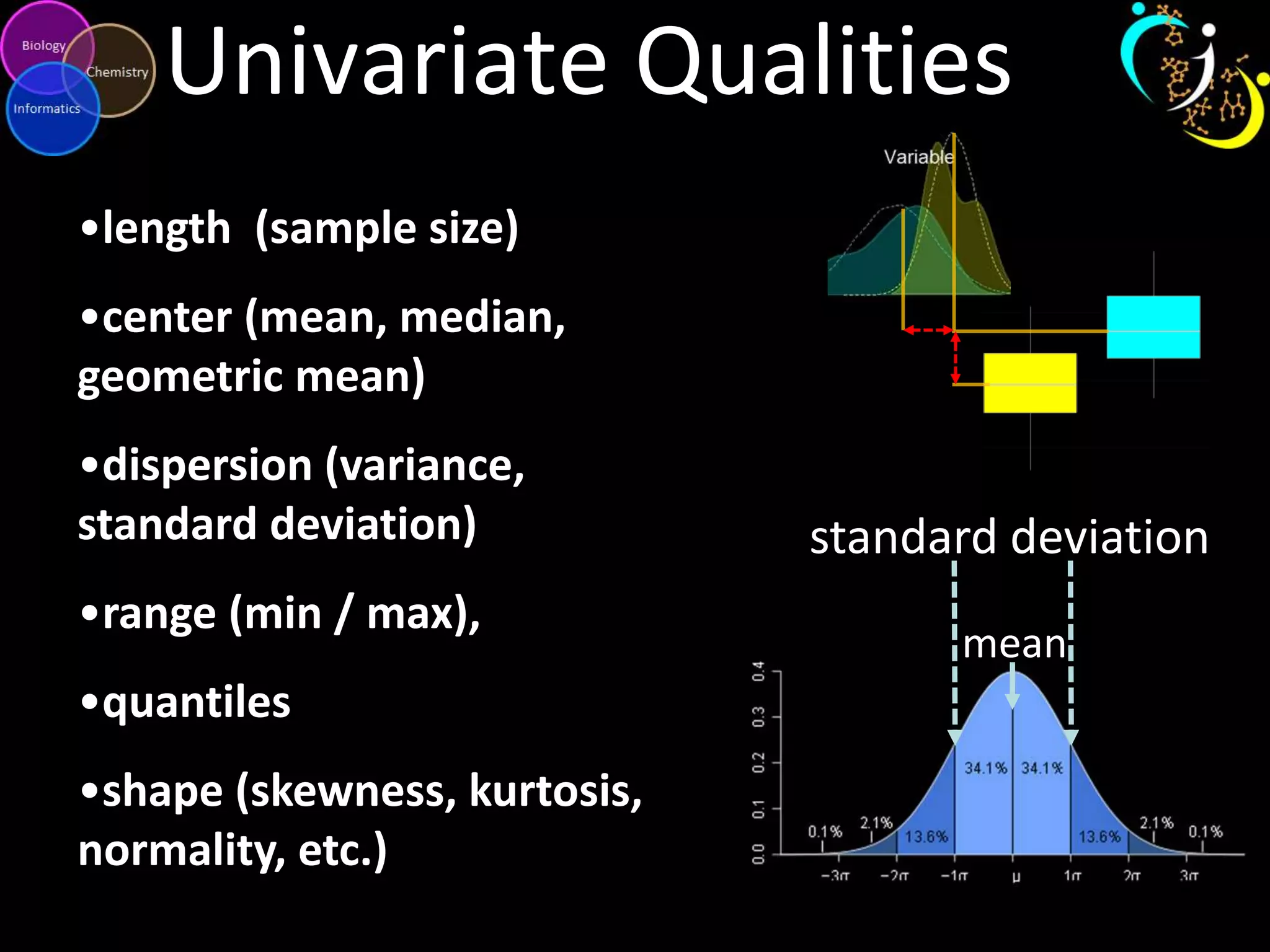
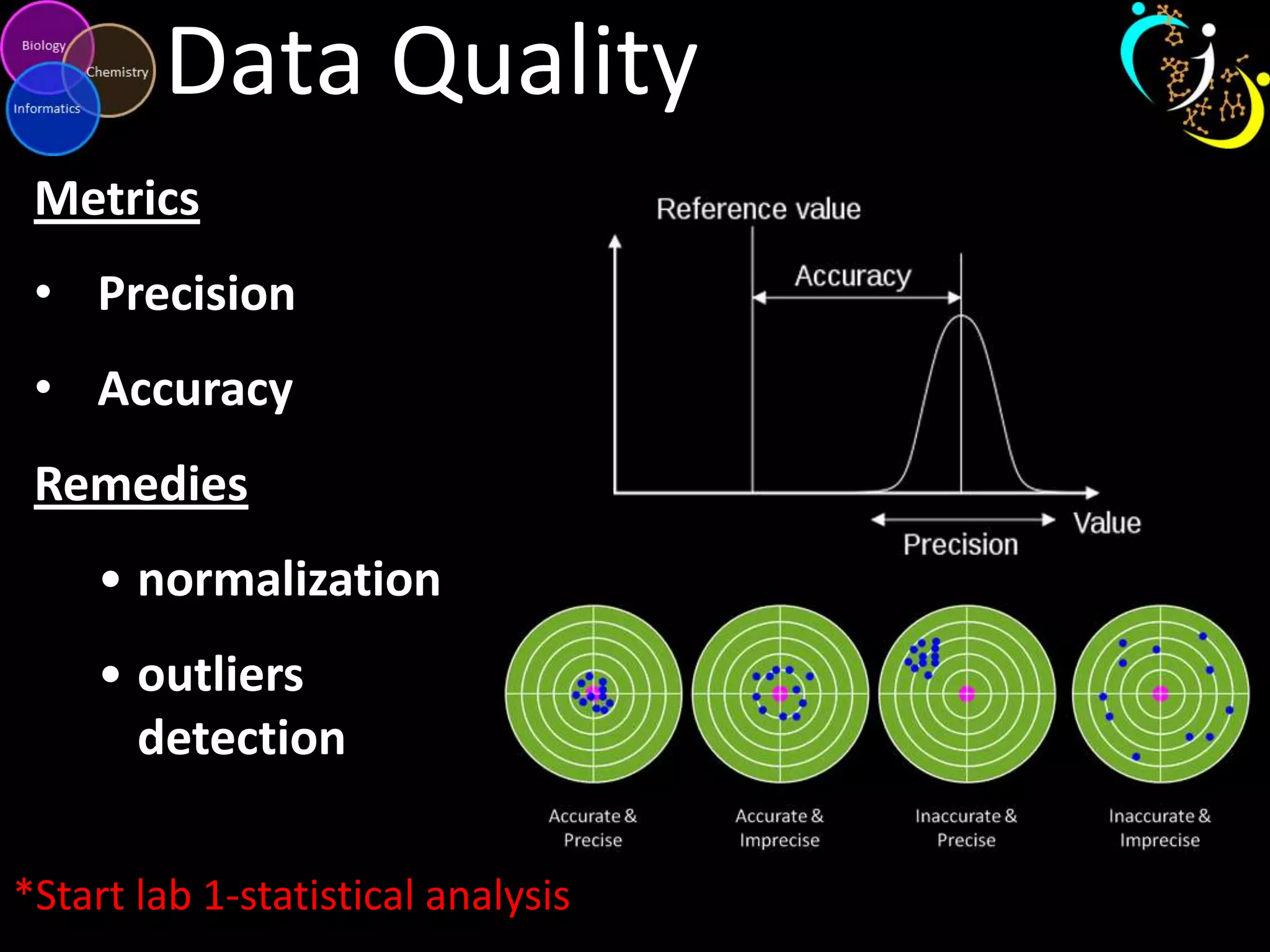
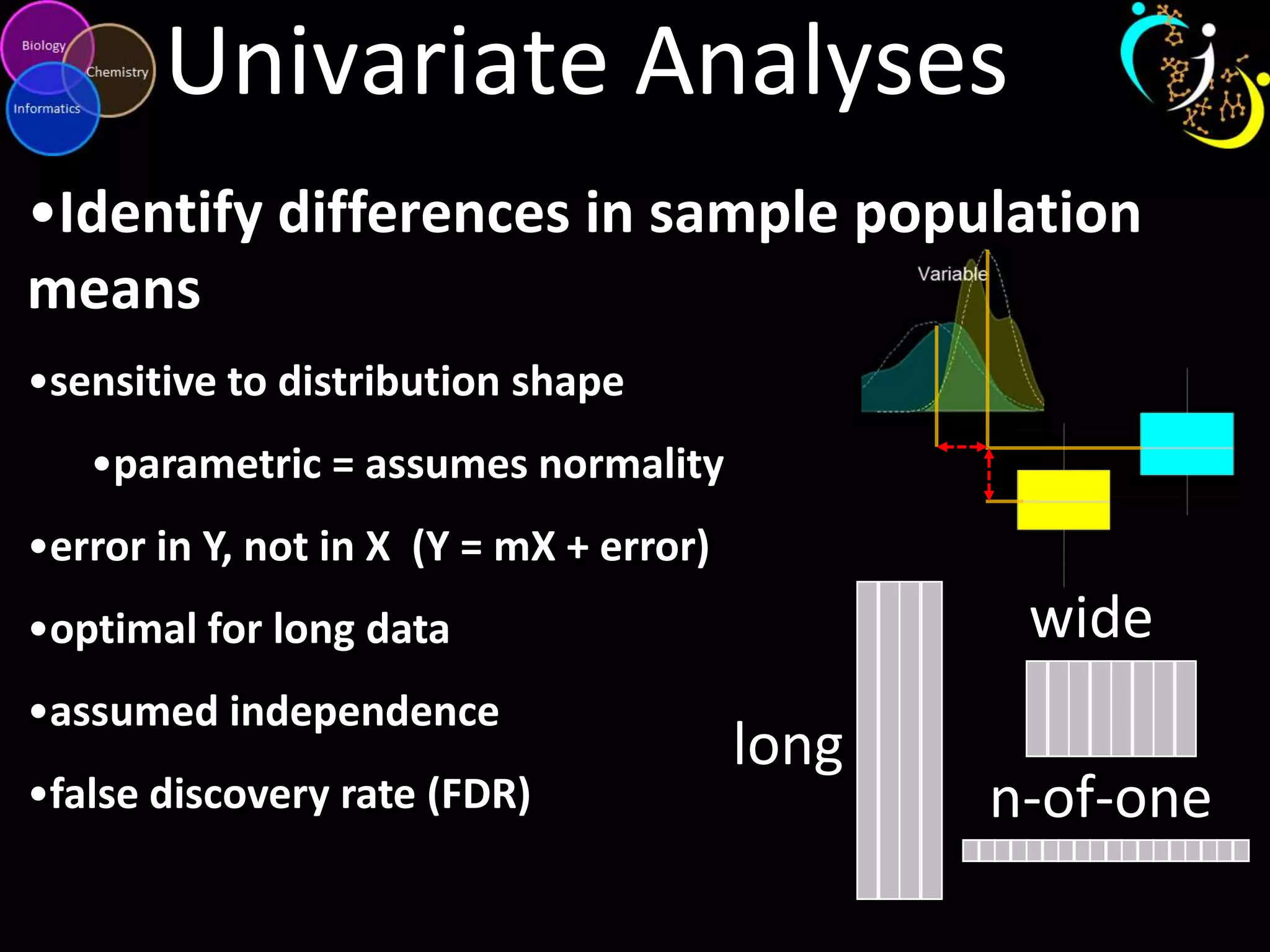
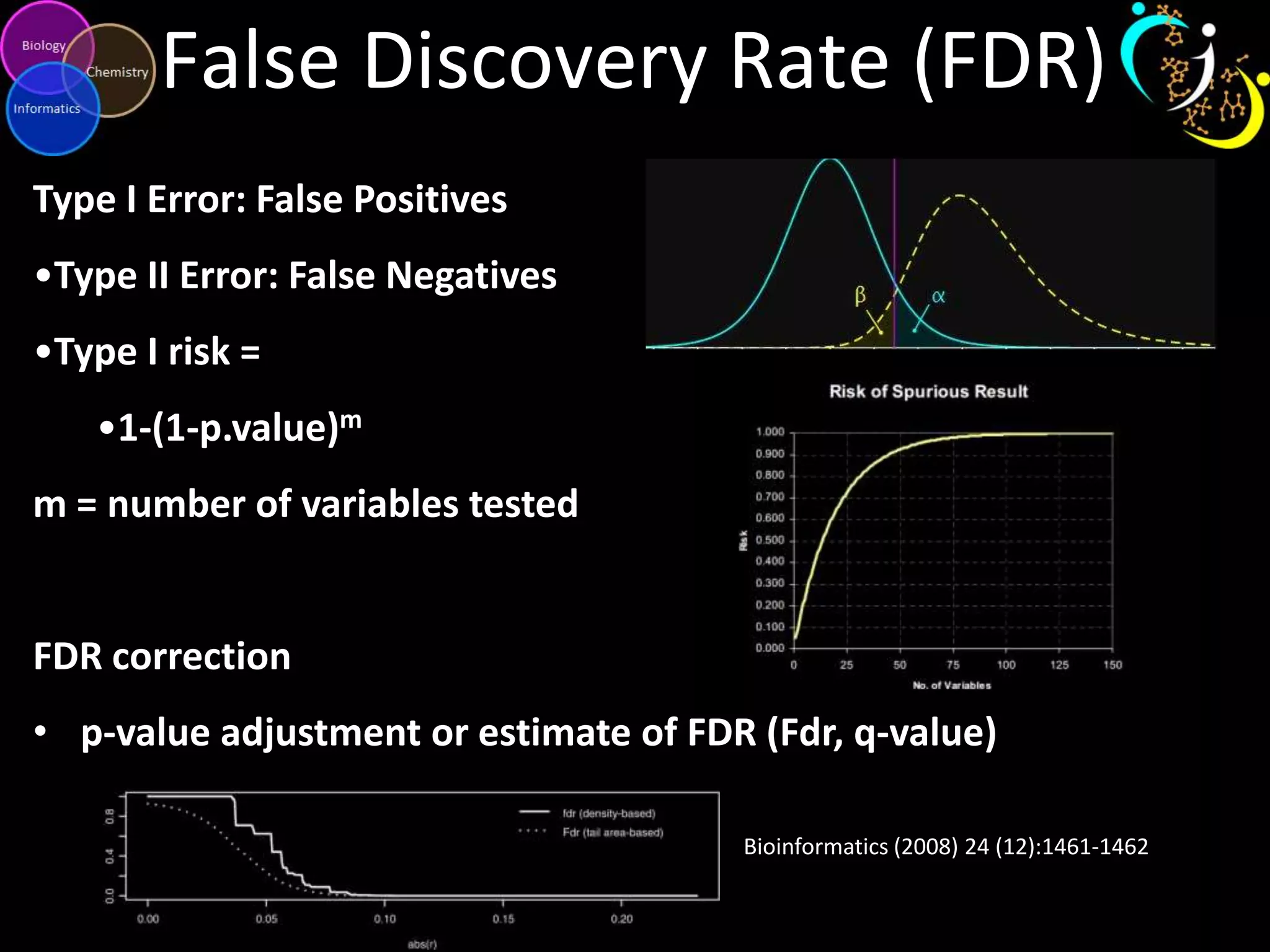
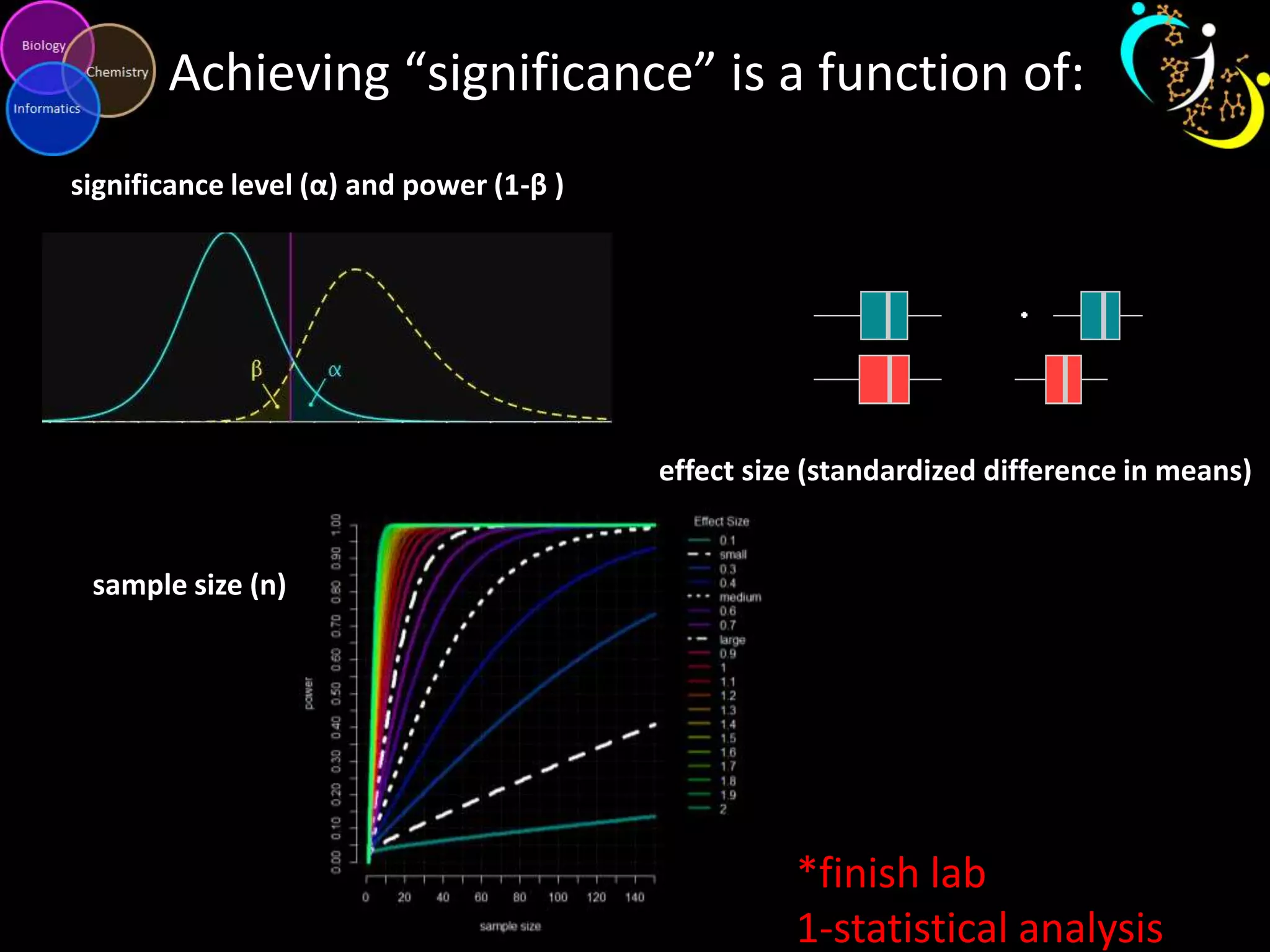
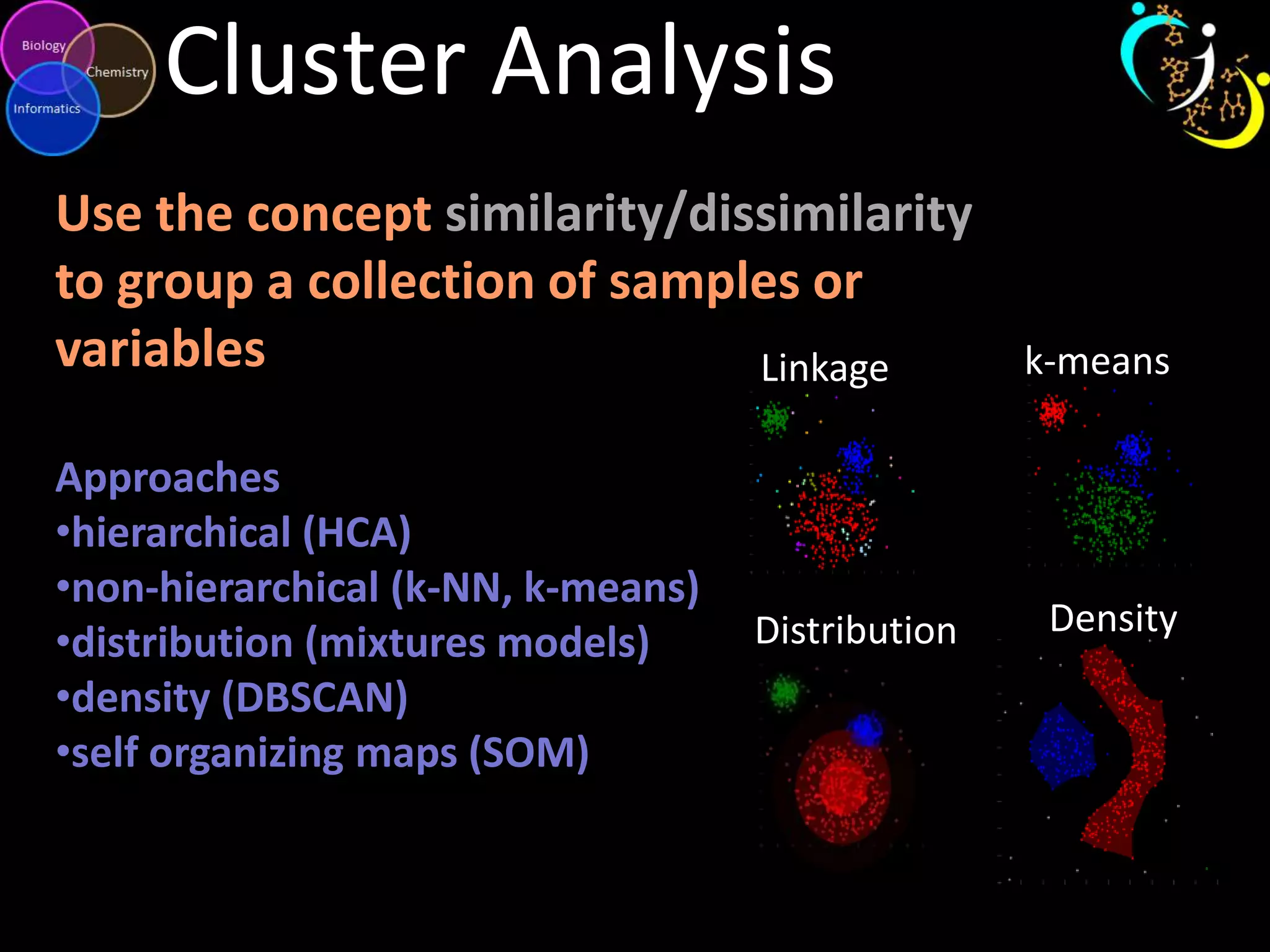
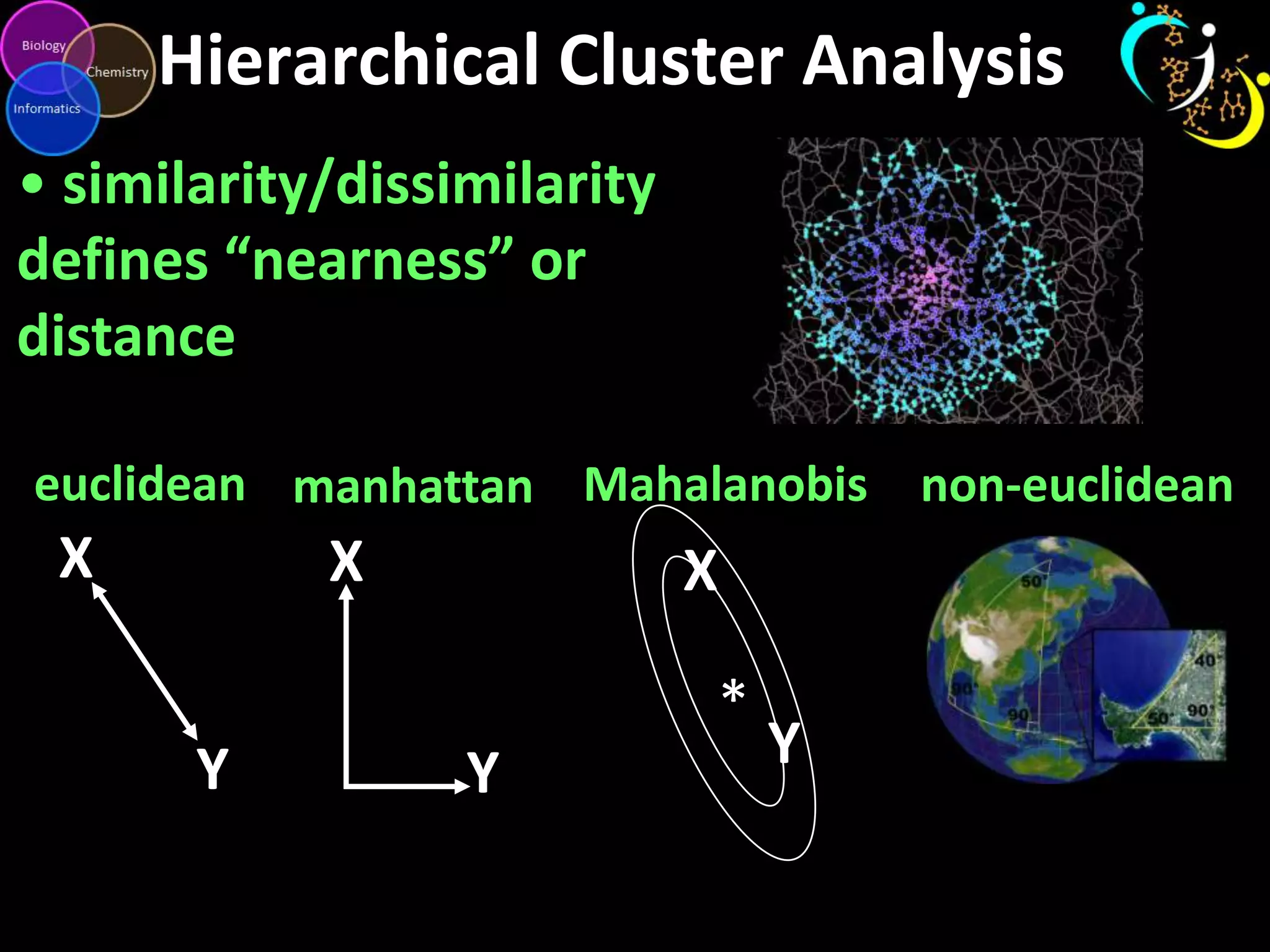
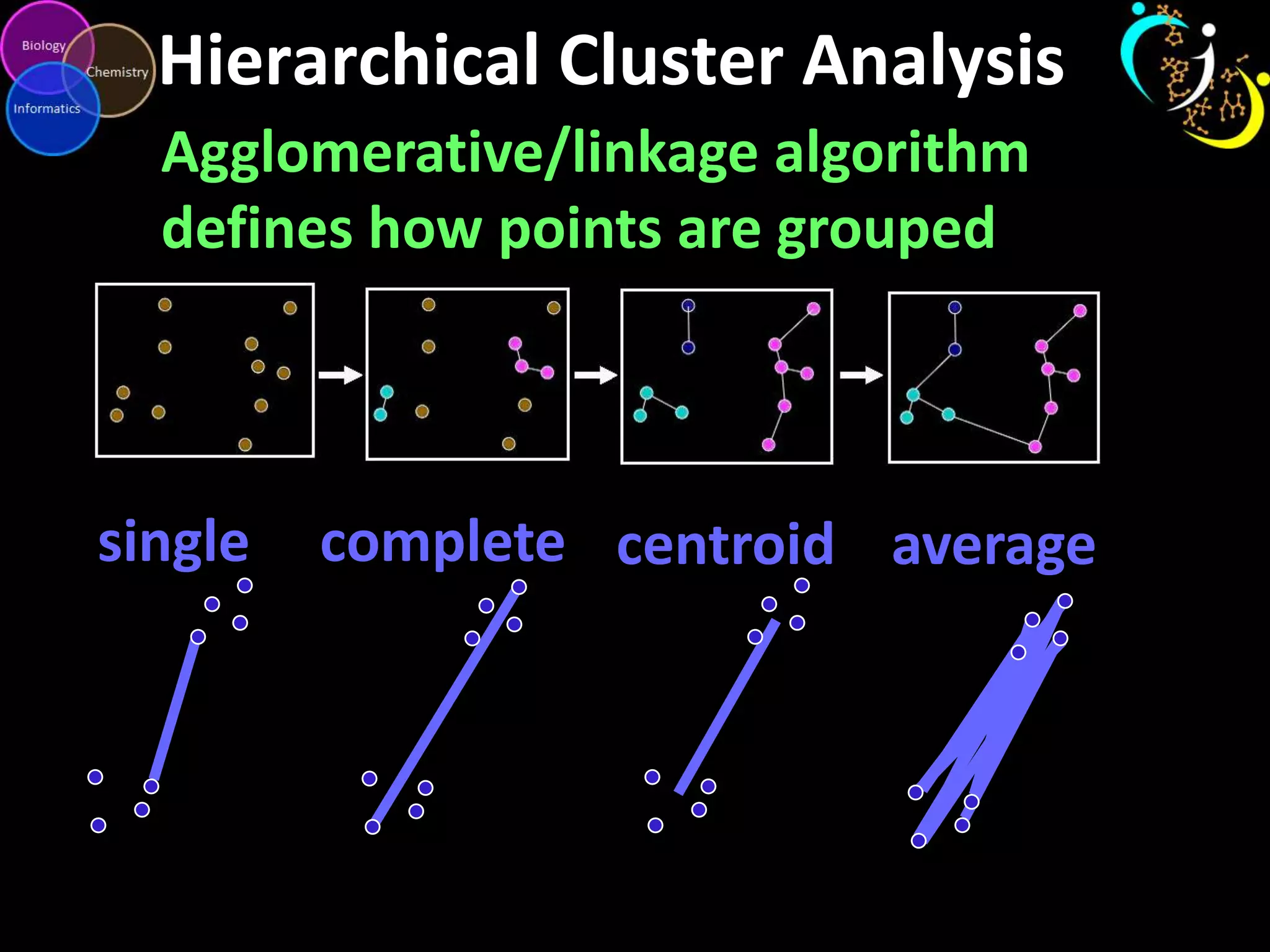
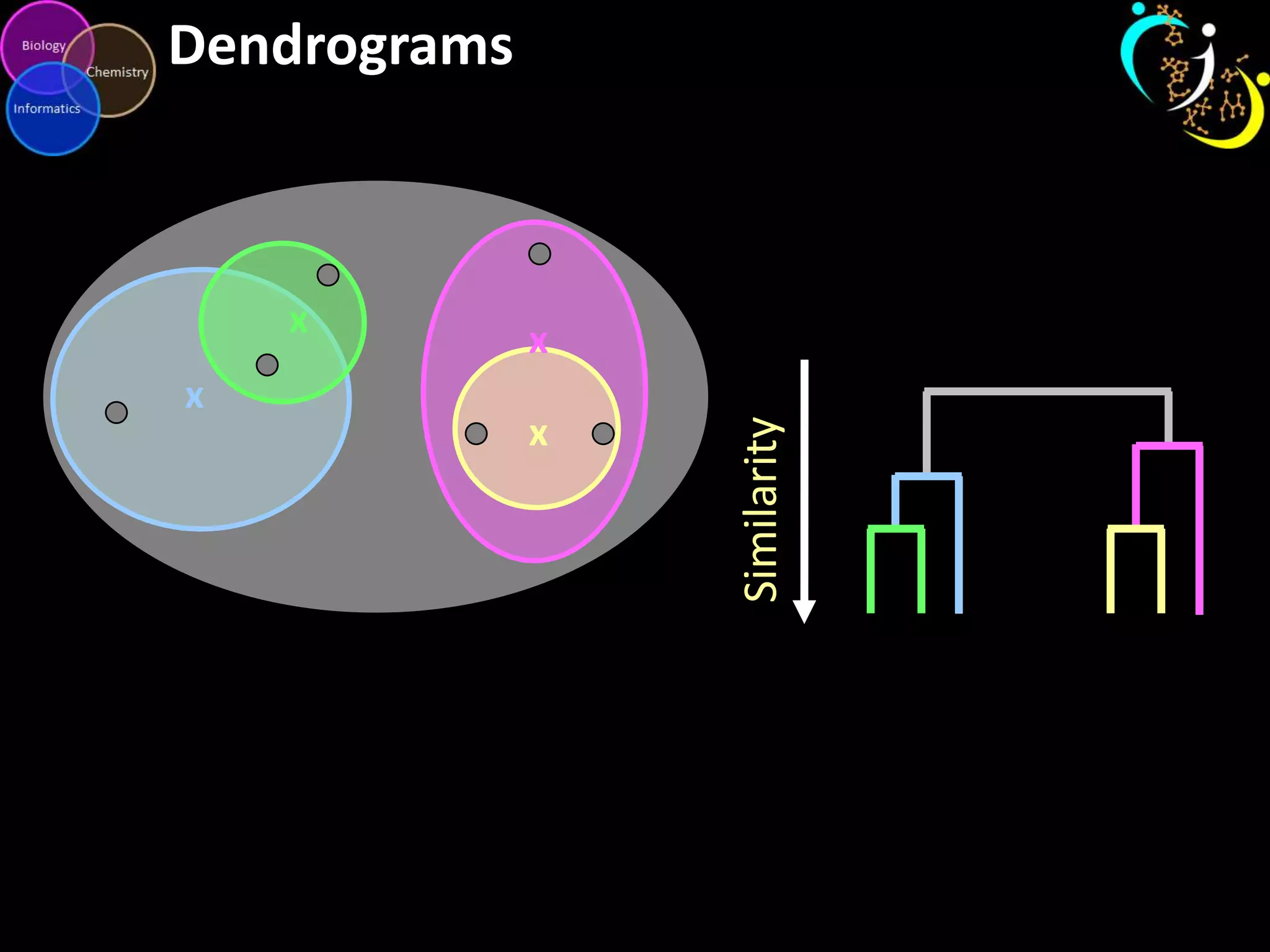
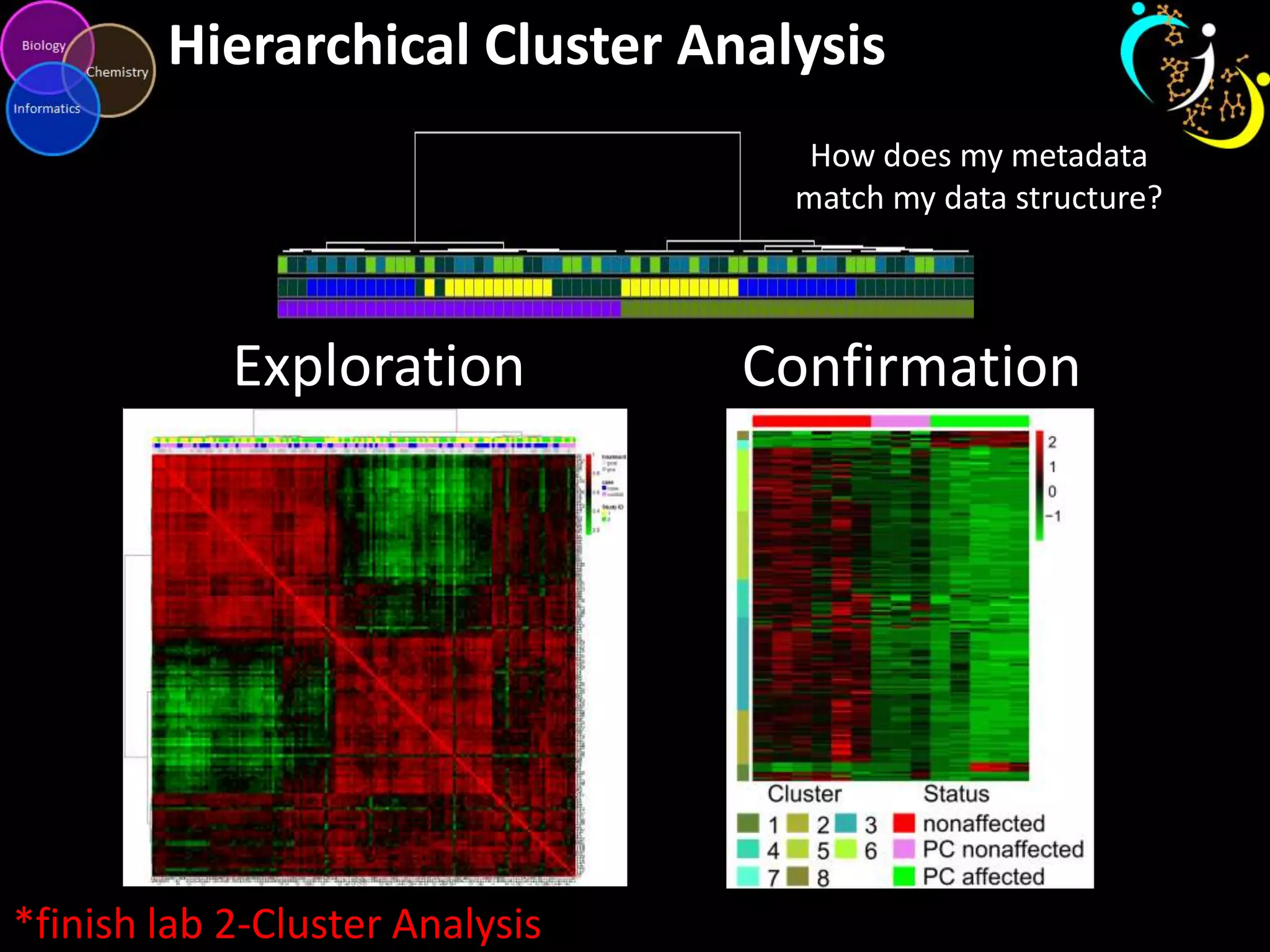
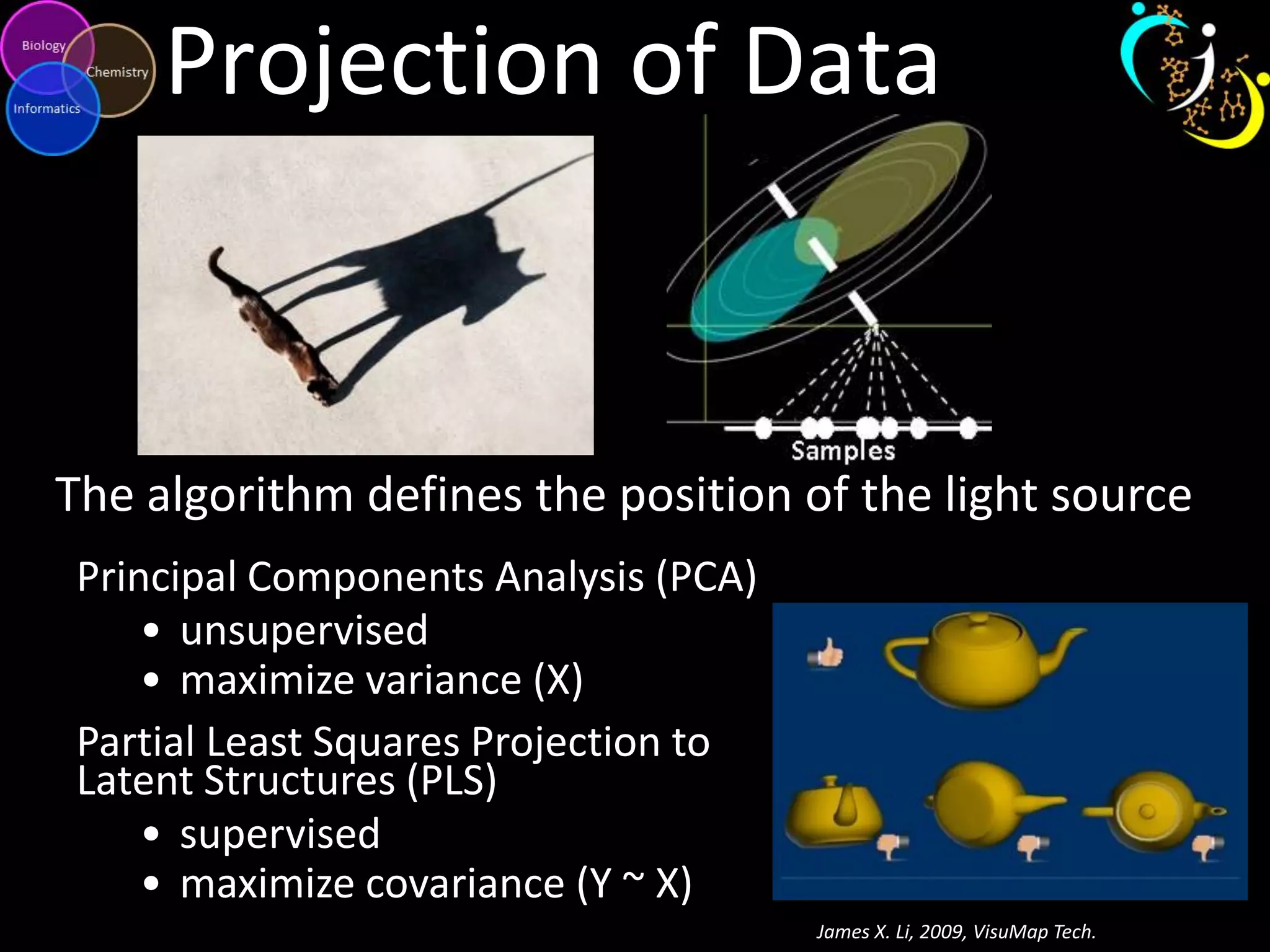
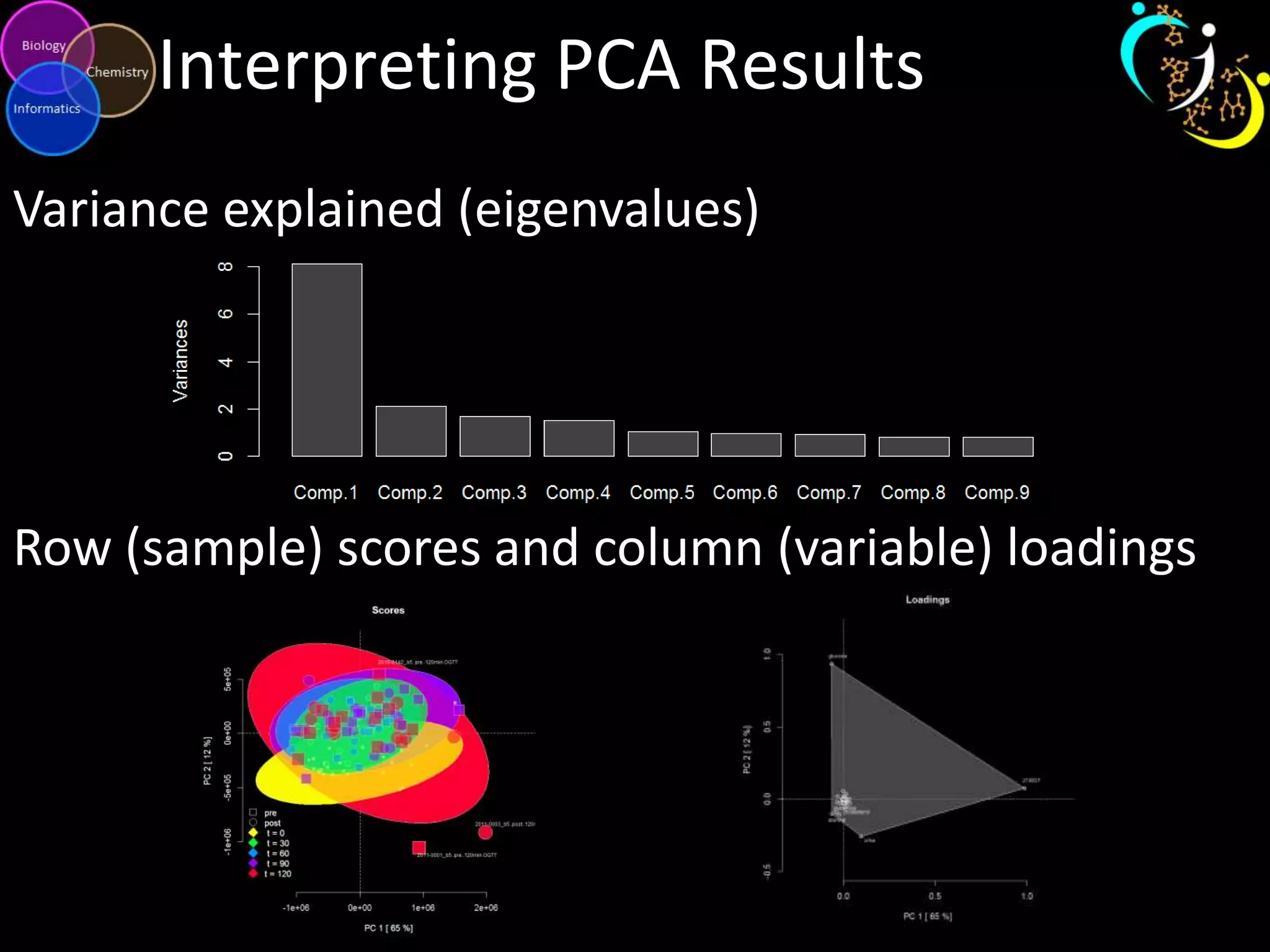
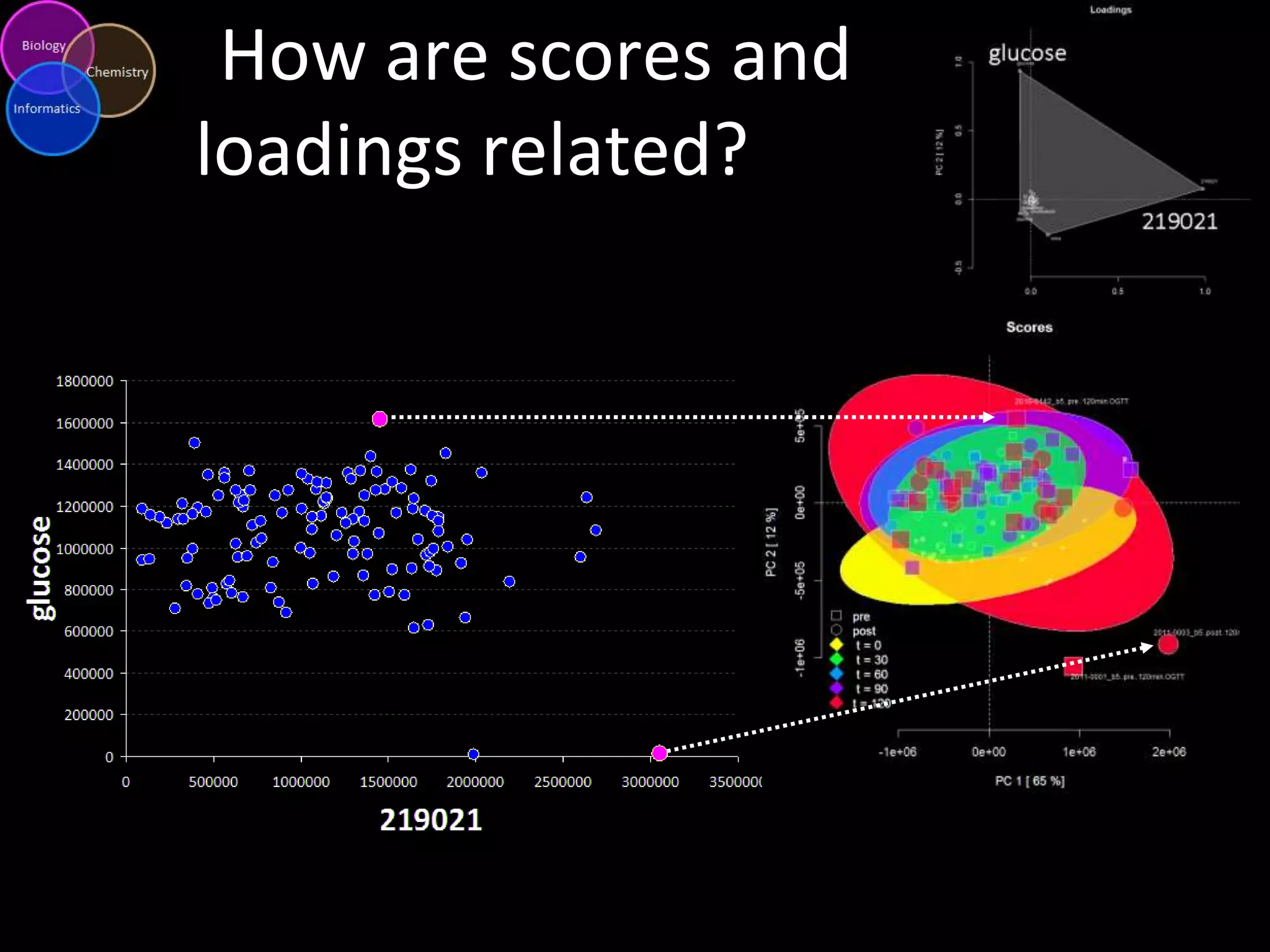
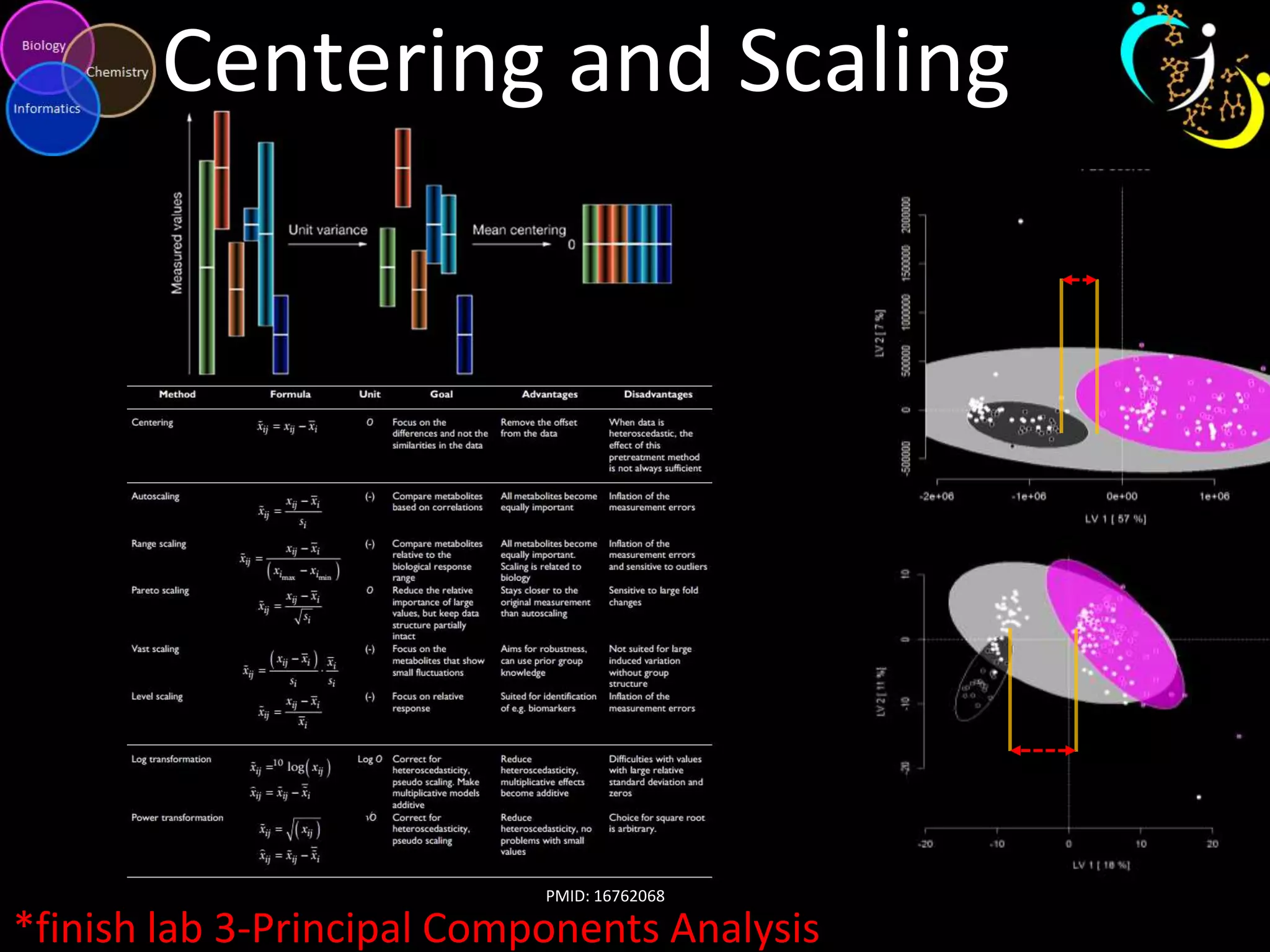
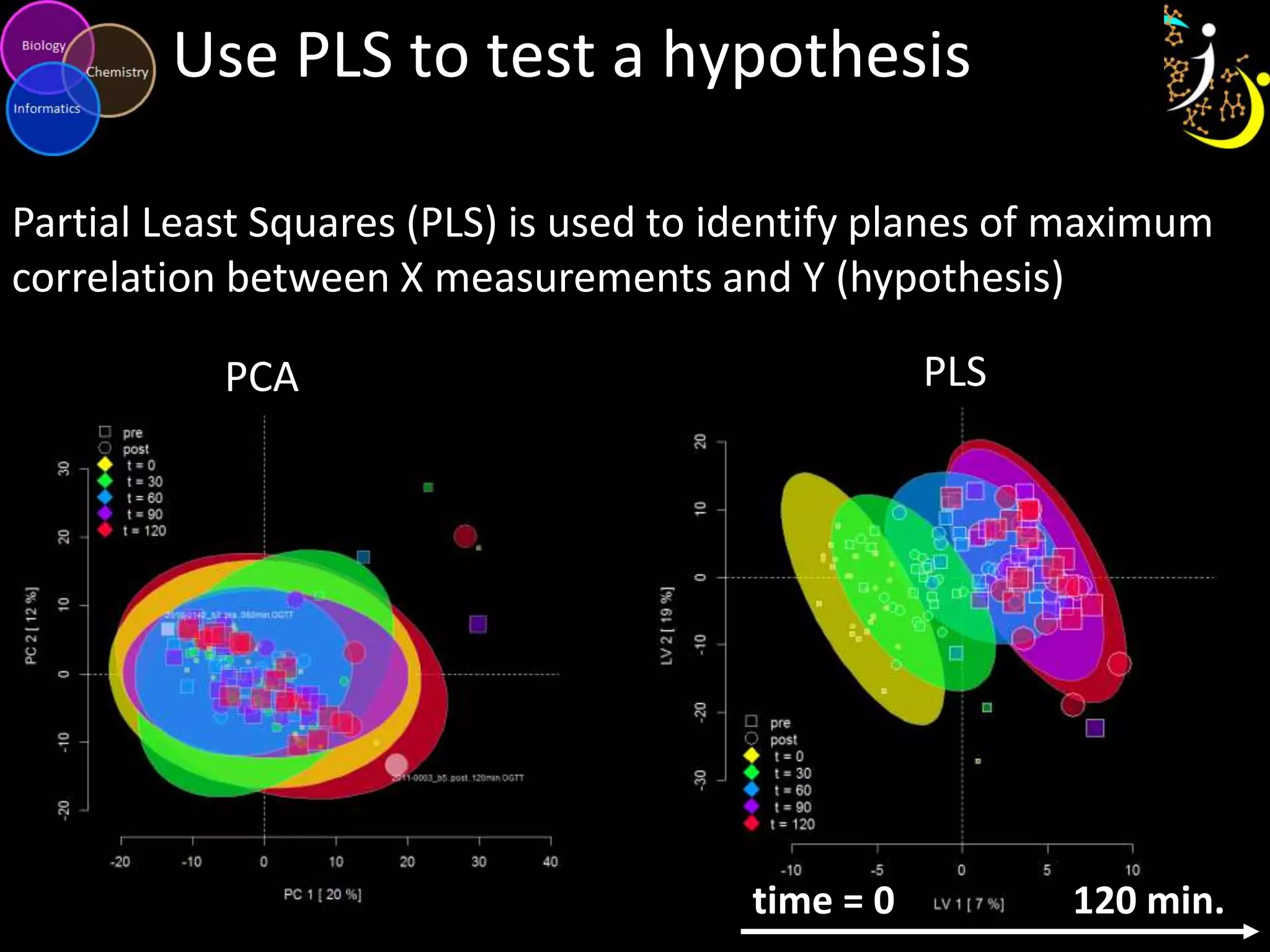
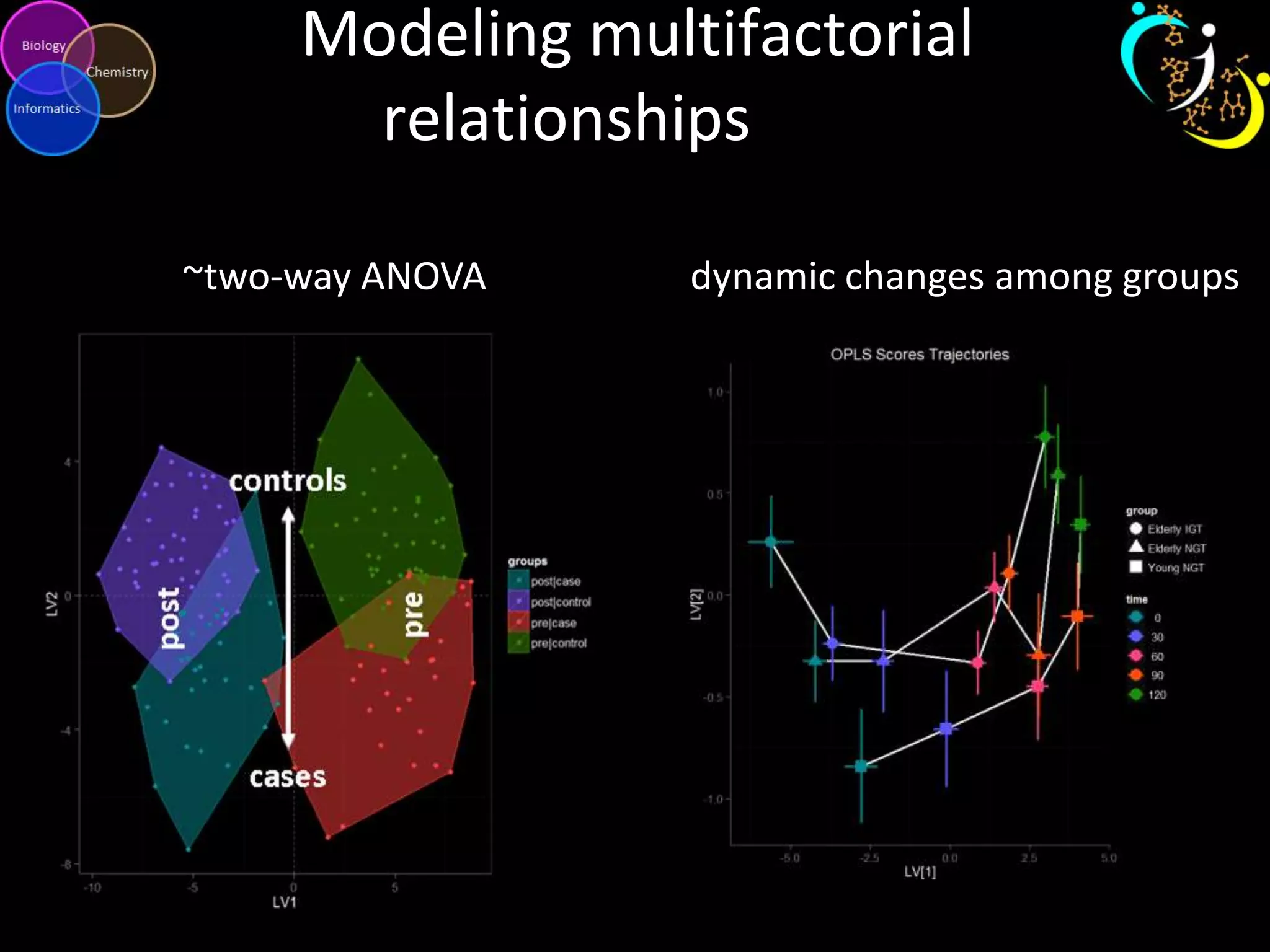
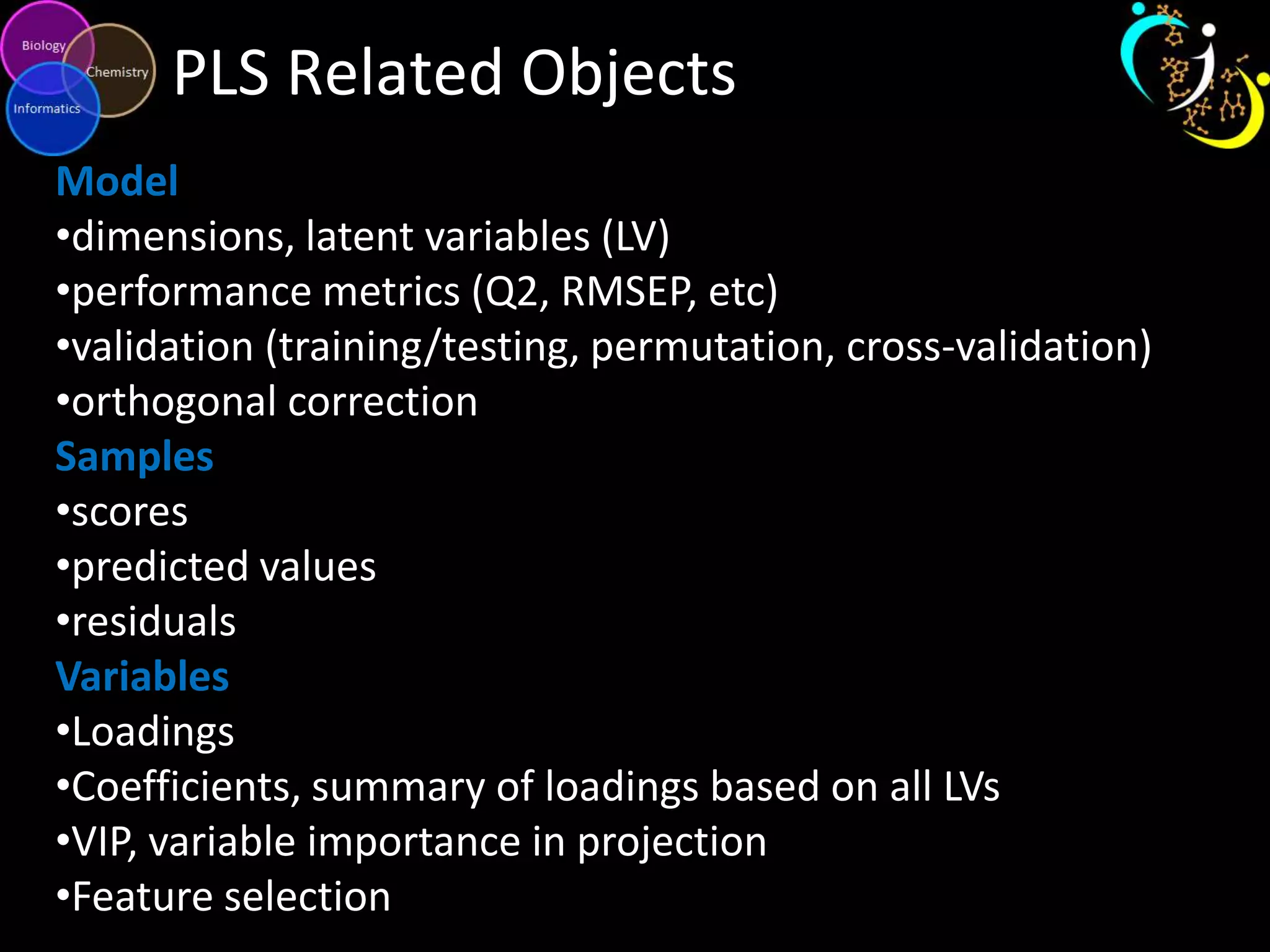
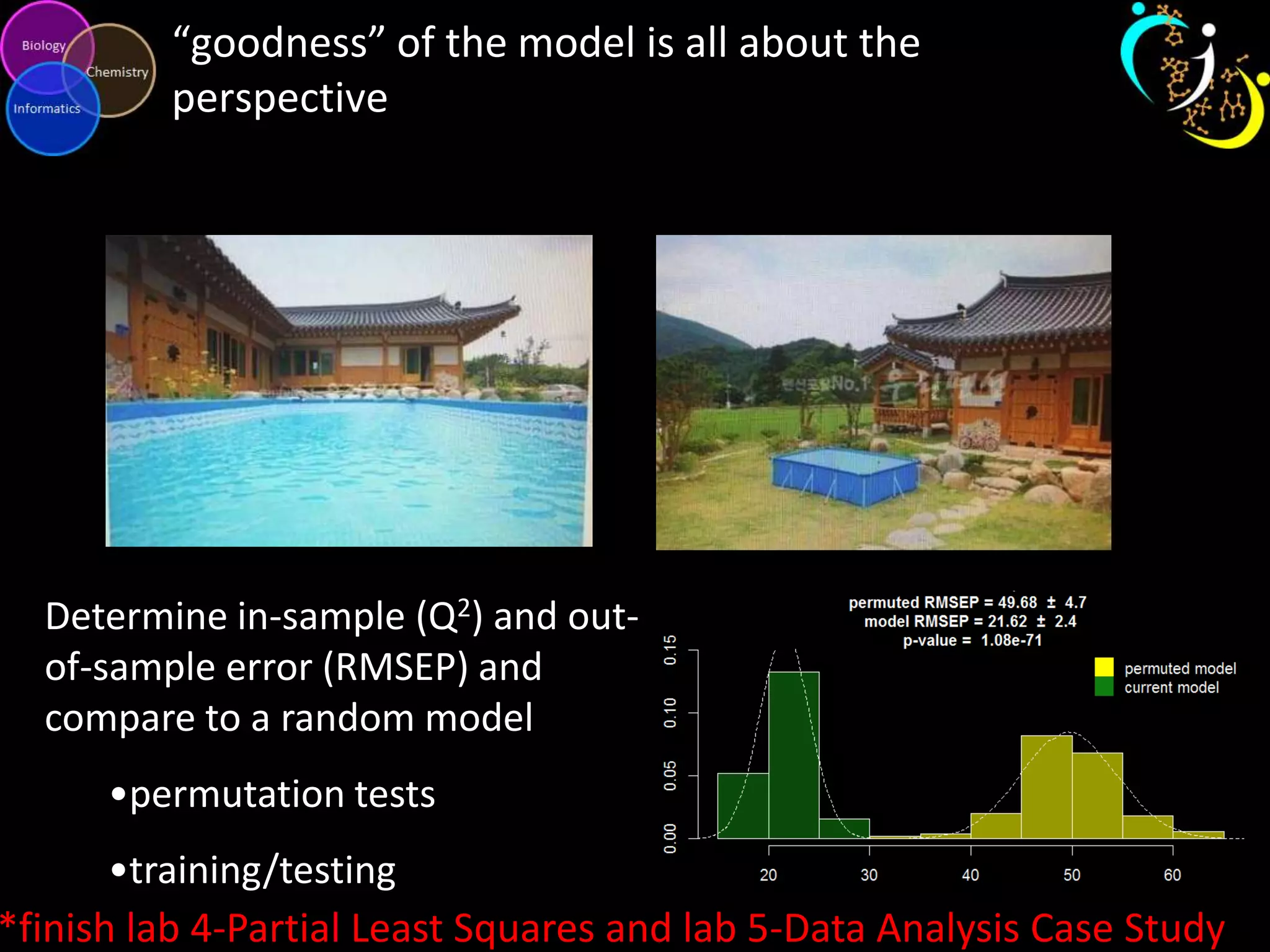
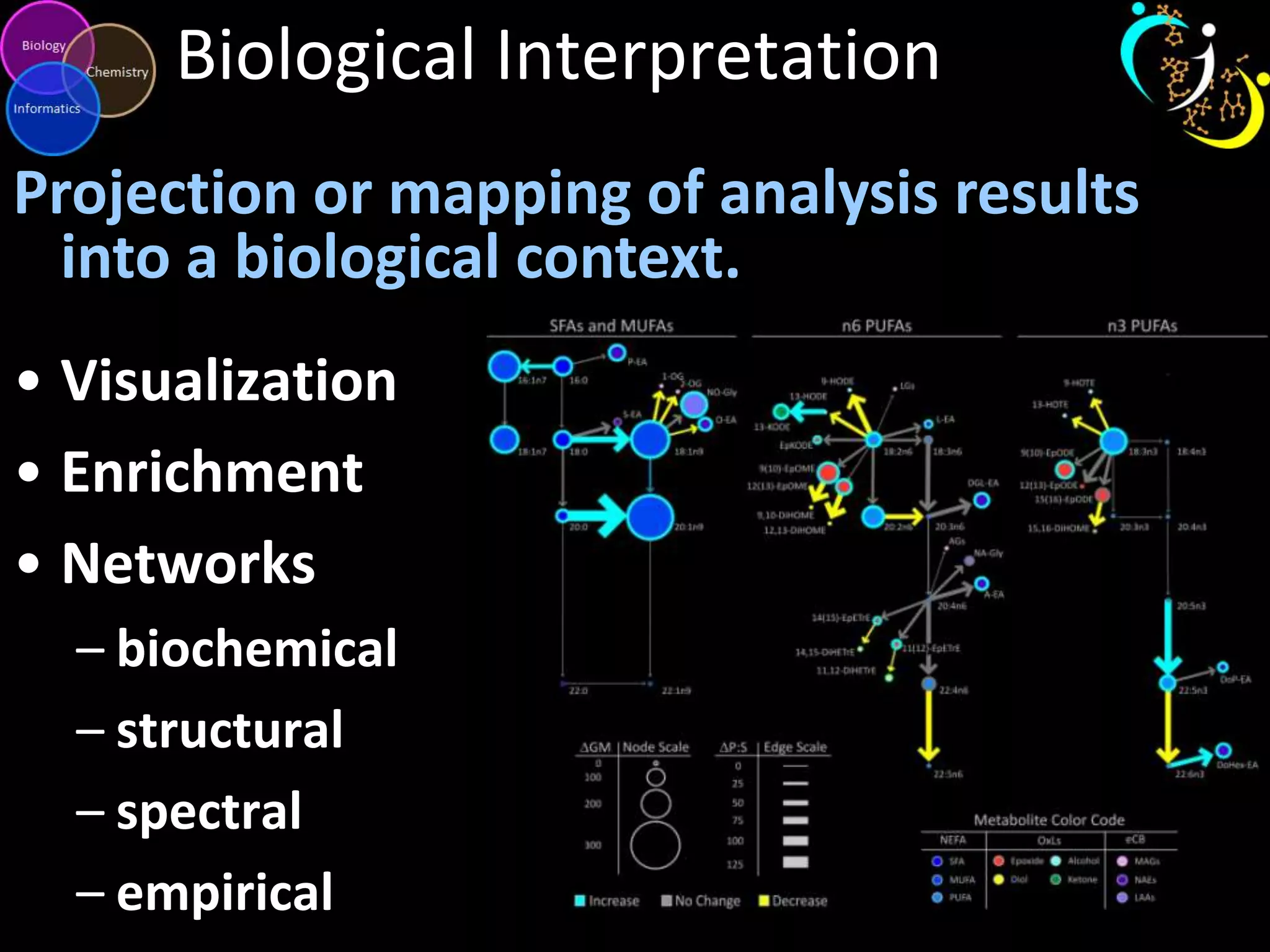
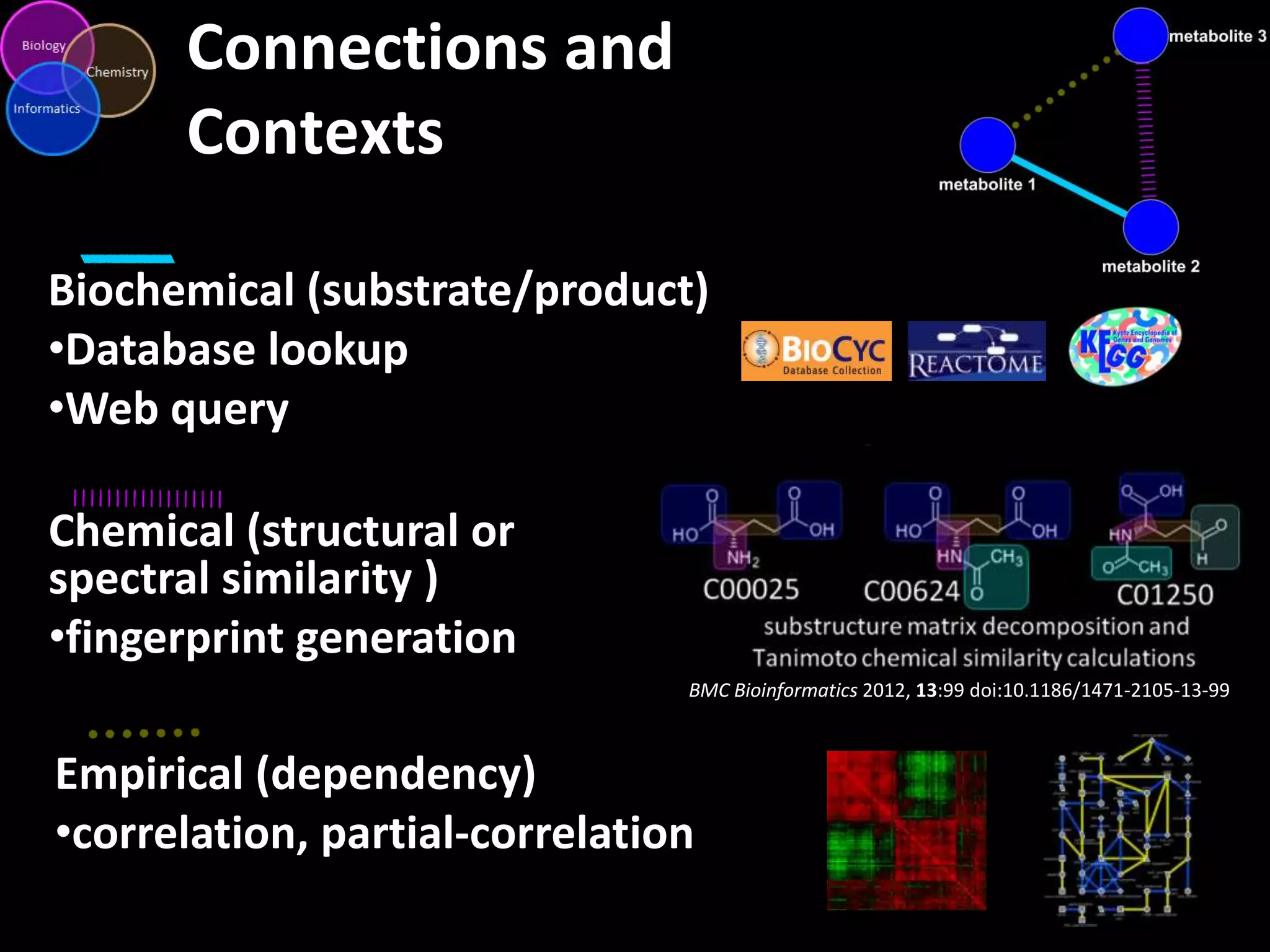
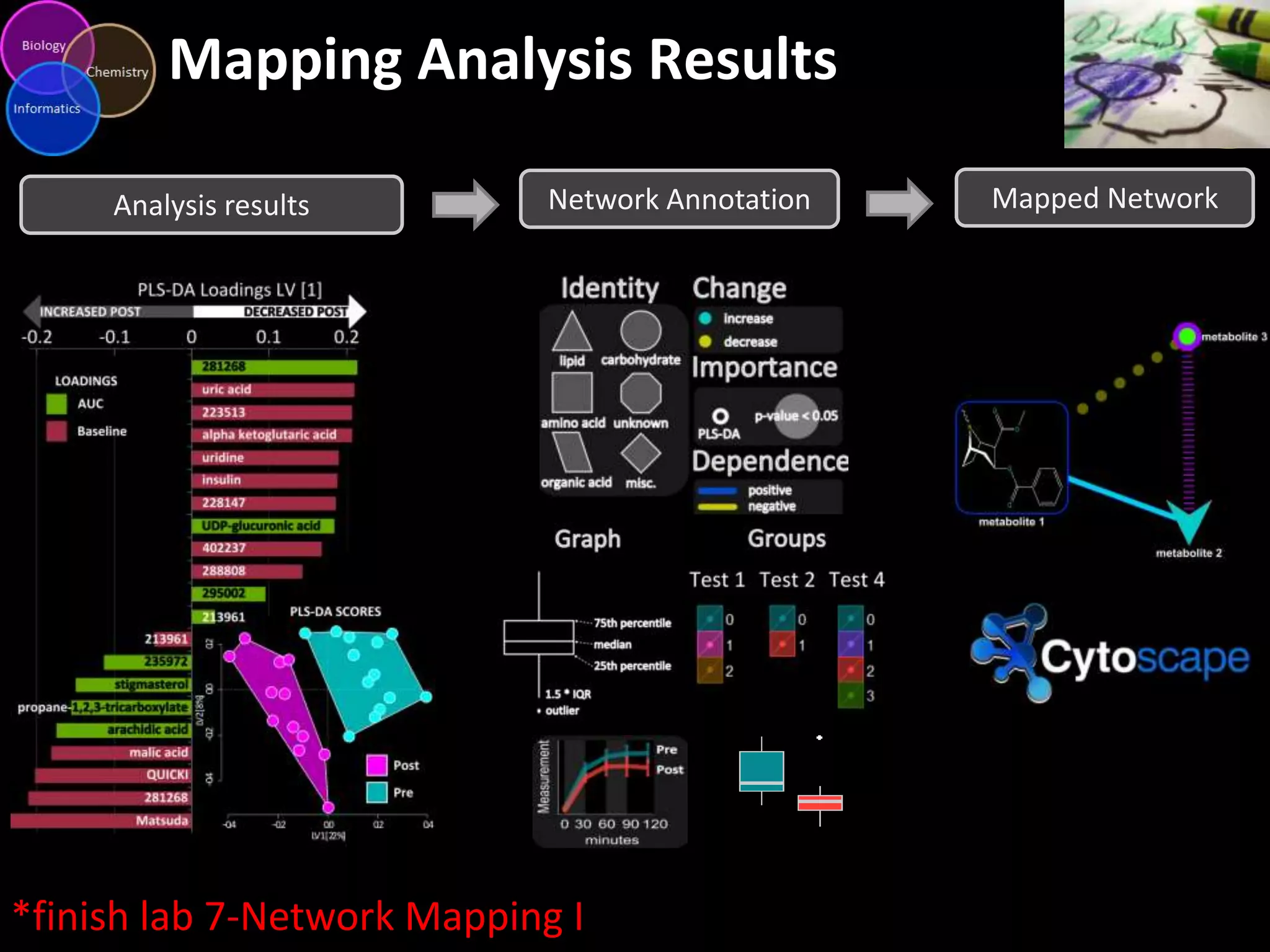
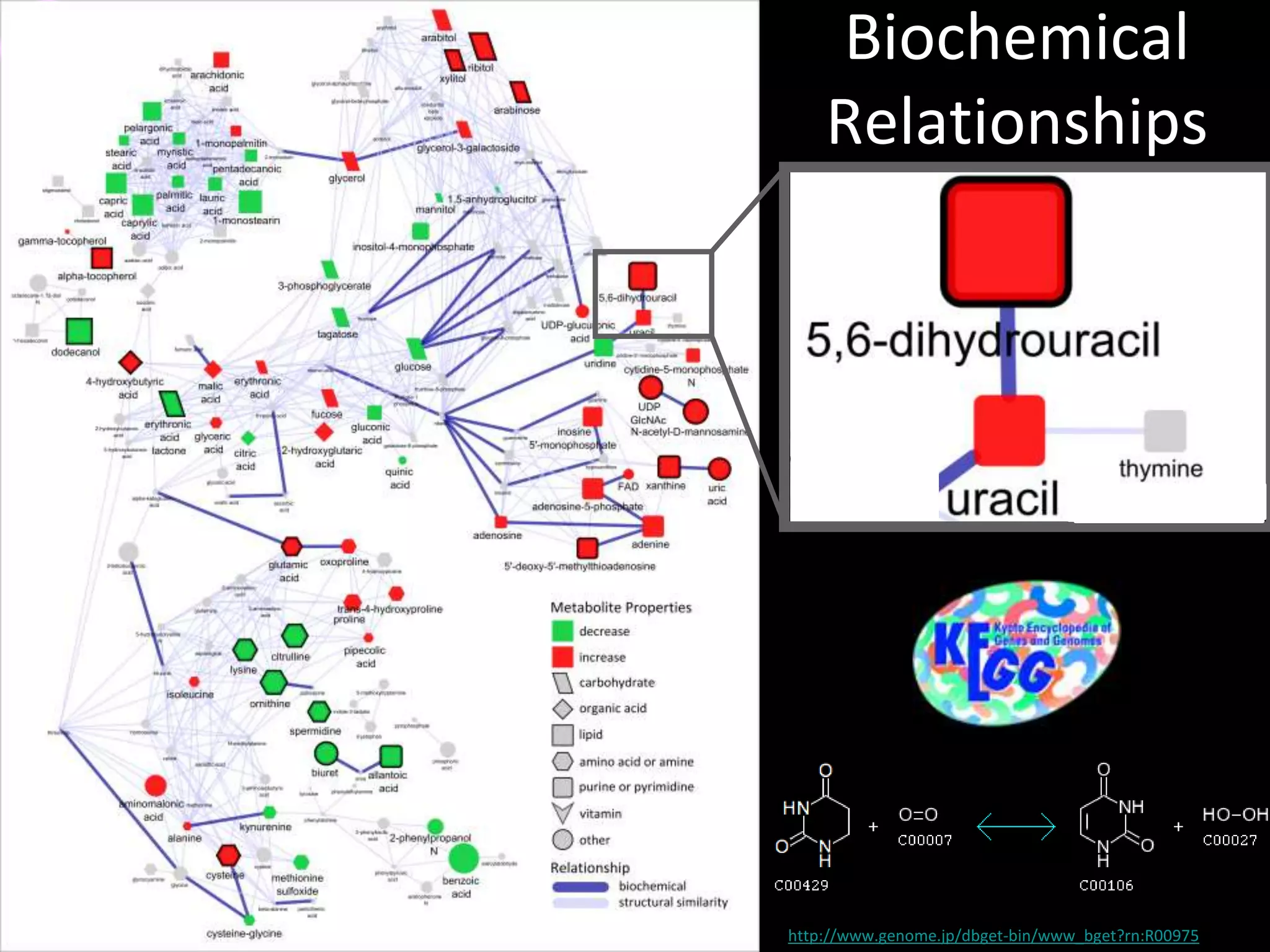
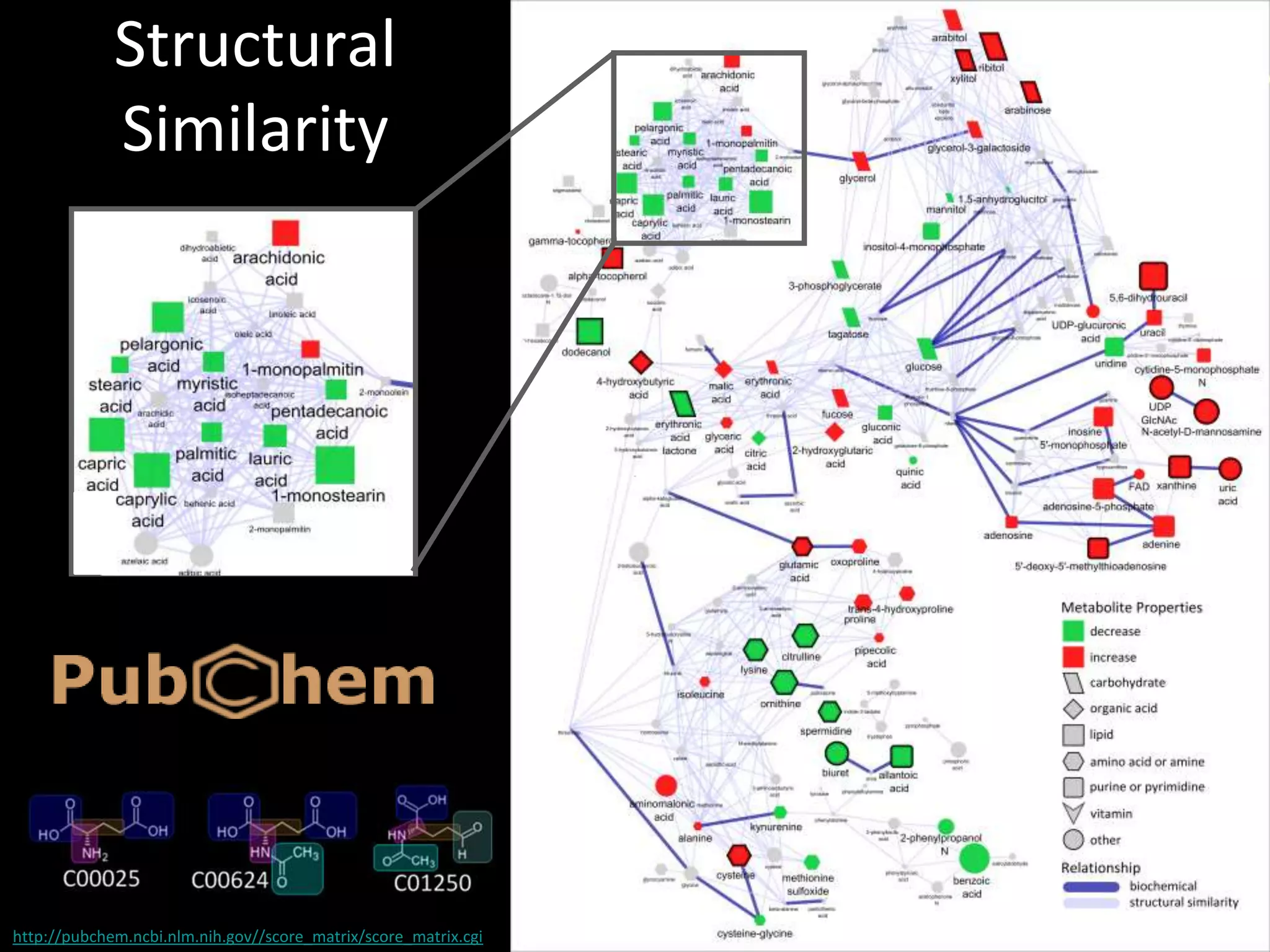
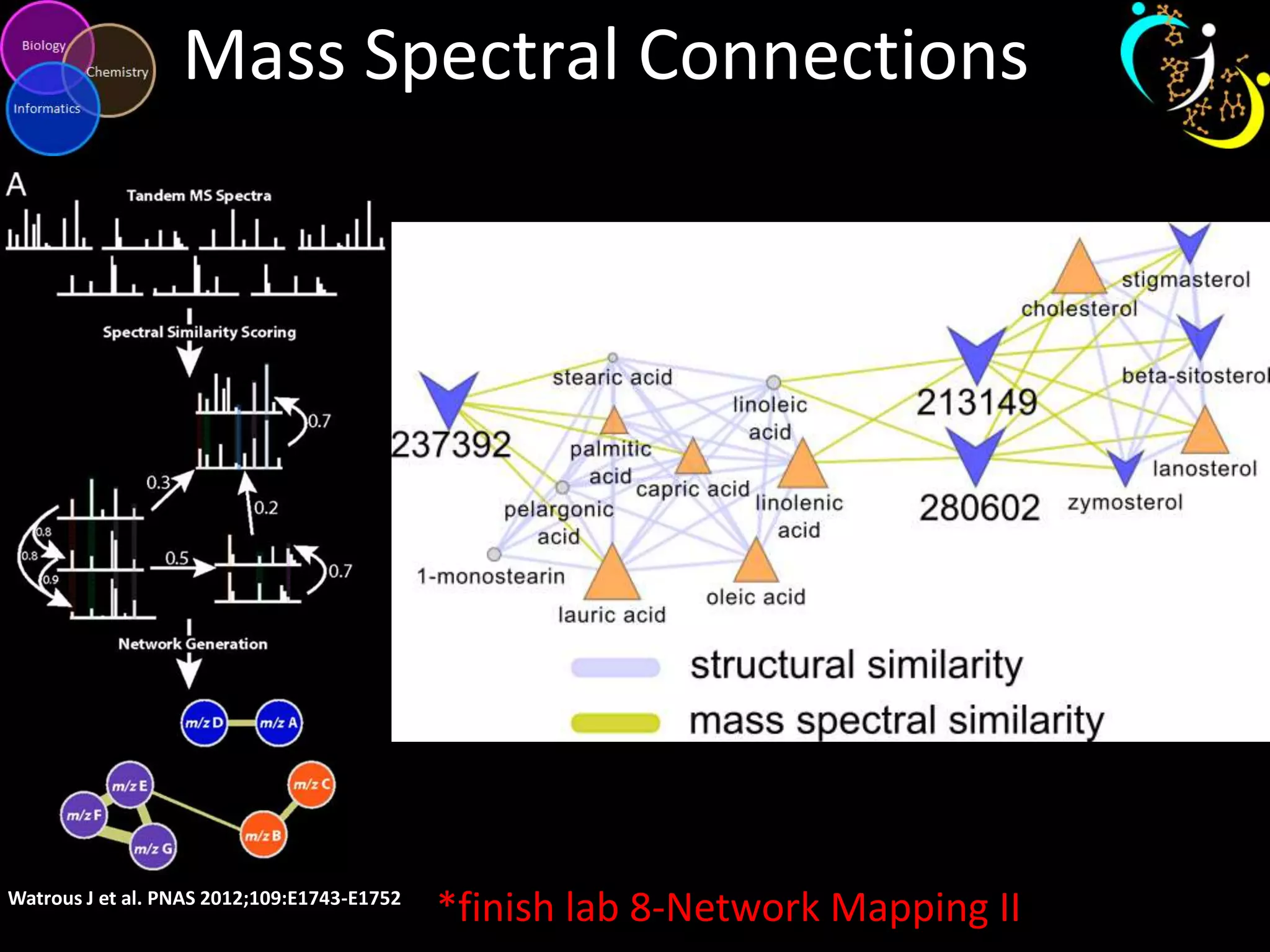
This document introduces a series of tutorials for metabolomic data analysis. It discusses important goals like hypothesis generation, data acquisition, processing, exploration, classification and prediction. It covers topics like univariate vs multivariate analysis, data quality metrics, clustering, principal component analysis, partial least squares modeling, and biological interpretation through metabolite enrichment and network mapping. The overall document provides a high-level overview of the key concepts and analytical approaches that will be covered in more detail in the tutorial series.