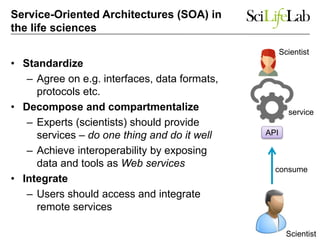
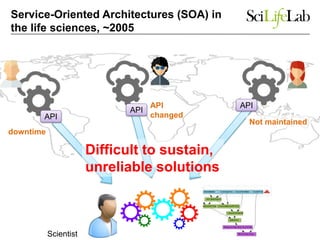
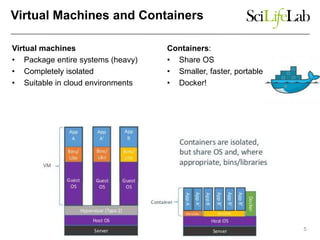
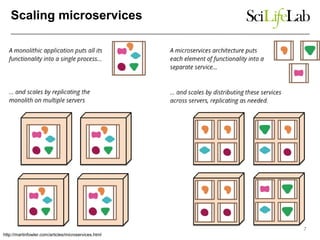
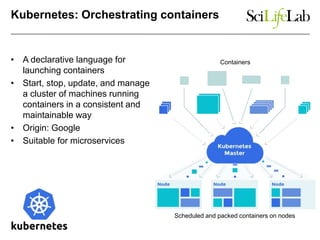
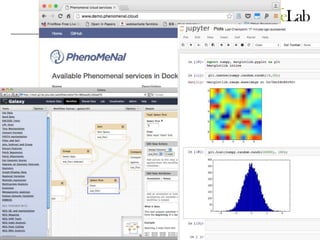
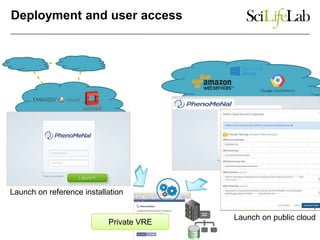
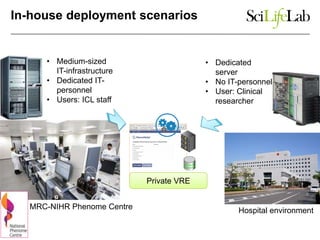
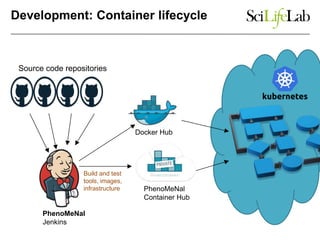
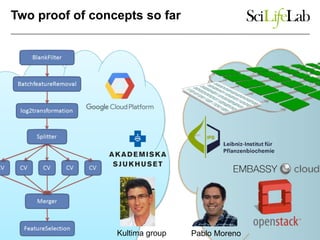
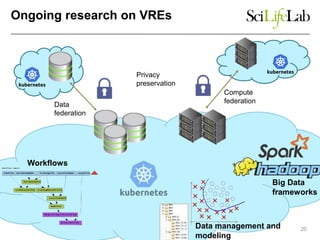
The document explores the benefits of service-oriented architectures (SOA) and microservices in life sciences, emphasizing the need for standardization, the advantages of cloud computing, and the role of containers in improving scalability and agility. It highlights the development of virtual research environments (VRE) that allow easy access to computational resources, promoting high availability and sustainability. However, it also points out that challenges regarding interoperability and data management still exist.