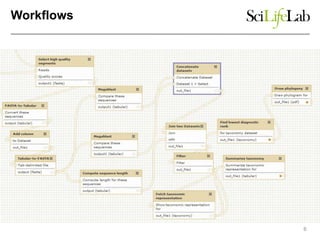
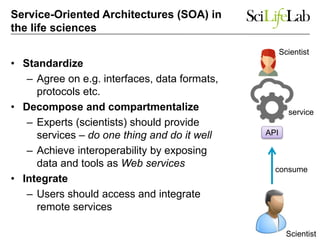
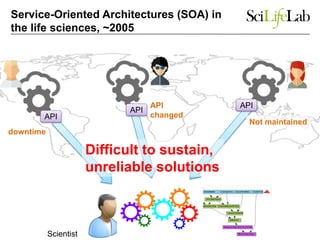
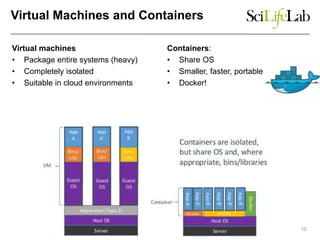
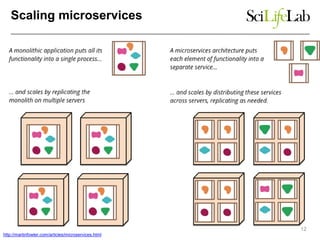
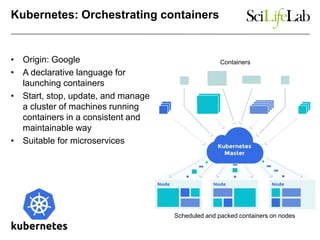
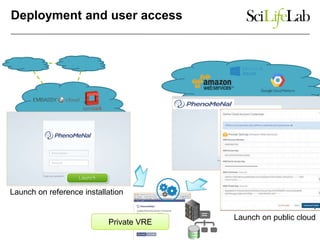
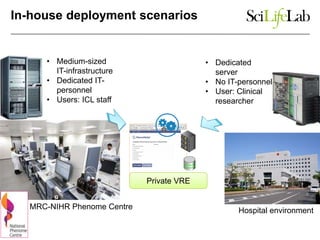
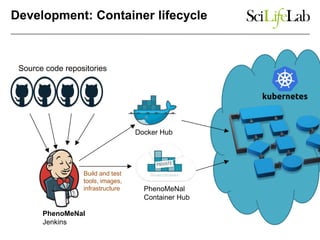
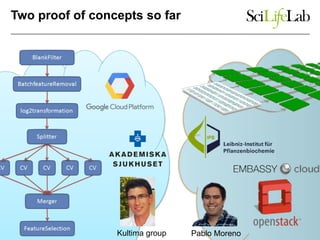
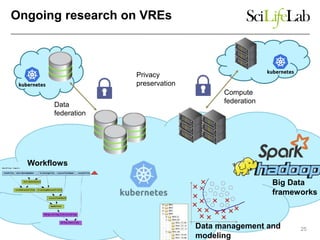
This document discusses analyzing big data in medicine using virtual research environments and microservices. It notes the vast amount of data being generated and challenges of data management, analysis and scaling. The European Open Science Cloud aims to enable access to shared scientific data across borders. Contemporary analysis uses high-performance computing but has limitations. Cloud computing, virtual machines, containers and microservices can help address these challenges by providing on-demand resources and decomposing functionality into independent services. The PhenoMeNal project is building a standardized e-infrastructure using these approaches to enable users to access tools and data. This improves sustainability, reliability, scalability and enables agile development and science.