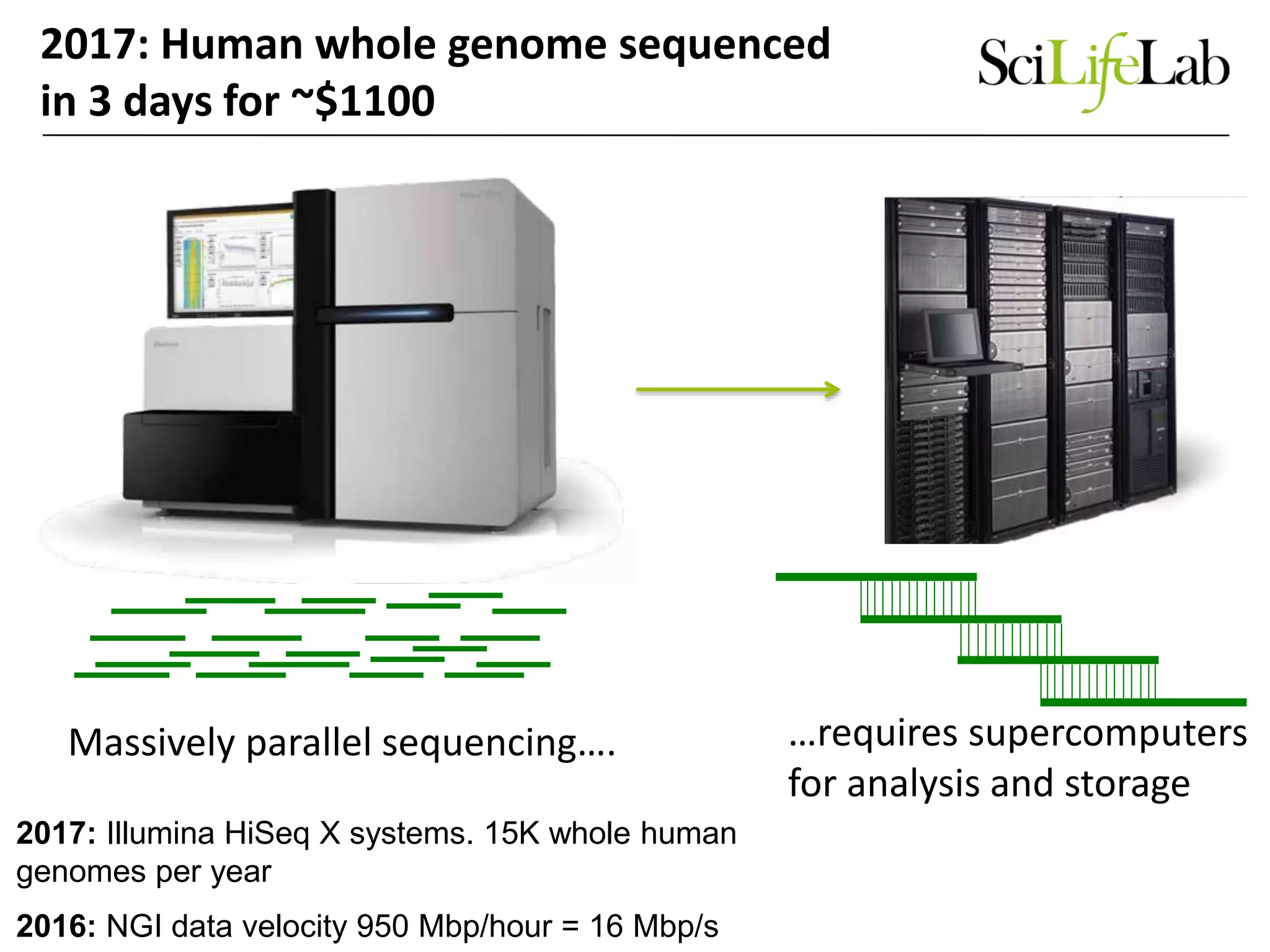
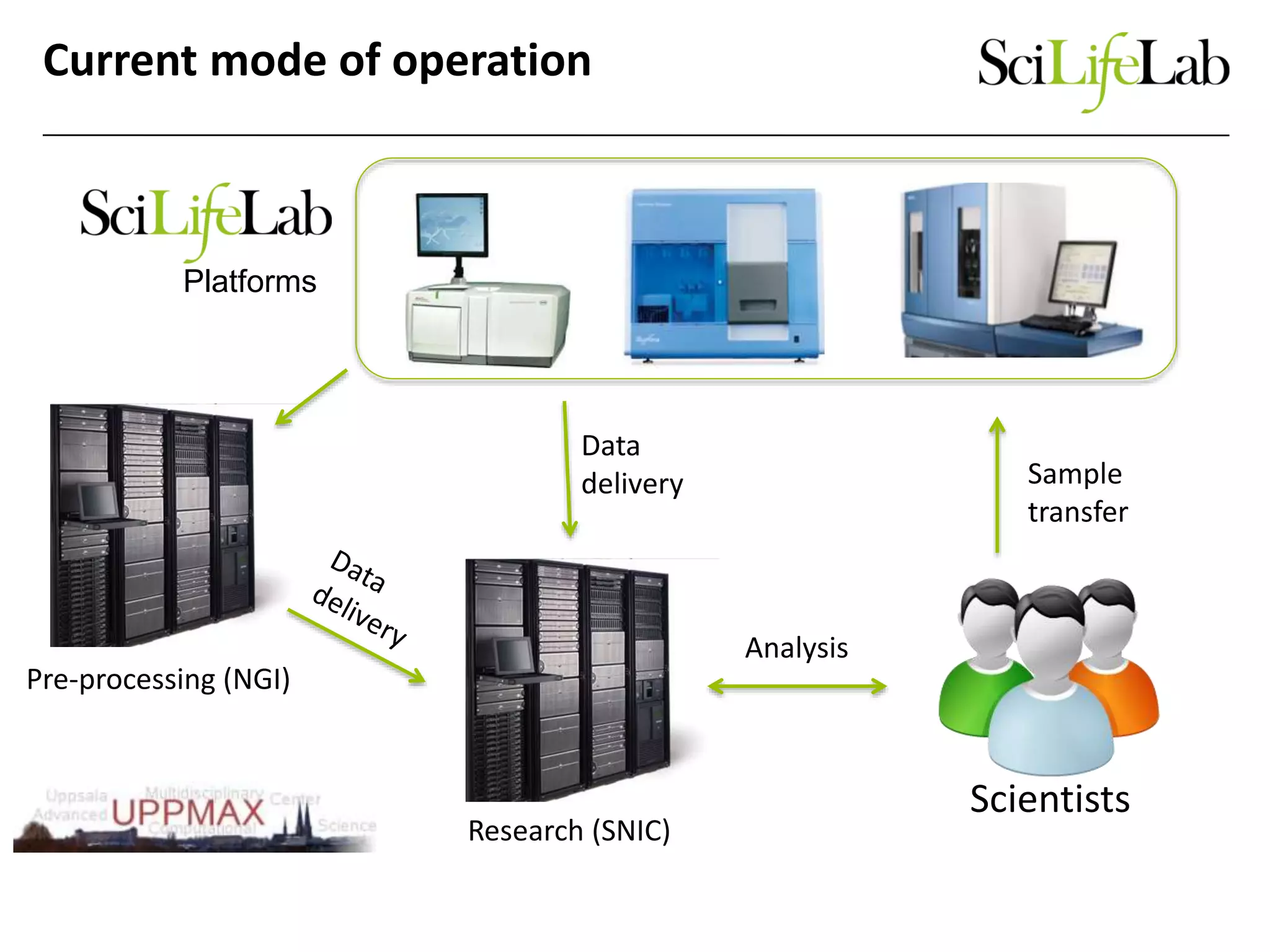
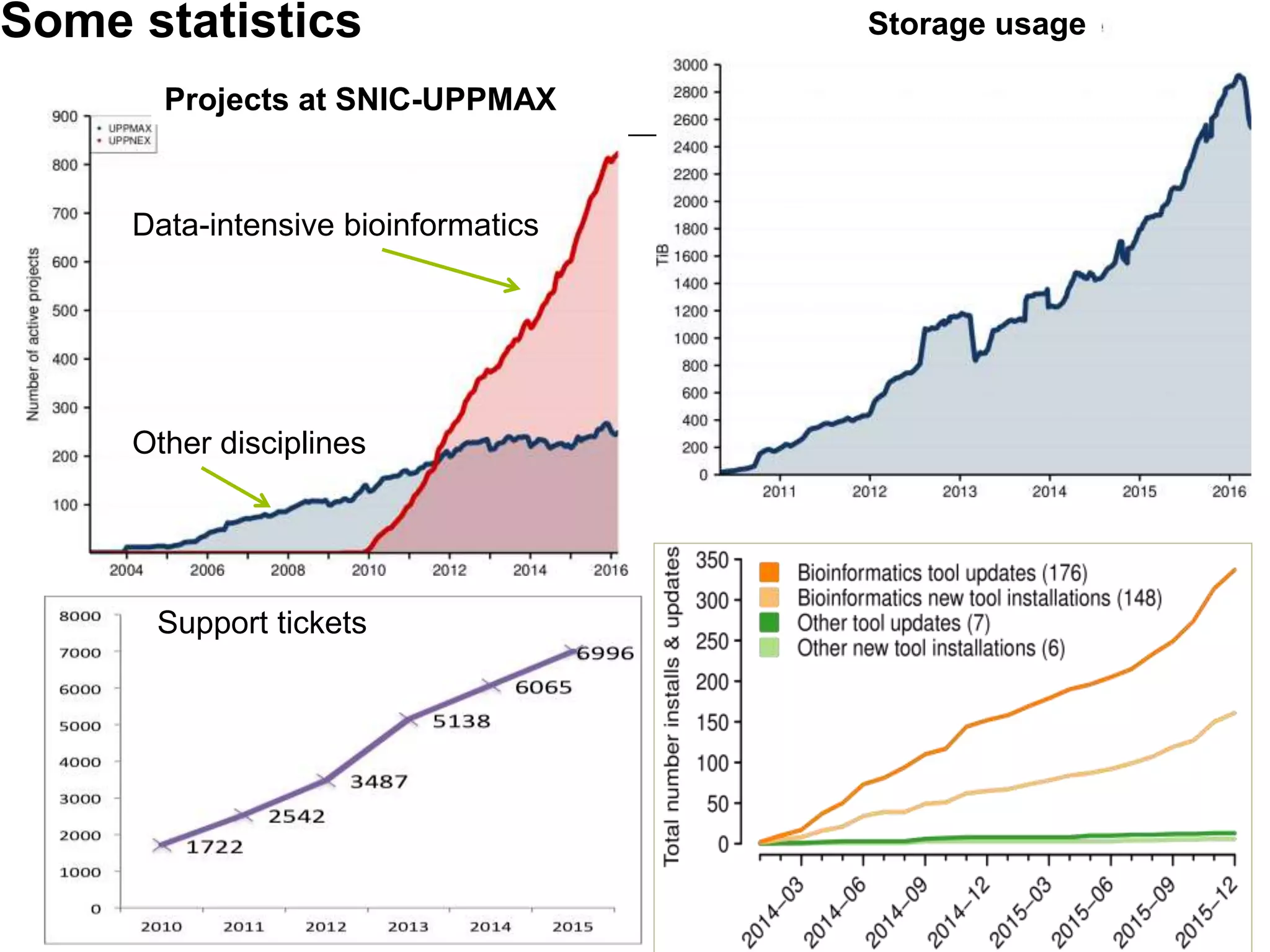
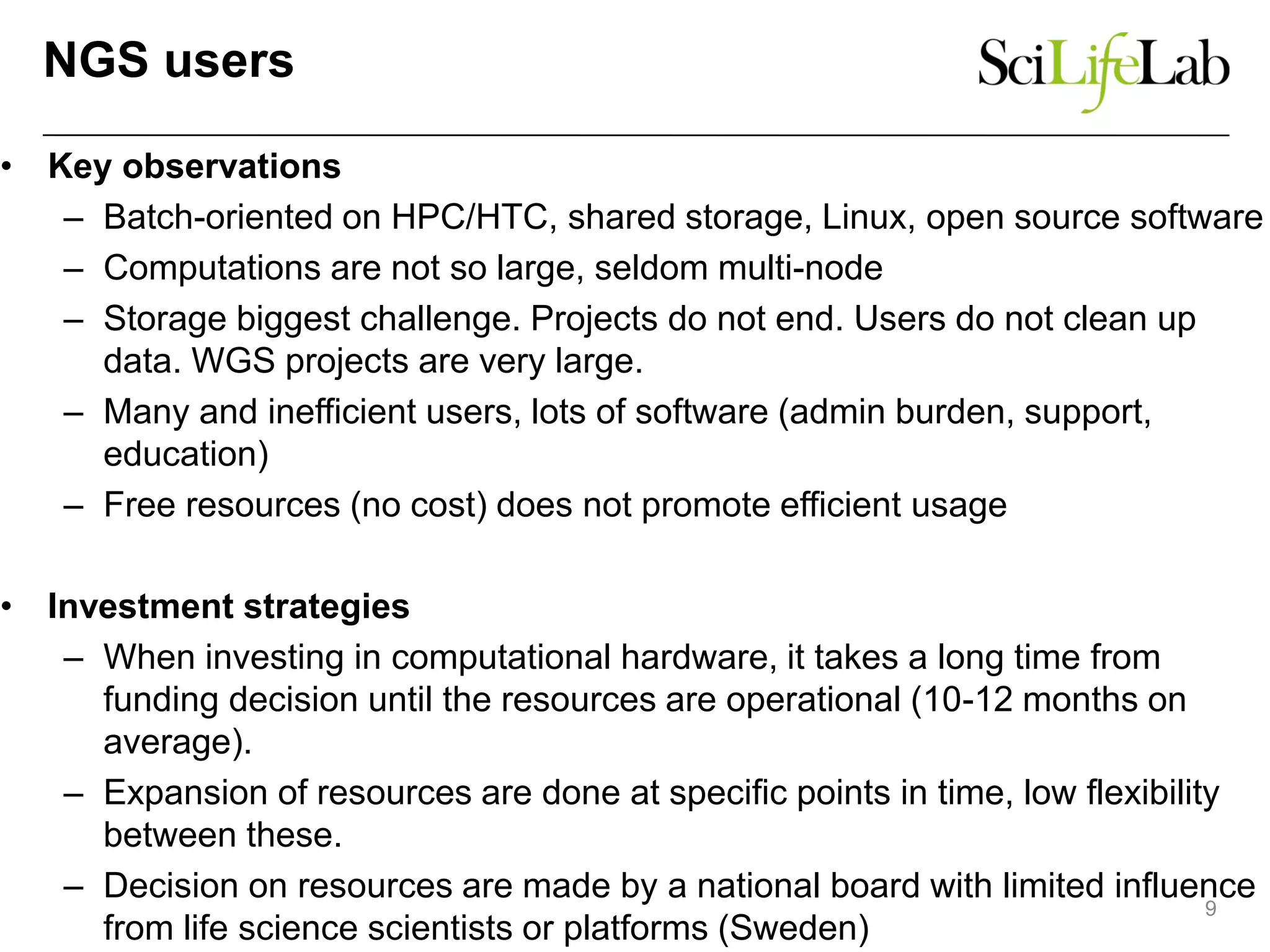
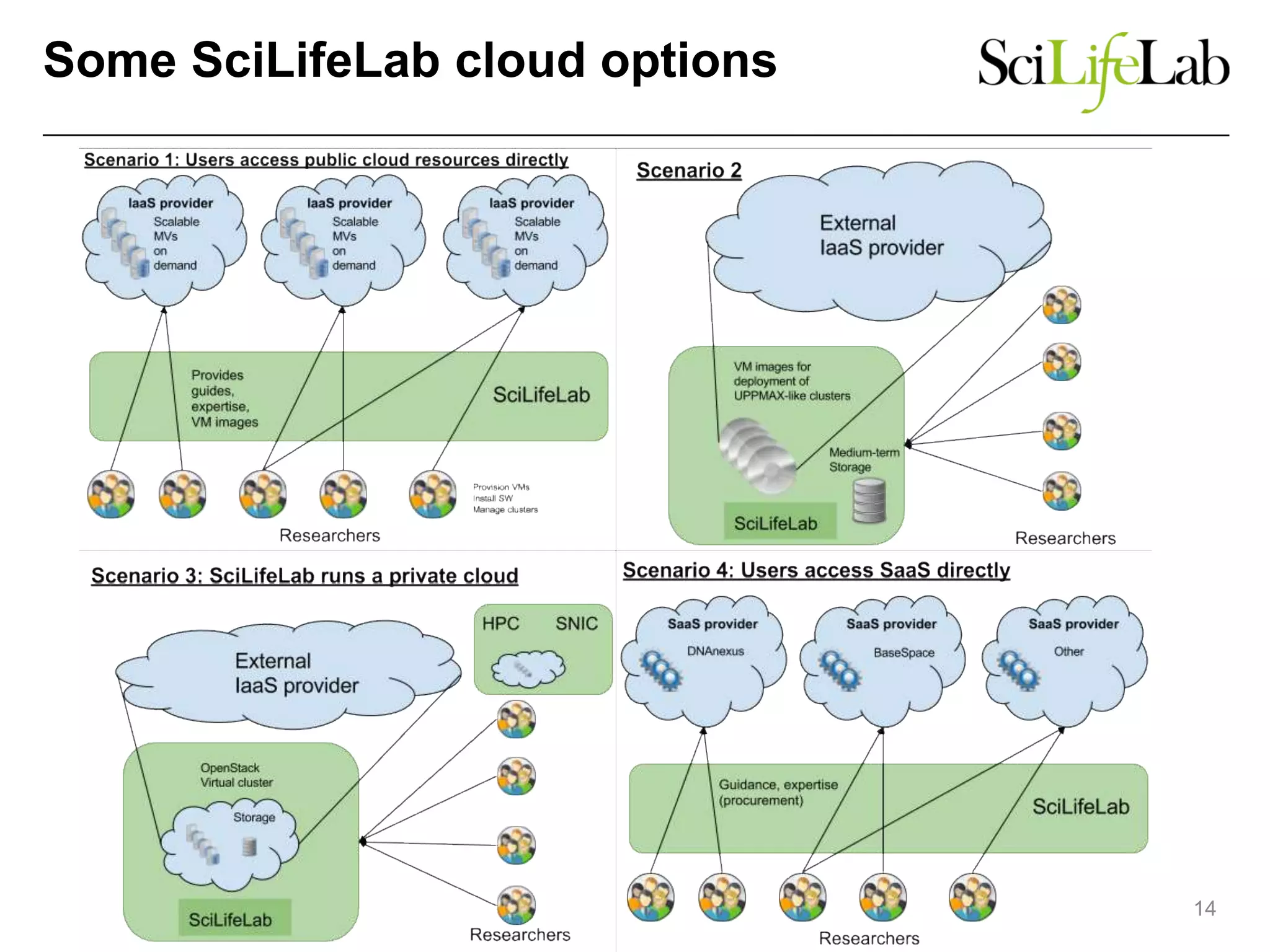
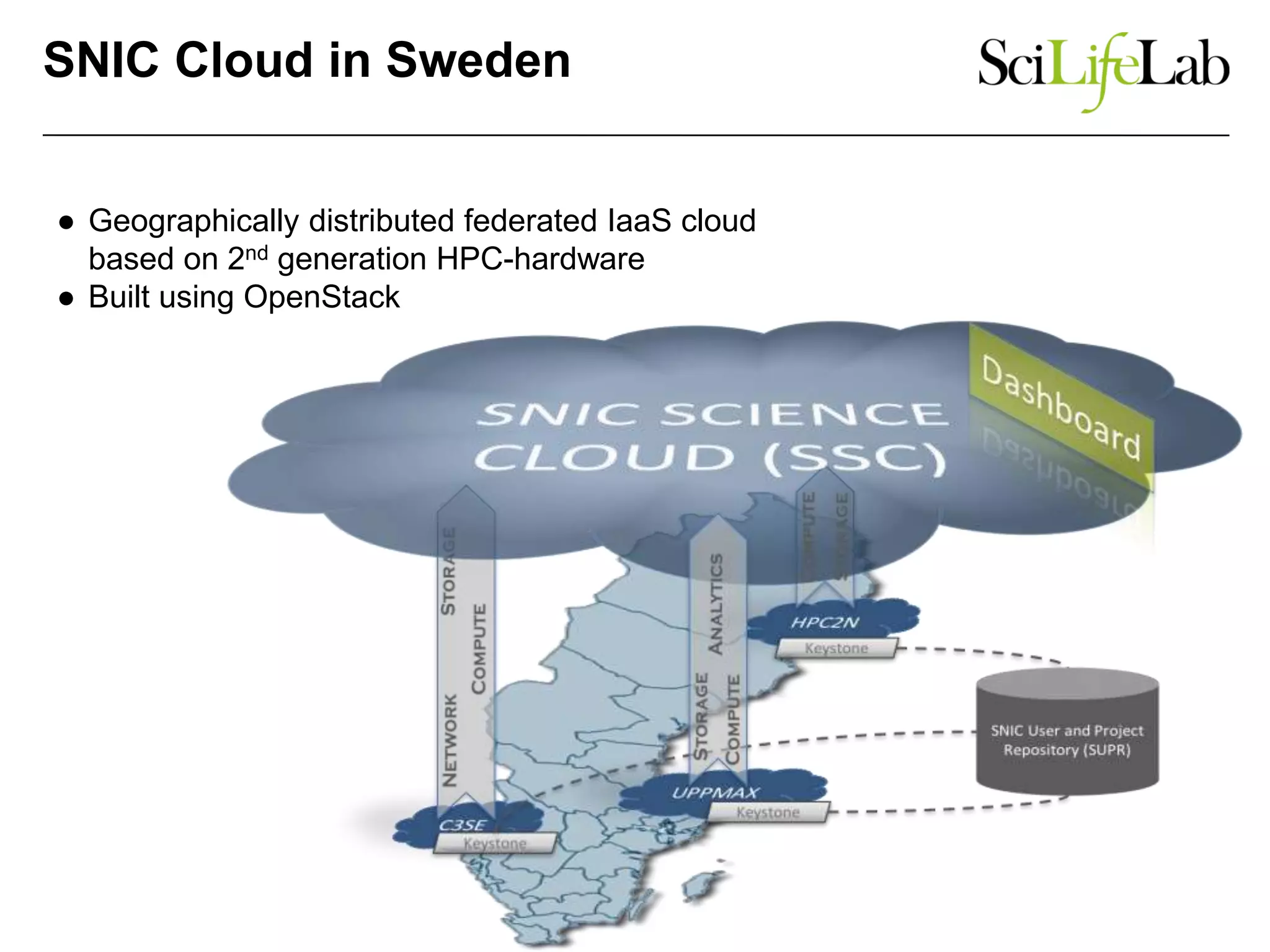
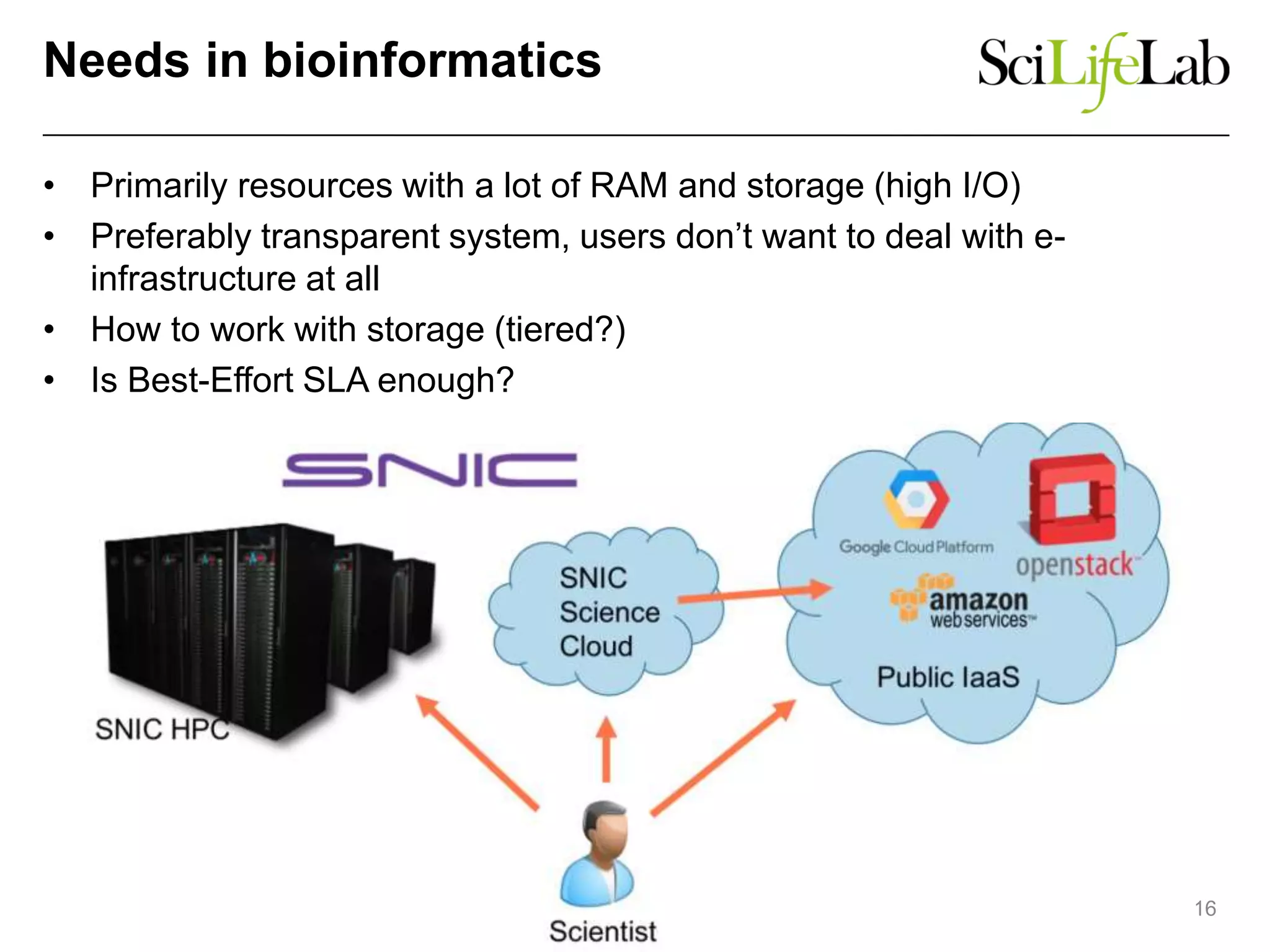
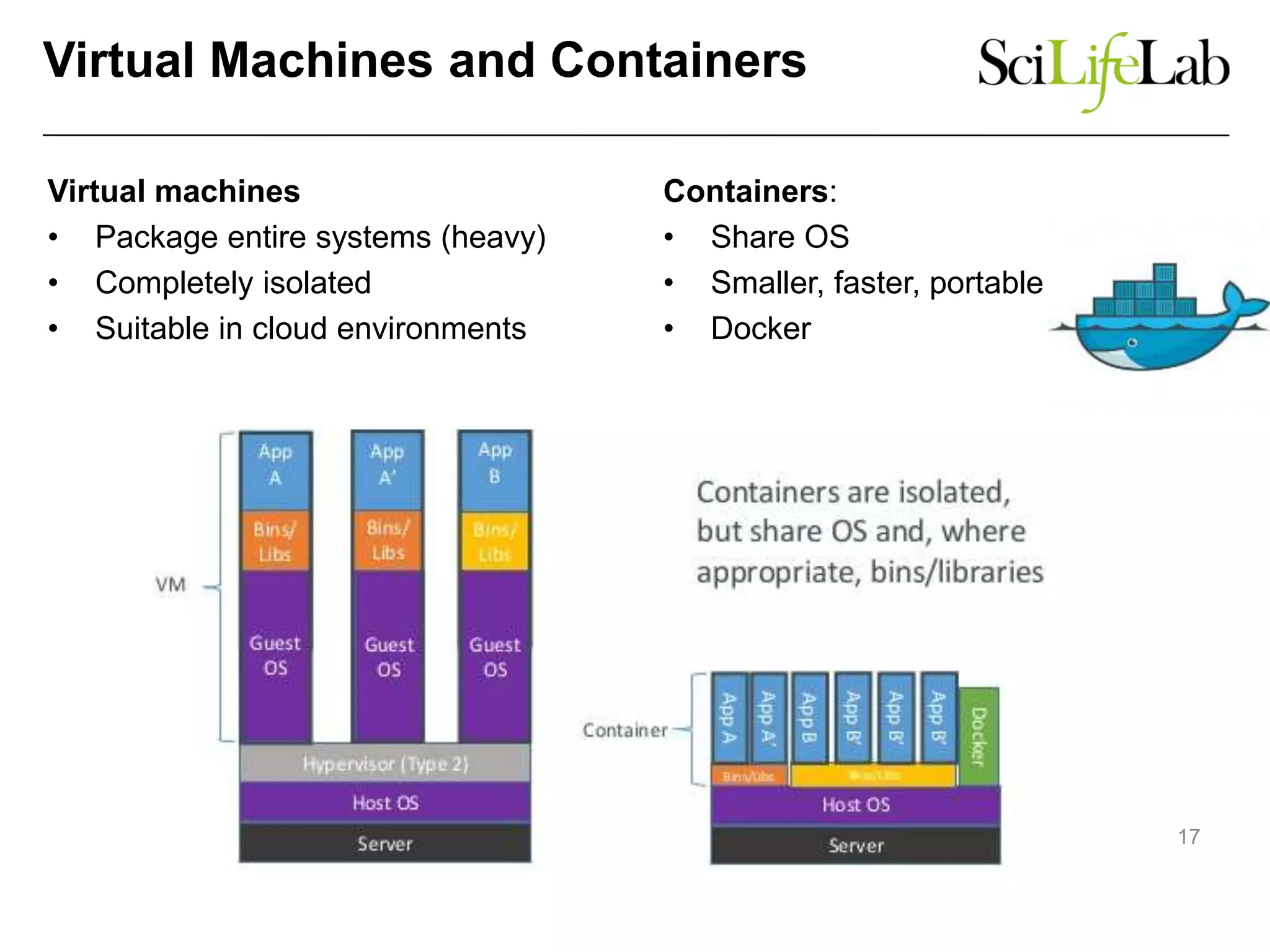
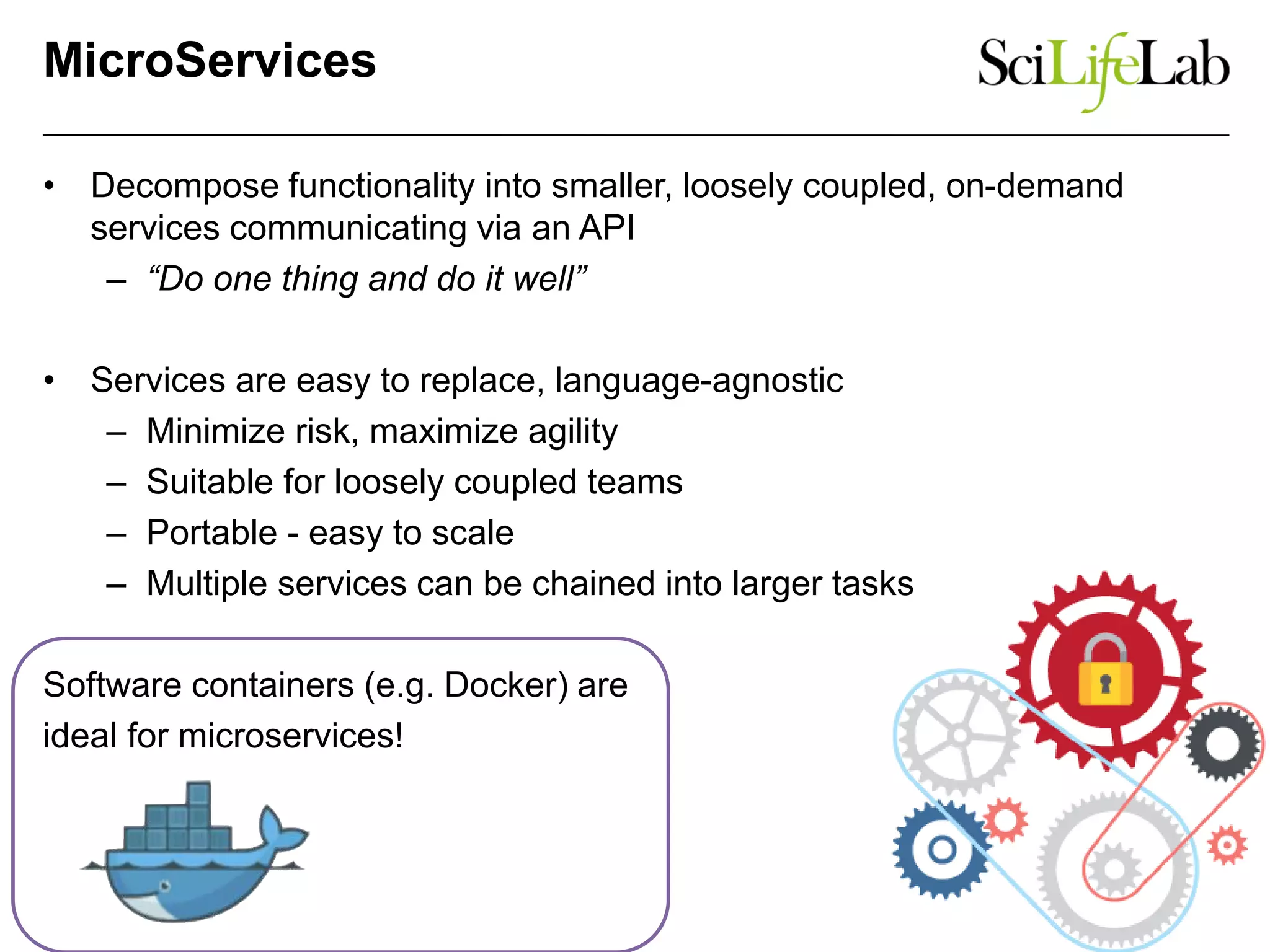
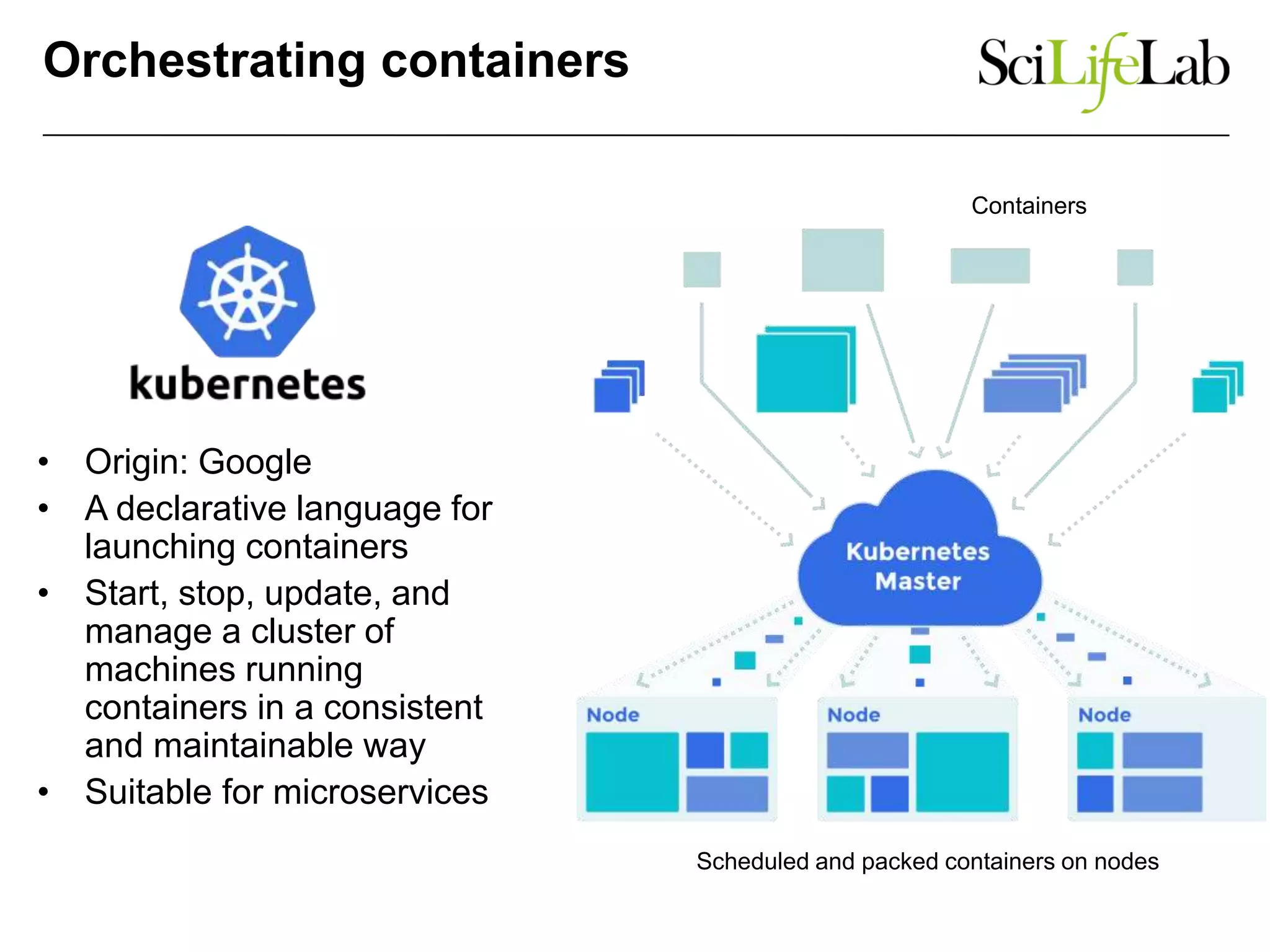
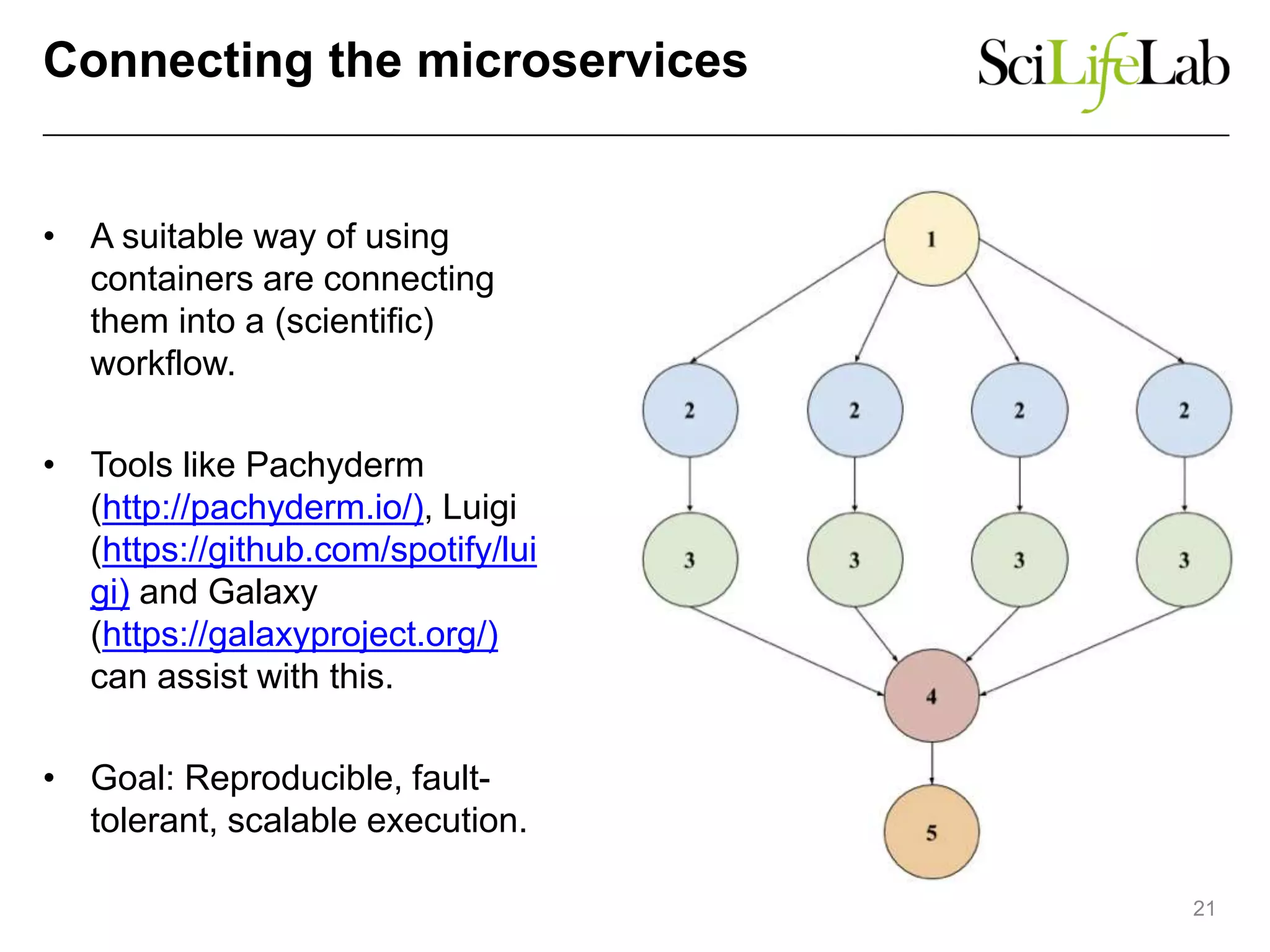
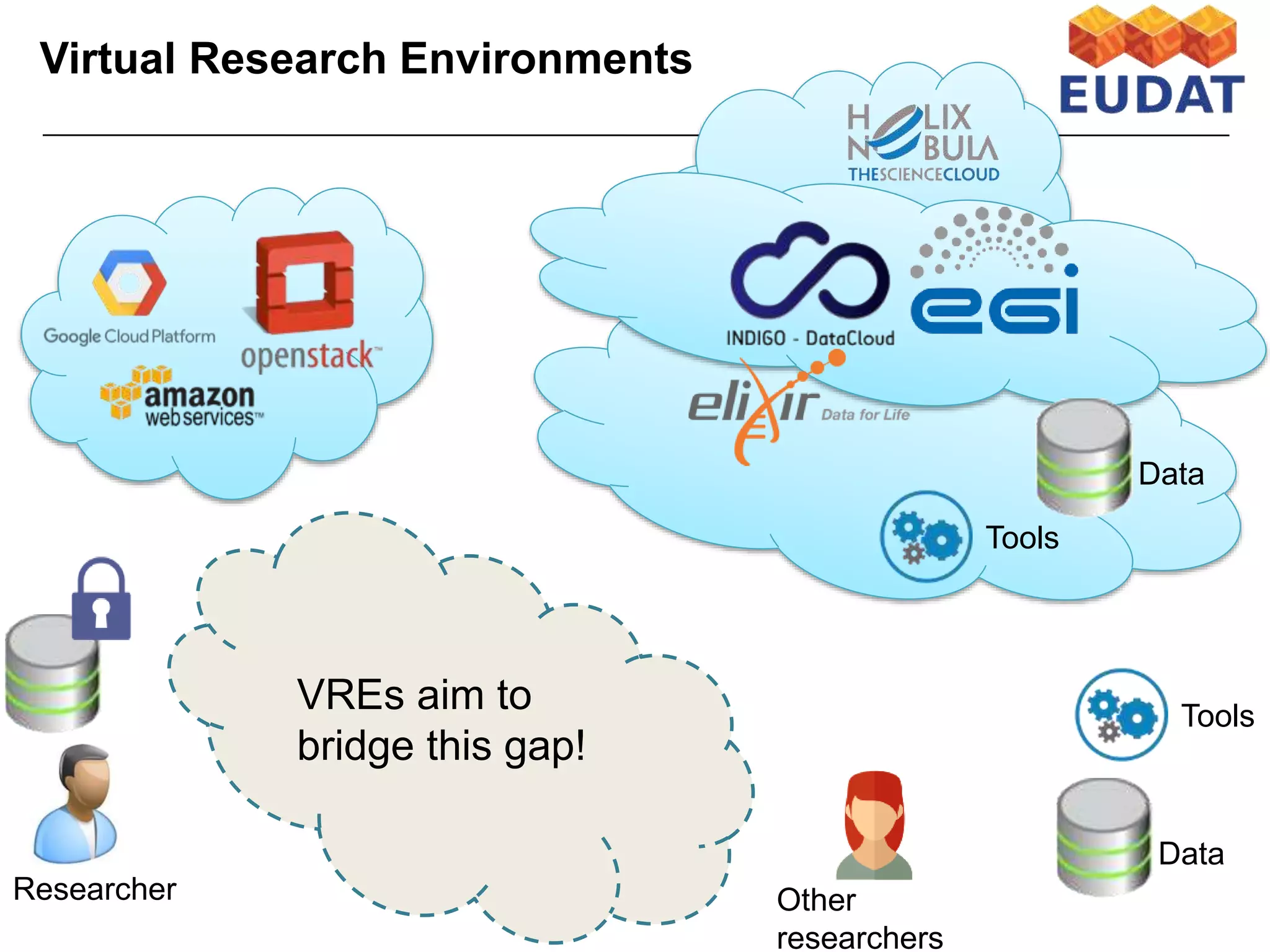
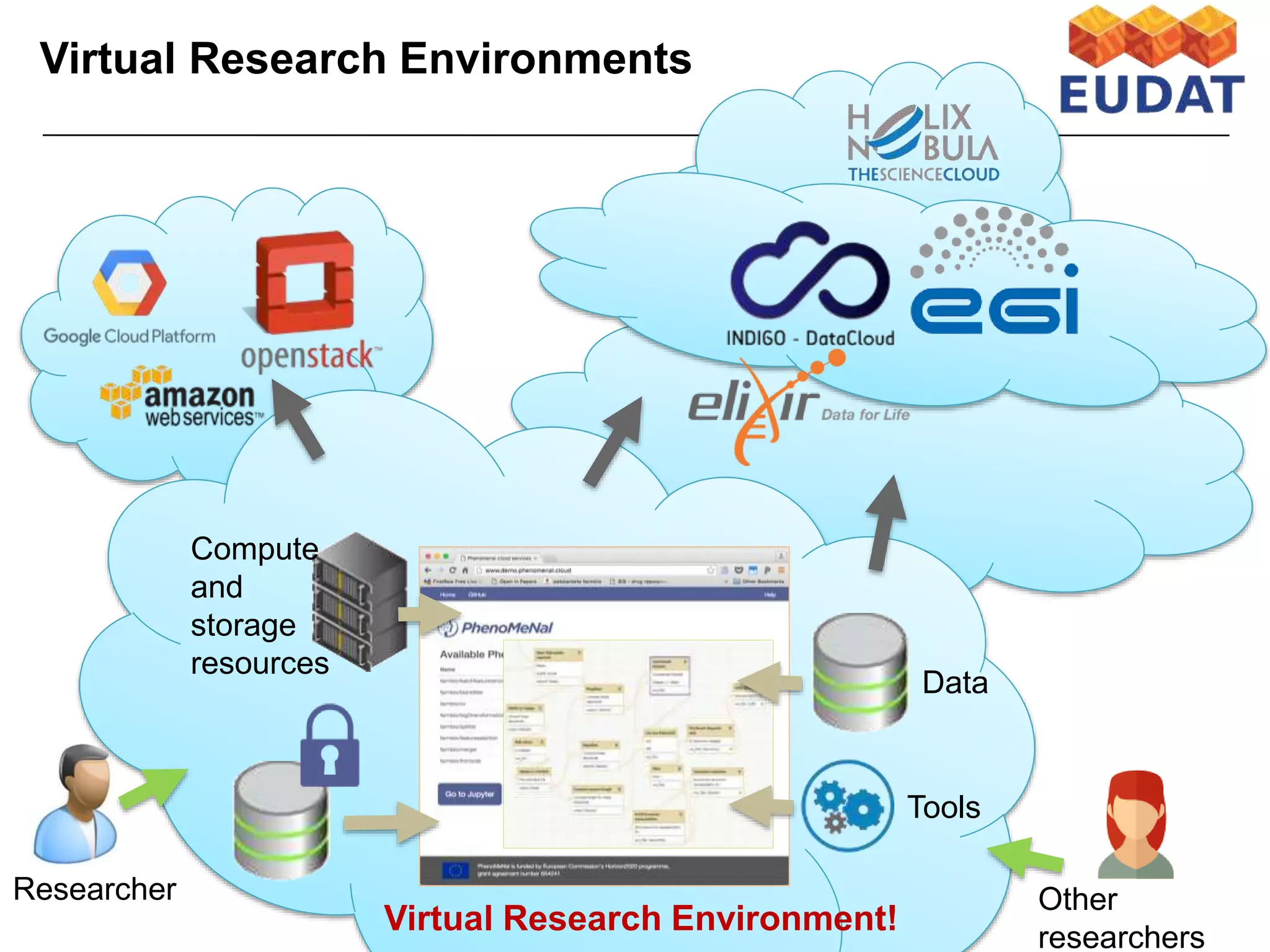
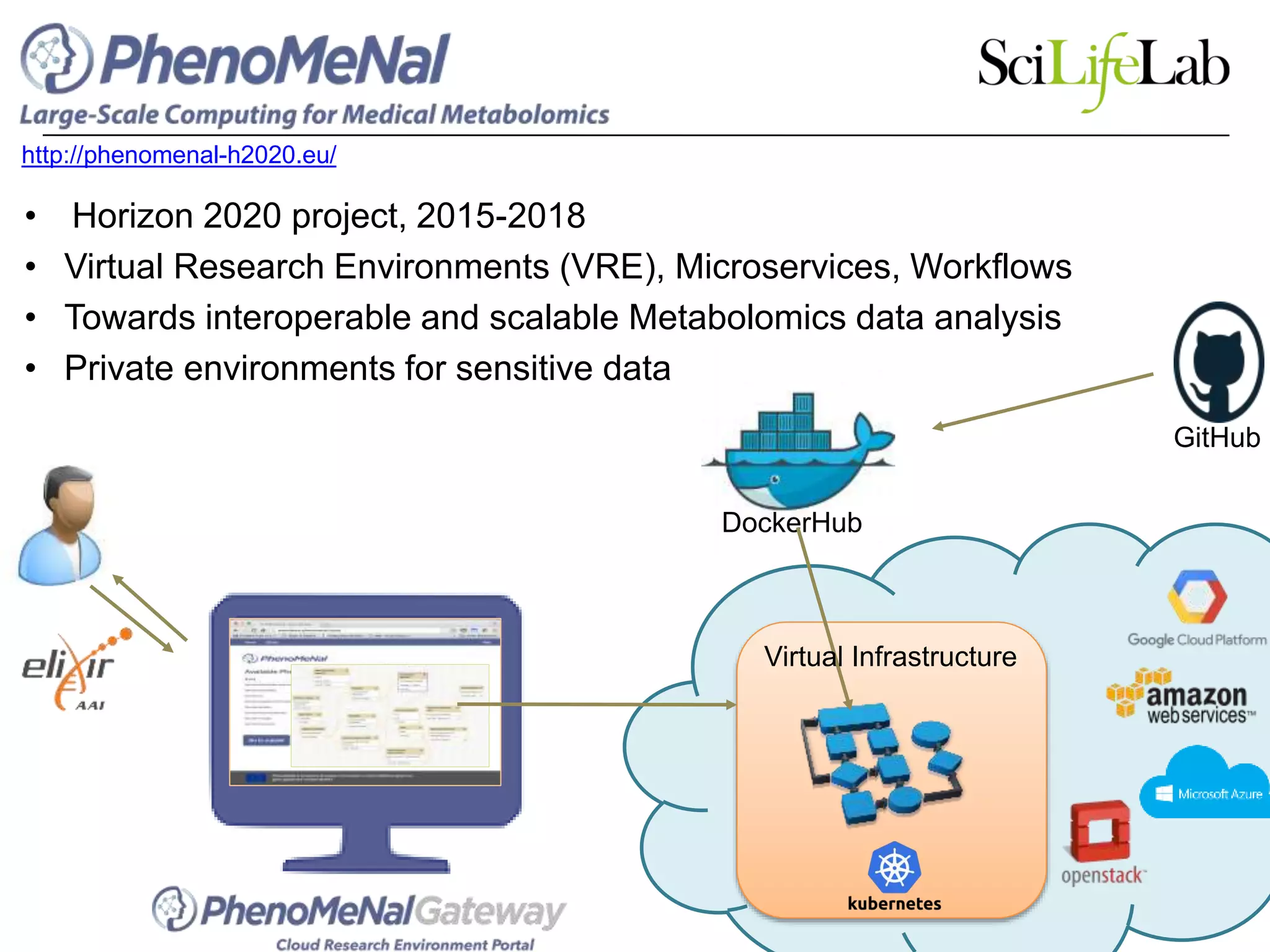
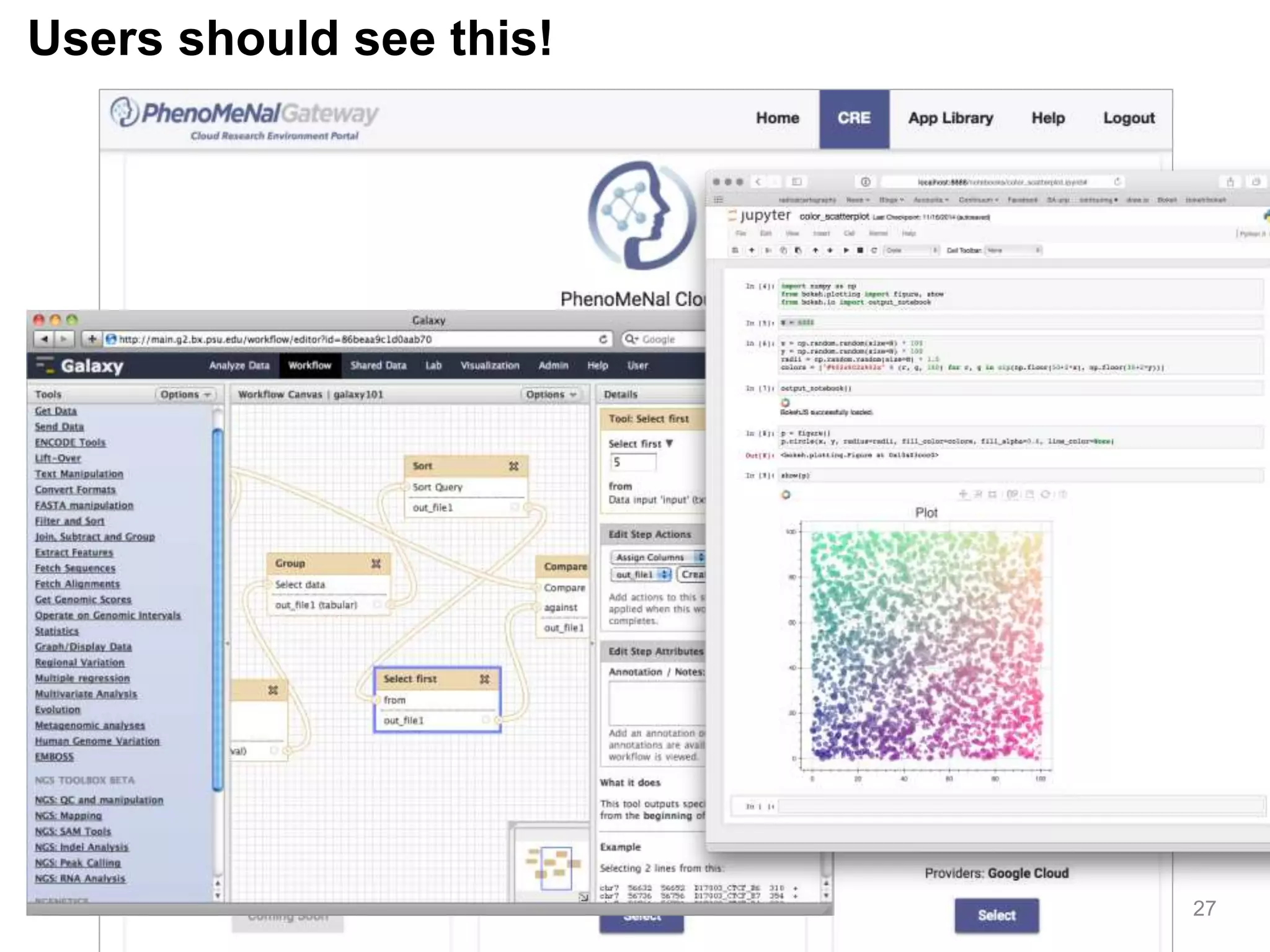
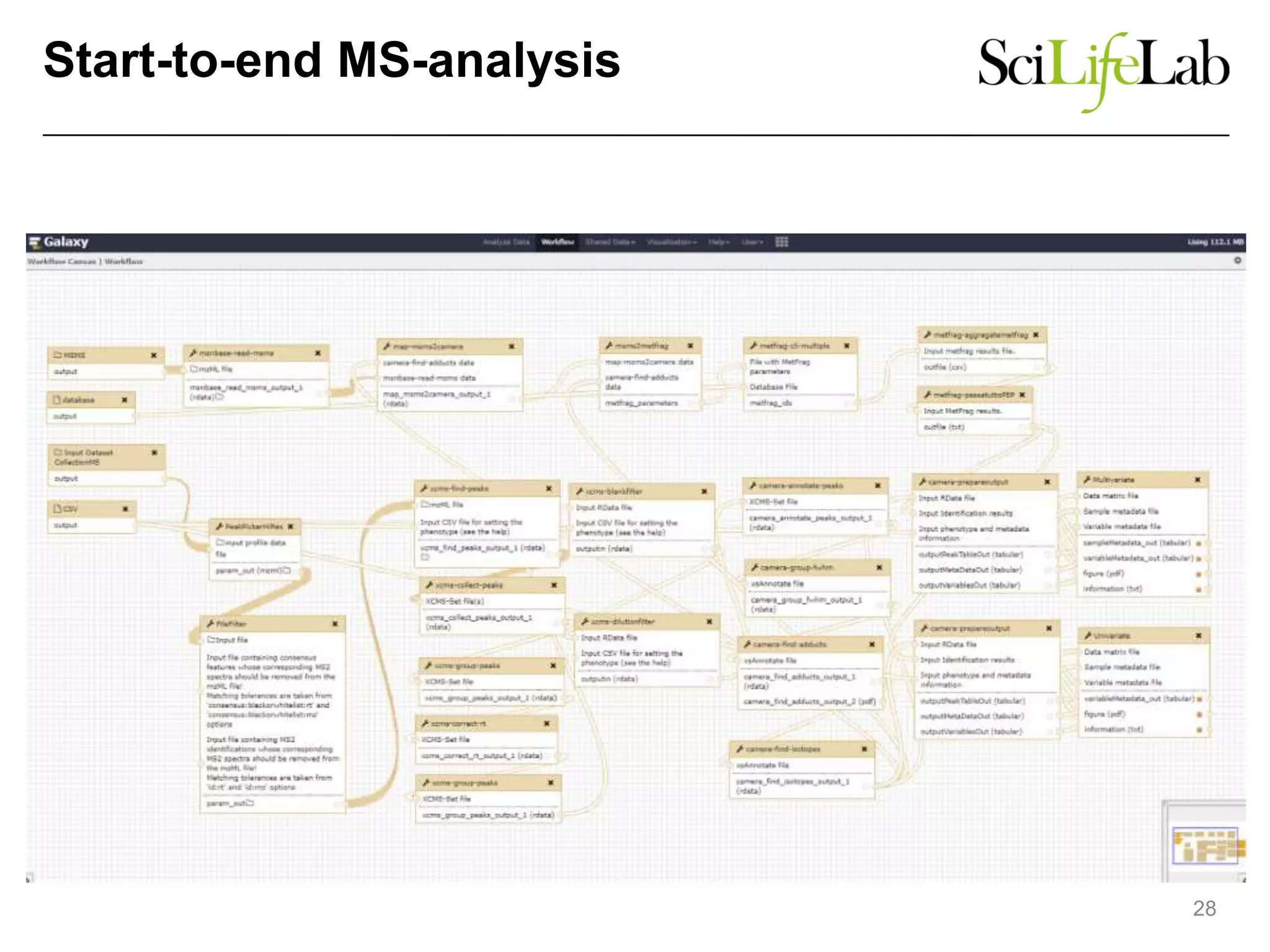
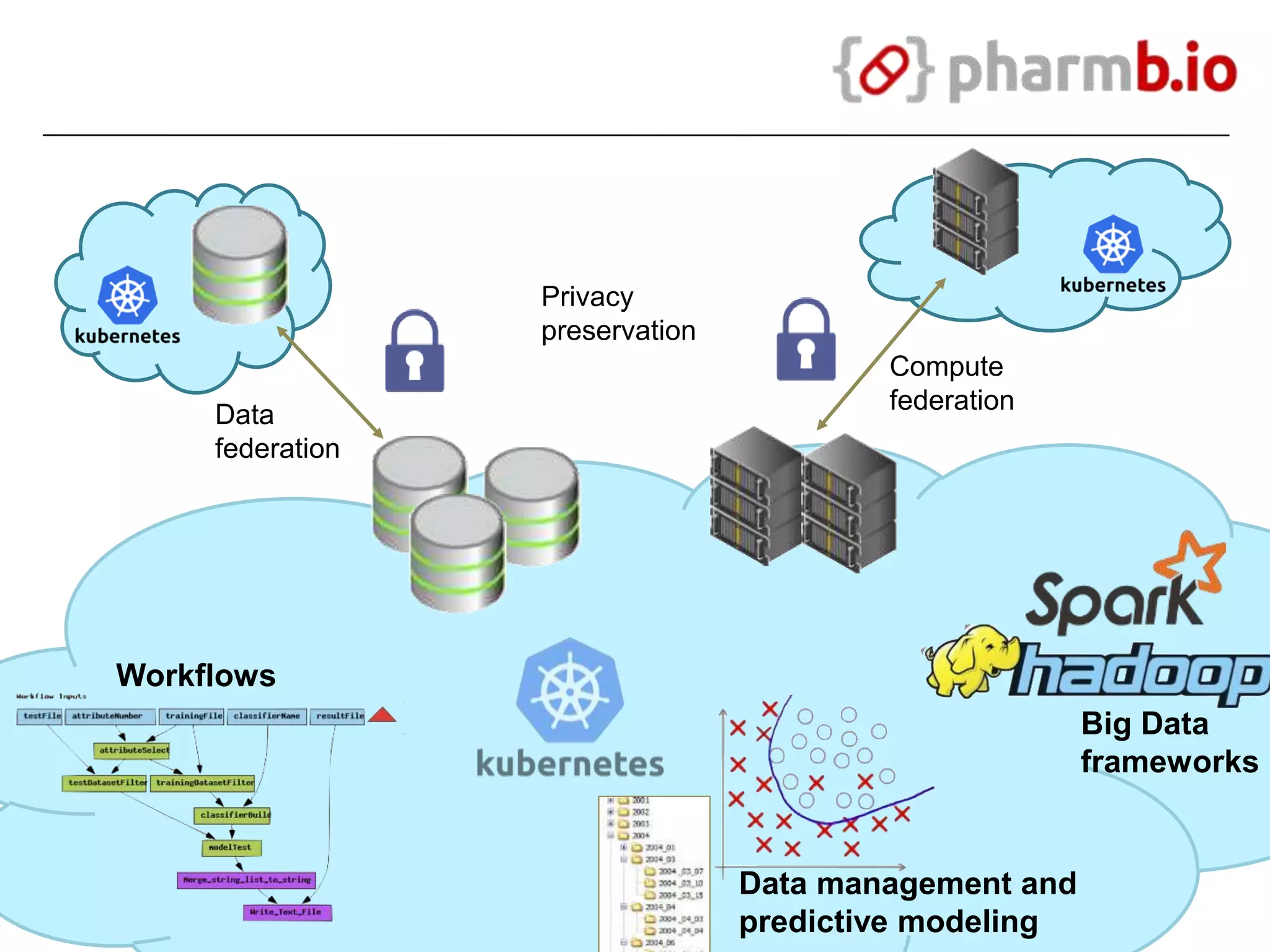
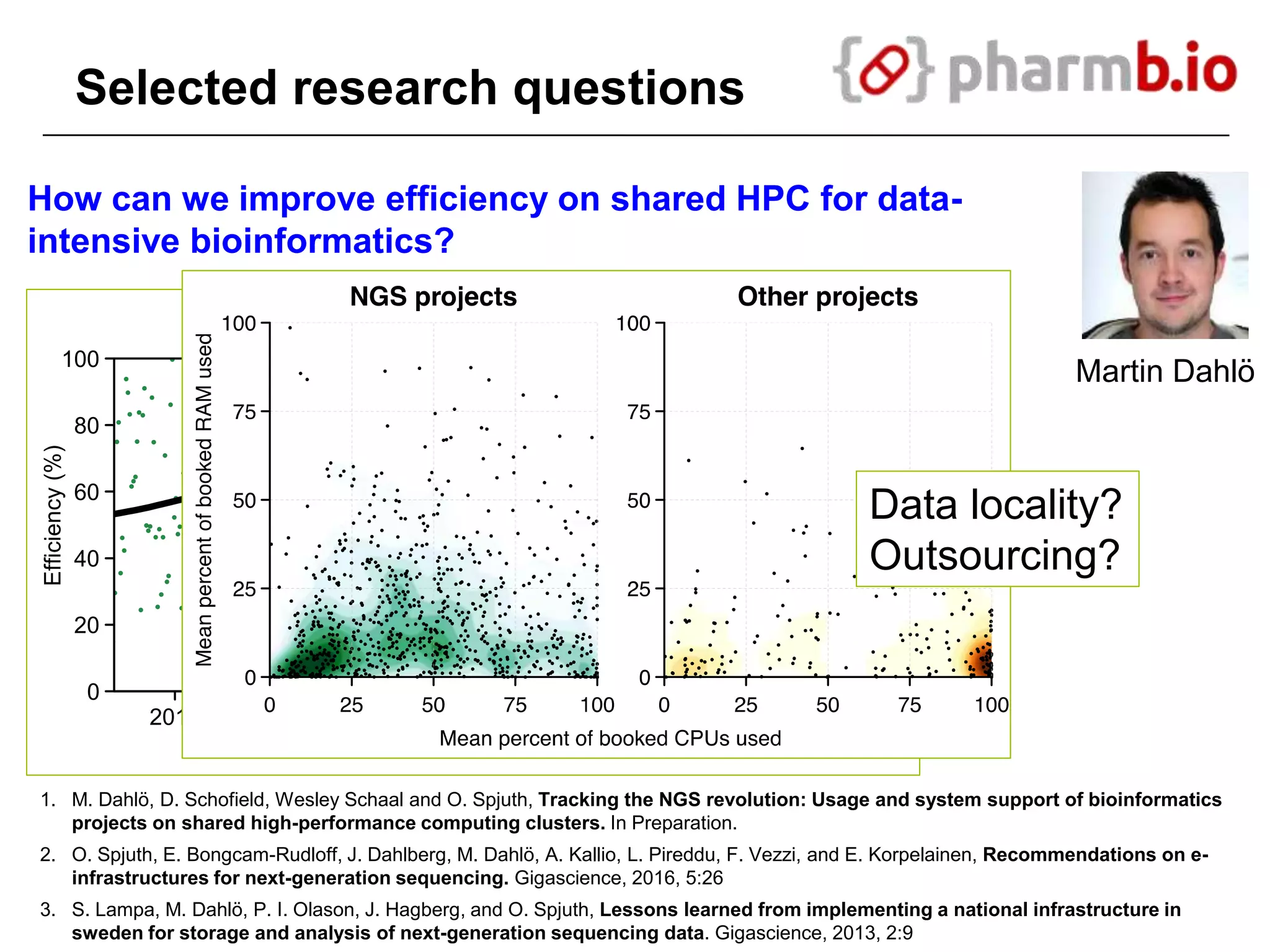
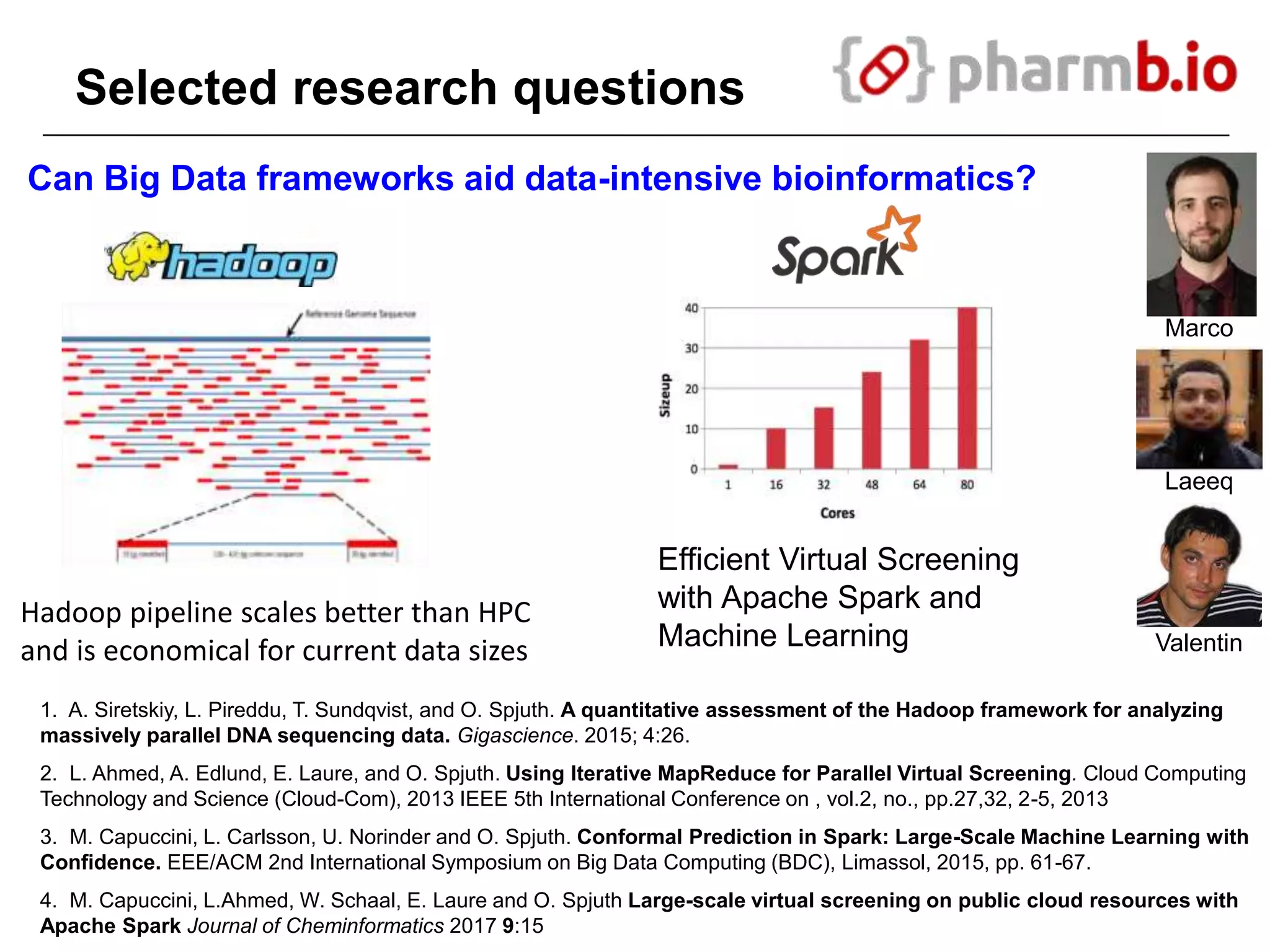
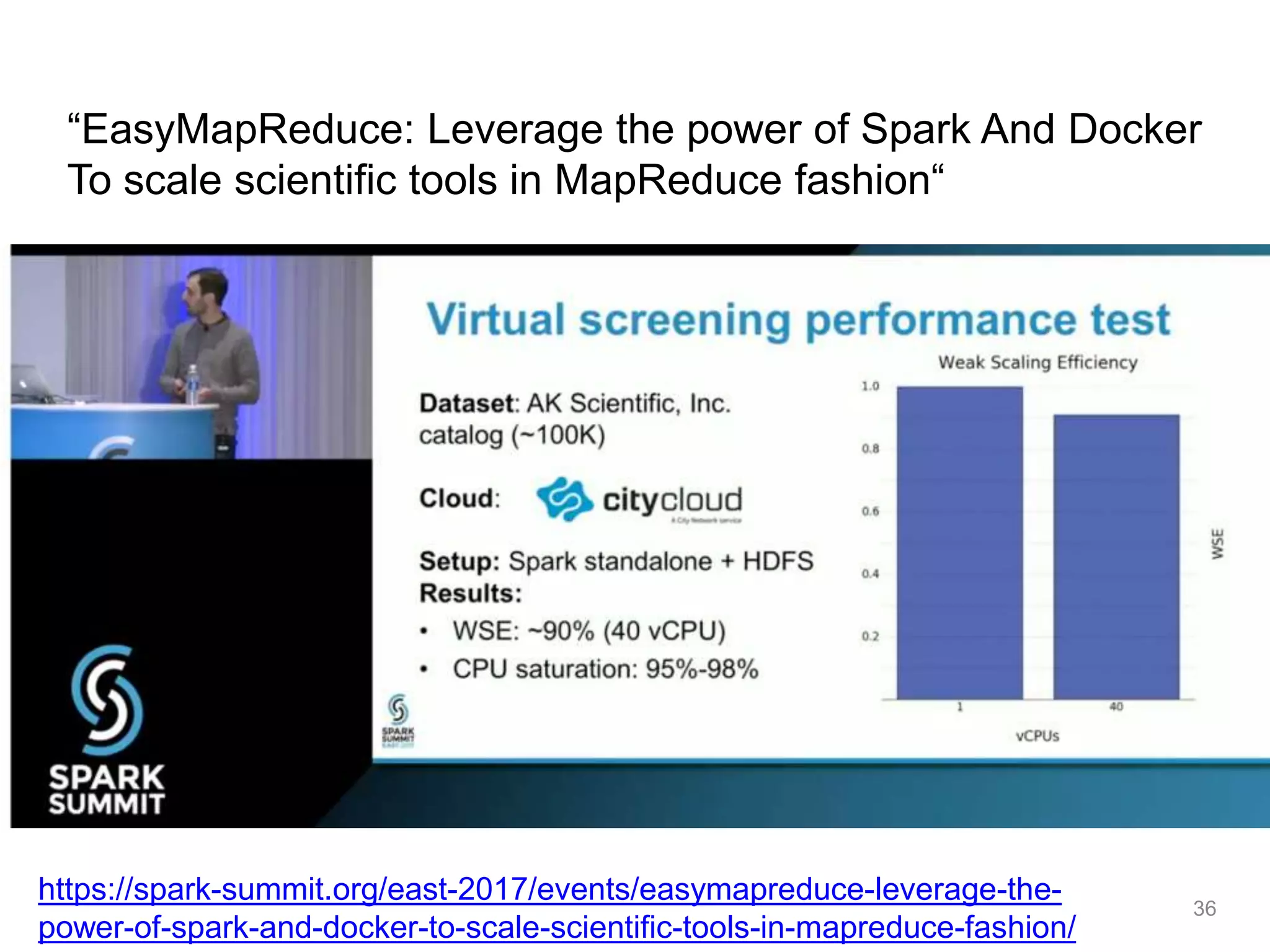
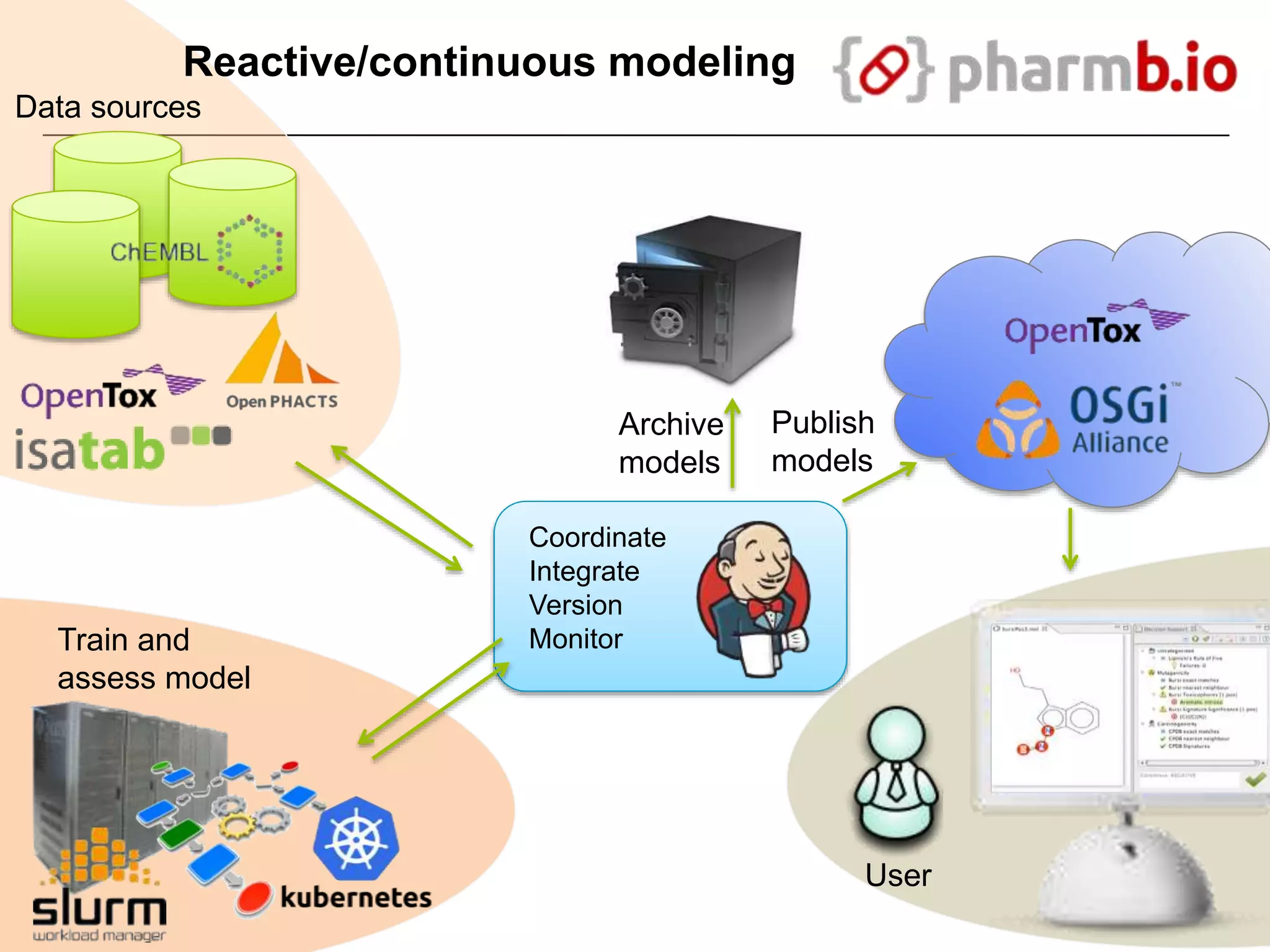
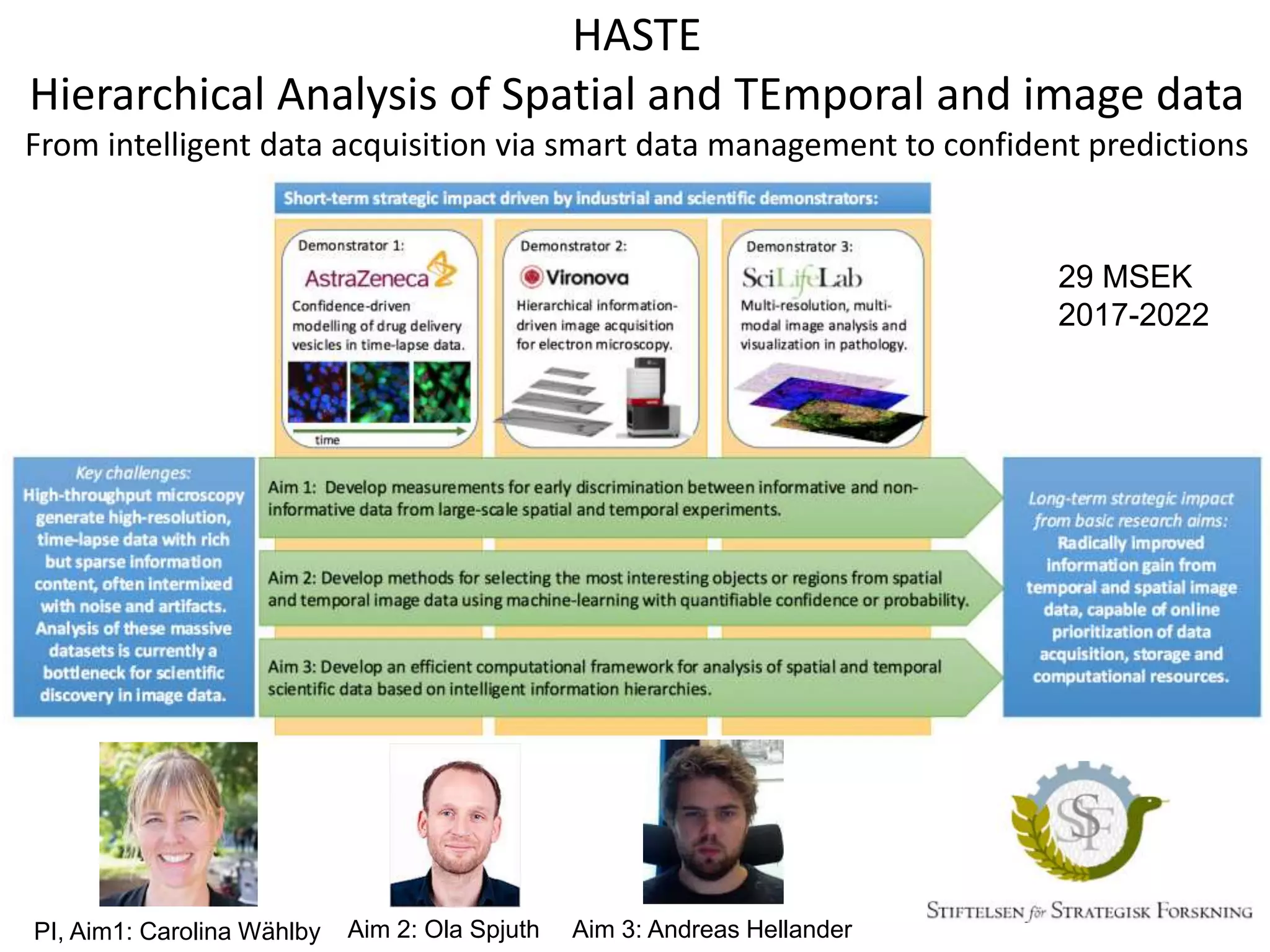
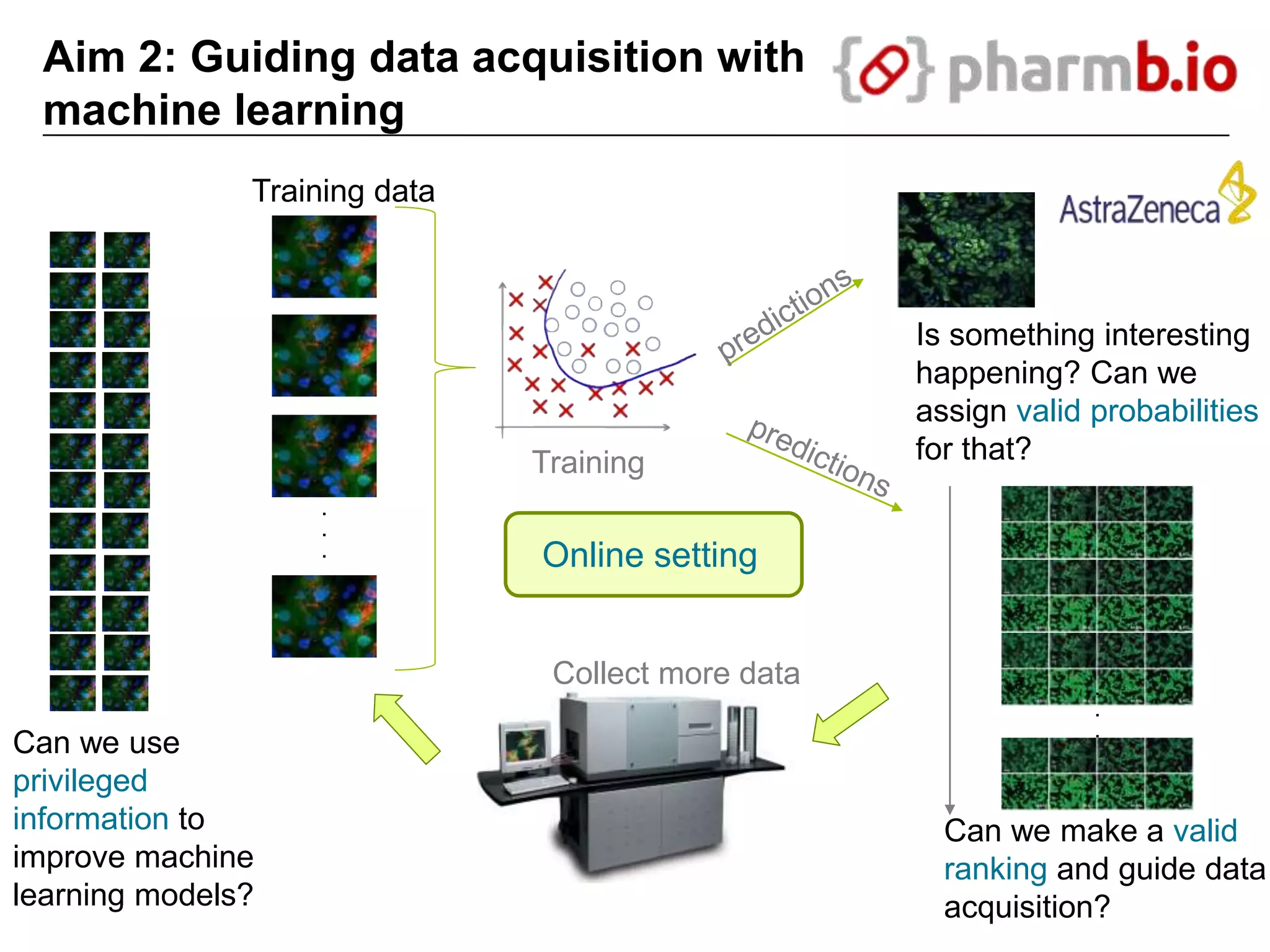
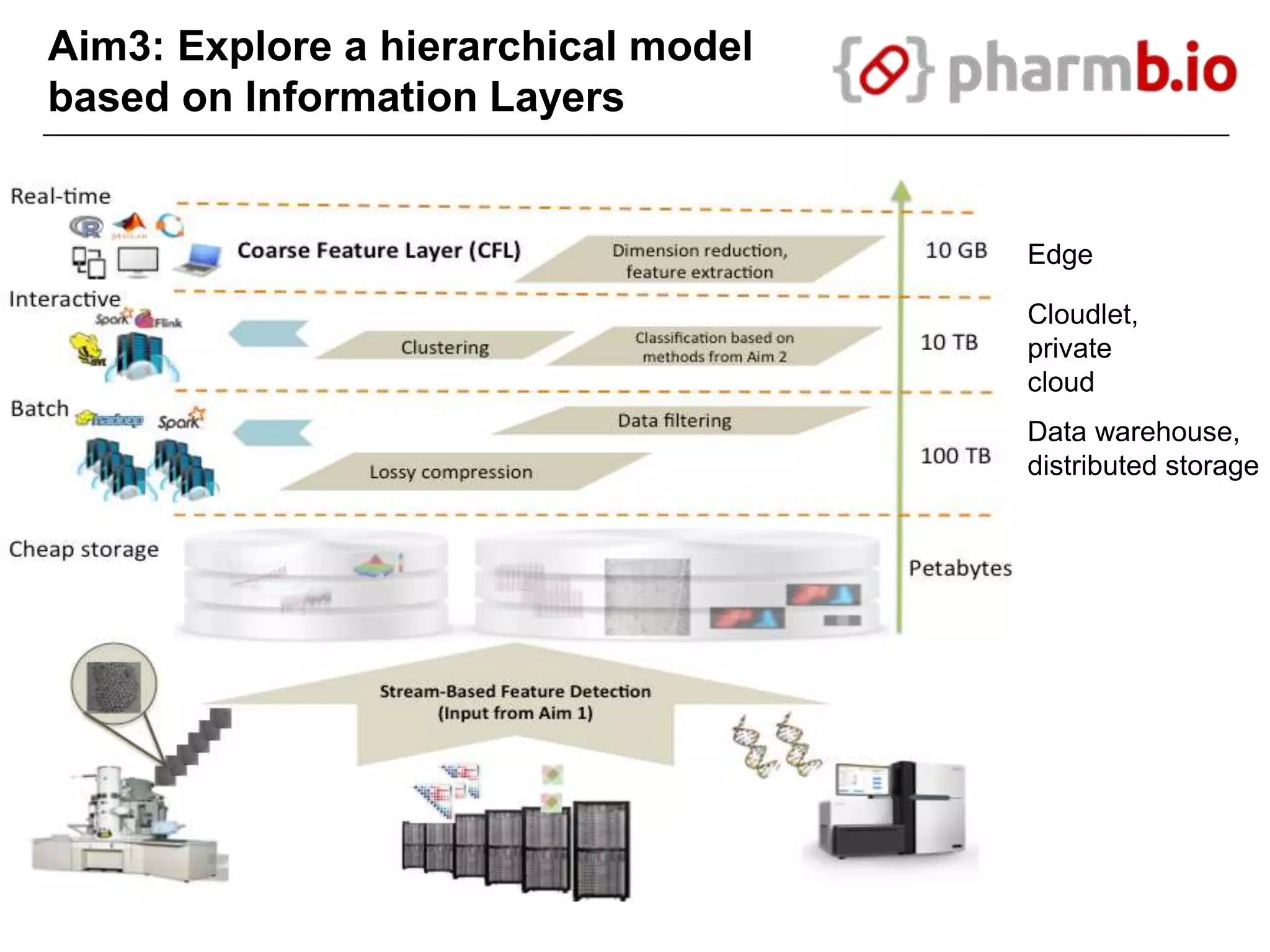
The document discusses the integration of cloud computing resources in data-intensive bioinformatics applications, particularly in life sciences. It highlights the challenges faced in traditional high-performance computing (HPC) environments, such as data storage and management inefficiencies, and emphasizes the benefits of cloud solutions like flexibility and collaboration across platforms. Furthermore, various strategies, tools, and research questions are introduced to improve the infrastructure and efficiency of bioinformatics workflows.