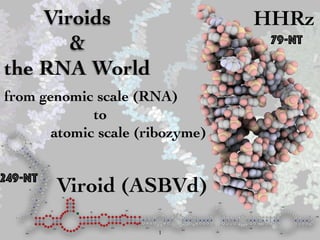
Viroids and the RNA World
- 1. Viroids & the RNA World from genomic scale (RNA) to atomic scale (ribozyme) Viroid (ASBVd) G G A A G A G A U U G A A G A C G A G U G A A C UAA U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U UUUGU A AAA A AACAAUGAAG AUA GAGGA A UAAAC C UUG CGA GAC UC AUCAGUGUU C UUCC CAU CUUUCC C U GAA G A GAC GAA GUG A UC 1 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 180 190 200 210220230 240 249 79-nt 249-nt HHRz 1
- 2. Introduction • evolution (RNA world)& genomics • RNA biology & RNomics (molecular and cellular functions) • structural biology & structural bioinformatics (structural basis for functions) • enzymology & computational enzymology (catalysis) 2
- 3. Prebiotic & RNA Worlds selfish elements, viroids, etc self-replication replication, amplification Horning & Joyce, PNAS, 2016.Attwater et al., Nat. Chem., 2013. ribozymes Martin et al., Life, 2015. 3
- 4. Virus World & RNA World « The ancient Virus World and evolution of cells » Pre-archaeal compartment Pre-bacterial compartment ocean selfish ribozymes (group I Introns) positive- strand, ds RNA viruses retrons, group II introns crust dsDNA and RCR viruses, plasmids Bacteria with plasmids, retrons, group I & II introns Archaea with plasmids, group I introns Escape of cells with their viruses and other parasitic elements RNA World RNA-DNA Retro World inorganic compartments RNA-Protein World DNA World Koonin et al., Biol. Direct, 2006 viroids 4
- 5. Viroids: « survivors from the RNA World » • small size • high GC • circRNA • periodicity • no protein- coding • ribozyme • error-prone replication • replication fidelity • replication • genome assembly • ribosome- free • replicationfeatures functions Holmes, J. Virol., 2011 5
- 6. Viroids: Families remain to be determined. A critical issue for all viroids is that bona fide promoter se- quences have not been characterized fully. This obviously is one of the most pressing issues that need to be addressed in order to fully understand how viroid RNA templates are recognized and transcribed by the cellular machinery. RNA motifs and protein factors for cleavage and ligation. The sequence and structural conservation of the CCR of several members of Pospiviroidae suggests its potential importance in Weak se some resea (Tabler and thought tha catalyzes t 1987a,b). R ing self-cle to the inter strates use Fig. 3. Asymmetric rolling circle replication of Potato spindle tuber viroid (PSTVd) and sym (ASBVd). The secondary structures of the genomic or circular RNAs are sketched to facilitate i an in vitro transcription system. With a combination of proto- cols to remove cellular RNAs and, thereby, enrich the de novo synthesized (–)-strand PSTVd RNAs from the circular (+)- RNA templates in potato nuclear extracts and primer extension, Kolonko and associates (2006) mapped the transcription initiate site on the circular (+)-RNA to U359/C1 of the left terminal loop (Fig. 1). Because of the low resolution of sequencing gels, it is not possible to determine precisely whether U359 or C1 is the exact initiation site. Site-directed mutagenesis in combination with infection studies in tomato revealed that the C1G mutation was maintained stably, whereas U359G reverted to wild type, suggesting that perhaps U359 is the bona fide initiation site. It is notable that these data are consistent with previous in vitro studies showing that Pol II binds to the terminal loop or loops of PSTVd (Goodman et al. 1984). These observations establish a basis for further investigations to determine whether the in vitro transcription initiation site is the same as that used in vivo. The transcription initiation sites on the (–)-strand template also remain to be determined. A critical issue for all viroids is that bona fide promoter se- quences have not been characterized fully. This obviously is one of the most pressing issues that need to be addressed in order to fully understand how viroid RNA templates are recognized and transcribed by the cellular machinery. RNA motifs and protein factors for cleavage and ligation. The sequence and structural conservation of the CCR of several members of Pospiviroidae suggests its potential importance in viroid processing during replication (Candresse et al. 1990; Diener 1986; Hashimoto and Machida 1985; Meshi et al. 1985; Tabler and Sänger 1985; Visvader et al. 1985). Extensive in vitro studies provided evidence to support this hypothesis (Baumstark and Riesner 1995). Furthermore, in vitro studies with longer than unit-length PSTVd transcripts mapped the cleavage and ligation site to between G95 and G96 (Baumstark et al. 1997). The first cleavage at the 5′ end of G96 occurs in a metastable tetraloop motif, which results in a conformational change to form a stable loop E that drives the second cleavage at the 3′ end of G95 and subsequent ligation (Baumstark et al. 1997). Recent work with a minicircle RNA showed that the CCR contains all the necessary elements for cleavage and liga- tion (Schrader et al. 2003). It is important to note that process- ing also can occur outside CCR, with the specific sites to be elucidated (Hammond et al. 1989; Tabler et al. 1992). A key question that remains to be answered is whether single or mul- tiple sites are used for processing in vivo. Weak self-cleavage of PSTVd RNAs has been reported by some researchers (Robertson et al. 1985) but not by others (Tabler and Sänger 1985; Tsagris et al. 1987a,b). It generally is thought that a cellular RNase which remains to be identified catalyzes the cleavage of concatemeric RNAs (Tsagris et al. 1987a,b). Reasoning that the general difficulty of demonstrat- ing self-cleavage of RNAs in Pospiviroidae could be attributed to the interference of nonribozyme RNA sequences in the sub- strates used during in vitro assays, Liu and Symons (1998) Flores et al., Arch. Virol., 1998. 6
- 7. Viroids: Plant Parasites Ding & Itaya, Mol Plant Microbe Interact, 2007. 10 / Molecular Plant-Microbe Interactions unsuccessful attempts to establish a transcription system for PLMVd in cell extracts of several plant species, Pelchat and associates (2002) tested whether the Escherichia coli DNA- dependent RNA polymerase would transcribe PLMVd in vitro. The observed transcription led to the suggestion that, in infected plant cells, the PEP catalyzes transcription of PLMVd. However, recent work suggests that NEP more likely is involved in the transcription of PLMVd in vivo (Delgado et al. 2005). Thus, further biochemical and genetic studies will be necessary to ments showed that this left loop is the binding site for the β and β′ subunits of the E. coli enzyme (Pelchat and Perreault 2004). The in vivo significance of these sites remains to be seen, in light of recent work that mapped the in vivo initiation sites of PLMVd to C51 in the (+)-strand RNA and A286 in the (–)-strand RNA, in similar 6- to 7-bp double-stranded motifs (Fig. 1) (Delgado et al. 2005). Mapping the transcription initiation sites for members of Pospiviroidae has been achieved only recently for PSTVd using Fig. 2. Distinct steps of systemic infection of Avocado sunblotch viroid (ASBVd) and Potato spindle tuber viroid (PSTVd), type members of the two viroid families. The mechanisms of the different trafficking steps for the family Avsunviroidae remain to be investigated. (Modified from Ding et al. 2005, with permission from Elsevier Ltd.) 7
- 8. Viroids: Replication Rz: hammerhead ribozyme Flores et al., Viruses, 20098
- 9. Viroids: 2D Structures Vol. 20, No. 1, 2007 . 1. Secondary structures of representative viroids from the two viroid families, Avsunviroidae: Avocado sunblotch viroid (ASBVd) and Peach latent mo id (PLMVd), and Pospiviroidae: Potato spindle tuber viroid (PSTVd). The transcription initiation sites on the viroid genomic RNAs are indicated. Note ASBVd and PSTVd, these sites are mapped to terminal loops. The transcription initiation site for the (–)-PSTVd RNA template remains to be determined. MVd, the dashed lines indicate kissing-loop interactions. For PSTVd, the five structural domains (Keese et al. 1985) are indicated. TL = left-terminal dom central domain, and TR = right-terminal domain. HPII′ and HPII indicate nucleotide sequences that base pair to form the metastable hairpin II structure. Ding & Itaya, Mol Plant Microbe Interact, 2007. Pospiviroids Avsunviroids 9
- 10. Viroids: Pospiviroids Figure 1. Structure and replication of Pospiviroidae. (A) Schematic representation of the consensus secondary structure of the 359 nt circular (+) PSTVd with the five functional domains. TL: Left terminal domain, P: pathogenicity-modulating domain, C: conserved central core, V: variable domain, TR: right terminal domain.11 (B) Replication follows an asymmetric rolling-circle mechanism.12 For details, see text. Hammann & Steger, RNA Biol., 2012. 10
- 12. Viroids & RNA silencing 12
- 13. ASBVd: 2D Structures esolving these issues is of great interest to broaden wledge of the molecular processes in these organelles rther our understanding of the molecular basis for the of infectious RNAs. zyme machinery for transcription. The DNA-depend- polymerase II (Pol II) is generally accepted to be in- the transcription of members of Pospiviroidae. Three plate in vitro (Rackwitz et al. 1981). Second, α-amaniti the replication of PSTVd (Mühlbach and Säng Schindler and Mühlbach 1992), Cucumber pale fr (Mühlbach and Sanger 1979), Hop stunt viroid (Yoshikawa and Takahashi 1986), and CEVd (Flor Flores and Semancik 1982; Rivera-Bustamante and S 1989; Semancik and Harper 1984). Low concentrati Ding & Itaya, Mol Plant Microbe Interact, 2007. 13
- 14. ASBVd(-): 2D Structures A G G A A G A G A U U G A A G A C G A G U G A A C UAA U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U UUUGU A AAA A AACAAUGAAG AUA GAGGA A UAAAC C UUG CGA GAC UC AUCAGUGUU C UUCC CAU CUUUCC C U GAA G A GAC GAA GUG A UC 1 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 180 190 200 210220230240 249 G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G 1a 10a 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230a 240a monomer 1 Rz gguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1 10 20 30 40 50 60 70 79 intra-molecular base-pairs tertiary or inter-molecular contacts nucleotide in tertiary contact cleavage site Rz 5' 3' 5' 3' (-) Leclerc et al., Sci. Rep., 2016. (-) 3bps (+) 2bps Hammerhead (HHR) 14 Flores et al., Adv, Virus Res., 2000.
- 15. HHR motifs in life forms Perreault et al., PLoS Comput. Biol., 2011 Gupta & Swati, Interdiscip. Sci., 2016. FIG. 1. Schematic representation of the ASBVd RNAs expressed from plasmids and produced by the replication process. Hammer ribozymes are represented as black boxes within the pFL61-ASBVd(ϩ) and pFL61-ASBVd(Ϫ) plasmids expressing, respectively, the A and ASBVd(Ϫ) DNA dimer (dASBVd), and in the different expected transcripts. Plasmid sequences are indicated by gray boxes. The A and ASBVd(Ϫ) sequences are represented in blue and green, respectively. In the first step, self-cleavage of the ASBVd dimeric form via the ribozymes lead to the linear monomeric form (lmASBVd) and to the circular monomeric form (cmASBVd). In the second RNA-dependent replication occurs, linear oligomers (loASBVd), lmASBVd, and cmASBVd of the opposite polarity are produced. Blue arrows encompassing the linker sequence (LK), indicated as a black line, represent primers used during strand-specific reverse transcrip During PCR amplification, the LK sequence alone is used with the PCR primers (blue or green arrows), in order to avoid any amplificati from the plasmid. It is important to note that the cDNA resulting from RT of dASBVd, cmASBVd, and loASBVd can be PCR amplifie VOL. 85, 2011 A VIROID REPLICATES IN YEAS Delan-Forino et al., J. Virol., 2011. yeast 15
- 16. A prototype for RNA Catalysis Scott et al., Science, 1996Scott, Q. Rev. Biophys., 1999 substrate/enzyme cleavage site Canonical 2D Structure folded 2D Structure “Minimum” Hammerhead Ribozyme 41-nt 16
- 17. Asp424 Glu357 O NiO O O P OR OS O H 5' 3' 2' 5' 4' O Ni+1 OO H 3'2' 4' R RO(-)H Mg2+ (OH-) Mg2+(H2O) Mg2+ Self-Cleaving / Self-Splicing O NiO O O P OR OS O H 5' 3' 2' 5' 4' O Ni+1 OO H 3'2' 4' R (H2O) Mg2+ (OH-) Mg2+ O NiO O O P OR OS O H 5' 3' 2' 5' 4' O Ni+1 OO H 3'2' 4' R RO(-)H Mg2+ (OH-) Mg2+(H2O) Mg2+ O NiO O O P OR OS O H 5' 3' 2' 5' 4' O Ni+1 OO H 3'2' 4' R AH(+) B(-) NHN His12 HN N His119 H ribozyme ribozyme RNase A 3’-5’-exonuclease SN2(P) reaction (“in-line”) 17
- 18. Catalytic Metal Ions: One/ Two ound water in the fully hydrated La3ϩ ion, the low kobs for cleavage reaction involving the La3ϩ ion in both positions not compatible with the observed correlation between the a of a water bound to a metal ion and the kobs produced by ferent divalent metal ions. That correlation has been inter- ted in the metal hydroxide model (Fig. 4) as an effect on concentration of the aqueous metal hydroxide, which then ves as a Brønsted base in the abstraction of the proton from 2Ј-oxygen. We have argued (12) that this logic is flawed, ause the metal hydroxide complexes formed with metal s with lower pKa values are weaker bases and, therefore, uld be less able to abstract the 2Ј-OH proton, despite their ater concentration. This conclusion is supported by the data sented in Fig. 3 because the pKa of the 2Ј-OH is two or more a units higher than those of any of the aqueous metal ions died, making the metal hydroxide poorly suited to the task deprotonating the 2Ј-OH. It has been convincingly shown t proton transfer does not occur in the rate-determining p of the ribozyme cleavage reaction (30). The observed pH pendence and the correlation between the pKa values of the ueous metal ions and kobs must, therefore, reflect the effects Mg2+/Mg2+ Mg2+/La3+ La3+/La3+ experimental: Pontius et al., 1997; Lott et al., 1998 theoretical: Boero et al., JCTC, 2005 X-ray (active) O NiO O O P OR OS O H 5' 3' 5' 4' O Ni+1 OO H 3'2' 4' RAH(+) B(-)O Mg2+ O Mg2+ 41-nt 18 theoretical: Leclerc & Karplus, JPC, 2006
- 19. Contribution of Metals to Catalysis -30 -25 -20 -15 -10 -5 0 5 10 15 20 25 30 I II III IV V VI VII VIII IX no metal 1 metal 2 metals RelativeFreeEnergy(kcal/mol) Reaction Coordinate B3LYP/6-31+G(d,p) B3LYP/6-311+G(2d,2p)//B3LYP/6-31G(d,p) B3LYP/6-31+G(d,p)//HF/3-21+G(d) 22.9 kcal/mol 20.8 kcal/mol 19.3 kcal/mol Lopez et al., 2006 Torres et al., 2003 Leclerc & Karplus, 2006 ΔG exp = 20.1 kcal/mol dianionic mechanism 19
- 20. Reaction Path Following: B3LYP/6-31+G(d,p)//HF/3-21+G(d) HHR: reaction mechanism 20
- 21. longest time courses (48–96 h). Each phase of the time course was 10-fold faster at pH 7.5 than at pH 6.5, as expected if each process were limited by the chemical step (15). Finally, purification of this phosphorothioate-substituted HH16 by anion exchange HPLC (8) re- sulted in partial separation of ribozyme forms such that the two phases had identical rate constants to those observed in the racemic mixture but different relative amplitudes (one fraction gave 0.8 of the fast component and 0.2 of the slow, whereas a second fraction gave 0.2 of the fast and 0.8 of the slow). Rates and relative amplitudes of the two phases for reactions in 10 mM Mg2⌅ did not change upon addition of 0.2 mM EDTA or 2 mM dithiothreitol to the reaction mixture, suggesting that neither kinetic process depended on the presence of contaminating metal ions. In reactions with added Cd2⌅ , the concentration of EDTA carried over from the ribozyme and substrate stocks was ⌃15 nM. RESULTS We have used two different hammerhead ribozyme con- structs, HH⇥1 and HH16 (Scheme 1), in testing the role and A Specific Metal Ion in the Hammerhead Ribozyme 26823 Folding Metal Ions Models experimental: Peracchi et al., 1997 X-ray (active) 20Å 41-nt 21
- 22. Metal Binding Sites in the Hammerhead Ribozyme ? unable to rescue activity for the A13 or A14 phosphoro- thioate substitutions (Ruffner & Uhlenbeck, 1990; Knoll et al+, 1997; Peracchi et al+, 1997; Scott, 1997)+ The A9 phosphate is part of a metal-binding site observed in the original X-ray structure of the hammerhead (Pley et al+, 1994), where a Mn2ϩ ion is ligated by the pro-RP high negative potentia also modeled metal b stead of the metal inte posed here (Fig+ 4), the with the N1 of G8+ We Brownian-dynamics sim FIG dem the high ing s ture The met pha to m pha gan resid colo illus and Hansen et al., RNA, 2008Chartrand et al., RNA, 1997 22
- 23. Minimum/Full-Length HHR Khvorova et al., Nat. Struct. Biol., 2003 de la Peña et al., EMBO J., 2003 Canny et al., JACS, 2004 Wang et al., Biochem., 1999 41-nt 50-nt 56-nt 50-nt 23
- 24. Variations in HHR motifs Perreault et al., PLoS Comput. Biol., 2011 24
- 25. p F l r e s o t d d p e t l h t FIGURE 7. A folding scheme for the hammerhead ribozyme. Schematic to show the two-stage folding scheme previously proposed for the hammerhead ribozyme. State U exists in the absence of added metal ions, in which the three helical arms extend from an open central core. Penedo et al. Cold Son November 10, 2016 - Published byrnajournal.cshlp.orgDownloaded from Penedo et al., RNA, 2004. 25 Loop-Loop interactions in HHR folding
- 26. Loop-Loop interactions in HHR catalysis 26
- 27. Minimum HHR, Tertiary Contact and Catalysis 45-nt O’Rourke et al., JMB, 2015 27
- 28. HHR morphing O’Rourke et al., JMB, 2015 28
- 29. O C17O O O P OR OS OR H 5' 3' 2' 5' 4' N1 N N N7 O6 H2N H G12 O G8 O O O H 5' O- H 2' Mg2+ A Nucleobase Catalyst experimental: Chi et al., PLoS Biol., 2008 theoretical: Lee et al., JACS, 2008 X-ray (active) O C17O O O P OR OS OR H 5' 3' 2' 5' 4' N1 N N N7 O6 H2N H G12 O G8 O O O H 5' OH H O- H 2' 29
- 30. Metal Catalysts in the 2’OH activation ? Chval et al., JPC, 2011 < O O O P OR OS OH3C H3' 2' 5' 4' N N N NO H2N H H O H O O O P OR OS OH3C H3' 2' 5' 4' N N N N O H2N H H O H H O O O P OR OS OH3C H3' 2' 5' 4' N N N N O H2N H H OHMg2+(VI) < < 30
- 31. Metal Catalysts in the Hammerhead Ribozymes ? O NiO O O P OR OS O H 5' 3' 2' 5' 4' O Ni+1 OO H 3'2' 4' RAH(+) B(-) Mg2+ N1 N N N7 O6 H2N H G12 O- H Osborne et al., Biochem., 2009 Osborne et al. e n e r n d + U al z 2 Scheme 1 31
- 32. Cooperative Models in Self-Cleaving ? O Ni O O O P O HRNA5' 3' 2' 4' O Ni+1O O OH RNA3' O B(-) AH(+) M/H-R R-H/M 5' 32
- 33. Cooperative Models in Self-Cleaving ? Leclerc, Molecules, 2010 O G O O O H 2' G-8 O C17 O O O P O HRNA5' 3' 2' 4' O N1.1 O O OH RNA3' O 5' N N N N -O NH2 R G-12 Mg2+ RNA3' RNA5' Mg2+ O G O O O H 2' G-8 O C17 O O O P O HRNA5' 3' 2' 4' O N1.1 O O OH RNA3' O 5' N N N NO- H2N R G-12 Mg2+ Mg2+ RNA3' RNA5' 33
- 34. Metal Ions back in the Hammerhead Catalysis Ward & DeRose, RNA, 2011 Cold Spring Harbor Laboratory Press11 - Published by and DeRose 2000; Boots et al. 2008). Moderate rates of catalysis can also be achieved in molar concentrations of monovalent cations, an important property that helped to uncover the critical roles of nucleobases in the HHRz re- action mechanism (Murray et al. 1998; O’Rear et al. 2001; Bevilacqua et al. 2004). At physiological ionic strengths, the HHRz requires divalent ions for appreciable rates of catal- ysis; therefore, it is reasonable to assume that the divalent metal-dependent channel is the primary mode of catalysis in nature (Khvorova et al. 2003). The HHRz was studied for years in its simplest active form, as three short helices meeting at a junction of con- served nucleotides that form the active site of the ribozyme (for review, see Blount and Uhlenbeck 2005). Studies using this ‘‘truncated’’ form of the HHRz (trHHRz) led to a model of catalysis in which a catalytic metal in the P9/ G10.1 site coordinates the pro-R oxygen of the scissile phosphate, presumably to stabilize the negative charge of the phosphorane transition state (Peracchi et al. 1997; Wang et al. 1999). Based on detailed metal-rescue exper- iments, Wang et al. (1999) predicted that the metal ion coordinates to the P9/G10.1 site in the ground state and bridges to the scissile phosphate in the transition state of the trHHRz reaction. A ground state that is very different from the transition state is consistent with structural studies of the truncated HHRz, which in general did not show catalytically relevant atoms within appropriate dis- tances of the active site (Blount and Uhlenbeck 2005). In these structures, the P9/G10.1 metal ion site is z20 A˚ away from its predicted ligand during catalysis, the pro-R oxygen of the scissile phosphate (Pley et al. 1994; Scott et al. 1995). FIGURE 1. (A) Secondary structure of the modified Schistosoma mansoni HHRz (MSL1L2) (Osborne et al. 2005) used in these studies. (B) Crystallographic active site of the S. mansoni HHRz (2OEU) 34
- 35. HHR: Active Conformation and Metal Ions 66-nt Insert Table of Contents artwork here Page 5 of 9 Biochemistry 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 Mir et al., Biochem., 201535
- 36. HHR: Metal Ions and Catalysis Mir & Golden, Biochem., 201536Leclerc, Molecules, 2010
- 37. ASBVd(-)/HHR(-): SANS StudyASBVd(-) HHR(-) Leclerc et al., Sci. Rep., 2016.37
- 38. HHR(-) dimerization/ catalysis 38 Leclerc et al., Sci. Rep., 2016.
- 39. HHR(-): 2D structures monomer 1 monomeHI HII HIII A E E Egguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1 10 20 30 40 50 60 70 79 g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c 1a 10a 20a 30a 40a 50a 60a 70a 79a 1b monomer 1 monomer 2HI HII HIII A B Emonomer (10ºC) = -26.9 kcal/mol Emonomer (25ºC) = -19.4 kcal/mol Emonomer (45ºC) = -9.6 kcal/molgguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1 10 20 30 40 50 60 70 79 g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b1b monomer 1 monomer 2HI HII HIII A Emonomer (10ºC) = -26.9 kcal/mol Emonomer (25ºC) = -19.4 kcal/mol Emonomer (45ºC) = -9.6 kcal/molgguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1 10 20 30 40 50 60 70 79 intra-molecular base-pairs tertiary or inter-molecular contacts nucleotide in tertiary contact cleavage site g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b 50b 60b 70b 79b1b 39
- 40. 3D Modeling of HHR(-) 40
- 41. HHR folding p F l r e s o t d d p e t l h t s FIGURE 7. A folding scheme for the hammerhead ribozyme. Schematic to show the two-stage folding scheme previously proposed for the hammerhead ribozyme. State U exists in the absence of added metal ions, in which the three helical arms extend from an open central core. On addition of metal ions, the minimal ribozyme undergoes folding in two steps, correspond- Penedo et al. Rg(I)/(F) ~ 1.1 ASBVd: Rg(I)/(F) ~ 1.241 Penedo et al., RNA, 2004.
- 42. HHR morphing Rg(I)/(F) ~ 1.1 ASBVd: Rg(I)/(F) ~ 1.2 O’Rourke et al., JMB, 2015 42
- 43. Modeling/SANS dexp = 96.0Å dcalc = 96.7Å Rg exp = 31Å Rg *calc = 31 (26)Å 43
- 44. HHR (-): dimerization monomer 1 monomer 2HI HII HIII A B Eint (10ºC) = -9.3 kcal/mol Eint (25ºC) = -8.5 kcal/mol Eint (45ºC) = -5.8 kcal/mol Edimer (10ºC) = -47.1 kcal/mol Edimer (25ºC) = -33.6 kcal/mol Edimer (45ºC) = -15.7 kcal/mol Emonomer (10ºC) = -26.9 kcal/mol Emonomer (25ºC) = -19.4 kcal/mol Emonomer (45ºC) = -9.6 kcal/molgguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1 10 20 30 40 50 60 70 79 gguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c gaaa c u ca g a g u c ggaaag uc ggaa ca gacc u g g u u ucgu c 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b 50b 60b 70b 79b 1b g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b 50b 60b 70b 79b1b monomer 1 monomer 2HI HII HIII intra-molecular base-pairs tertiary or inter-molecular contacts nucleotide in tertiary contact cleavage site g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b 50b 60b 70b 79b1b 44
- 45. HHR (-): dimerizationmonomer 1 monomer 2HI HII HIII B Eint (10ºC) = -9.3 kcal/mol Eint (25ºC) = -8.5 kcal/mol Eint (45ºC) = -5.8 kcal/mol Edimer (10ºC) = -47.1 kcal/mol Edimer (25ºC) = -33.6 kcal/mol Edimer (45ºC) = -15.7 kcal/mol Edimer (10ºC) = -53.7 kcal/mol Edimer (25ºC) = -38.9 kcal/mol Edimer (45ºC) = -19.2 kcal/mol gguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c gaaa c u ca g a g u c ggaaag uc ggaa ca gacc u g g u u ucgu c 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b 50b 60b 70b 79b 1b g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b 50b 60b 70b 79b1b monomer 1 monomer 2HI HII HIII gguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c aa guc g a a a c u c a g a g u c ggaaag uc ggaa ca gacc u g g u u ucgu c 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b50b 60b 70b 79b 1b C g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b 50b 60b 70b 79b1b 45
- 46. HHR(-) dimer 46
- 51. Mir et al., Biochem., 2015 HHR: Monomer & Dimer Figure S2. Crystal contacts involve intermolecular base pairs. A. In crystals of the RzB PDB ID: 51
- 52. Rg & self-association HIII HII HI/HII core R(exp)g ~ 50Å Rg*=50.4Å Rg*=44.6Å Rg*=46.1Å52 HI HI/HI
- 53. MD setup for HHR dimer 53 •~ 300 000 atoms • K+ & Cl- (0.15M) • T = (283K, 313K) • NAMD
- 54. Dynamics of HHR(-) dimer RMSD vs time (ns) Rg vs time (ns) Histogram of Rg Histogram of RMSD Rg (monomer 1) vs time (ns) Rg (monomer 2) vs time (ns) Histogram of Rg (monomer 1) Histogram of Rg (monomer 2) RMSD(Å) 0 5 10 15 Rg(Å) 25 30 35 40 45 time (ns) 0 10 20 30 40 50 0 2500 5000 7500 10000125001500017500 54
- 55. HHR dimer: MD trajectory 55
- 56. G G A A G A G A U U G A A G A C G A G U G A A C UAA U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U UUUGU A AAA A AACAAUGAAG AUA GAGGA A UAAAC C UUG CGA GAC UC AUCAGUGUU C UUCC CAU CUUUCC C U GAA G A GAC GAA GUG A UC 1 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 180 190 200 210220230240 249 G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A 1a 10a 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230a monomer 1 A Emon Emon Emon G G A A G A G A U U G A A G A C G A G U G A A C UAA U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U UUUGU A AAA A AACAAUGAAG AUA GAGGA A UAAAC C UUG CGA GAC UC AUCAGUGUU C UUCC CAU CUUUCC C U GAA G A GAC GAA GUG A UC 1 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 180 190 200 210220230240 249 G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C 1a 10a 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230a 240a 249a monomer 1 monomer 1 monomer 2HI HII HIII A B Eint (10ºC) = -9.3 kcal/mol Eint (25ºC) = -8.5 kcal/mol Eint (45ºC) = -5.8 kcal/mol Edimer (10ºC) Edimer (25ºC) Edimer (45ºC) Emonomer (10ºC) = -26.9 kcal/mol Emonomer (25ºC) = -19.4 kcal/mol Emonomer (45ºC) = -9.6 kcal/molgguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c 1 10 20 30 40 50 60 70 79 gguu c uucc cau cuuucc c u gaa g a gac ga a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a aa c u ca g a g u c ggaaag uc ggaa ca gacc u g g u u ucgu c 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 50b 60b 70b 79b 1b g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b 50b 60b1b intra-molec tertiary or i nucleotide cleavage si g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a c c u g g u u u c g u c g g u u c u u c c c a u c u u u c c c u g a a g a g a c g a a g c a a g u c g a a a c u c a g a g u c g g a a a g u c g g a a c a g a 1a 10a 20a 30a 40a 50a 60a 70a 79a 10b 20b 30b 40b 50b 60b1b HHR (-) ASBVd(-): self-association 56
- 57. Edimer (10ºC) = -163 kcal/mol Edimer (25ºC) = -120 kcal/mol Edimer (45ºC) = -63.6 kcal/mol Eint (10ºC) = -9.35 kcal/mol Eint (25ºC) = -7.27 kcal/mol Eint (45ºC) = -5.61 kcal/mol G G A A G A G A U UG A AG A C G A G U G A A C U A A U U U U U UU A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U CAC AA GUC G A AA C U CA G A G U C GGAAAG UC GGAA CA GACCUGGU UU C GUC AA A CAAA GUUUA A U CA U A UCC U C AC U U C UUGUU C UAAU A A ACAAG A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A U U G A A G A C G A G U G A A C U AAU U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U UUUGU A A AA A AACAA UGAAG AUA GAGGA A UAAAC CUUG C GA GAC UC AUCAG UGUU C UUCC C AU CUUUCC C U GAA G A GAC G A A G U G AUC1a 10a 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 60a 170a 180a 190a 200a 210a 220a 230 240 249a 10b 20b 30b 40b 50b 60b 70b 80b 90b 100b 110b 120b 130b 140b 150b 160b 170b 180b 190b 200b 210b 220b 230b 240b 249b 1b C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230a 240a 249a 10b 20b 30b 40b 50b 60b 70b 80b 90b 100b 110b 120b 130b 140b 150b 160b 170b 180b 190b1b C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230a 240a 249a 10b 20b 30b 40b 50b 60b 70b 80b 90b 100b 110b 120b 130b 140b 150b 160b 170b 180b 190b1b omer 1 monomer 2 B monomer Emonomer (25ºC) = -68.7 kcal/mol Emonomer (45ºC) = -37.9 kcal/mol G G C A C A A G U C G G A A A G G G A A G A C C U G G U G U C C A A G U U U A U C C U C C U U C U U G U U A C A A A U UUUGU A AAA A AACAAUGAAG AUA GAGGA A UAAAC C UUG CGA GAC UC AUCAGUGUU C UUCC CAU CUUUCC C U GAA G A GAC GAA GUG A UC 1 170 180 190 200 210220230240 249 Edimer (10ºC) = -163 kcal/mol Edimer (25ºC) = -120 kcal/mol Edimer (45ºC) = -63.6 kcal/mol Eint (10ºC) = -9.35 kcal/mol Eint (25ºC) = -7.27 kcal/mol Eint (45ºC) = -5.61 kcal/mol monomer 1 monomer 2 G G A A G A G A U UG A AG A C G A G U G A A C U A A U U U U U UU A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U CAC AA GUC G A AA C U CA G A G U C GGAAAG UC GGAA CA GACCUGGU UU C GUC AA A CAAA GUUUA A U CA U A UCC U C AC U U C UUGUU C UAAU A A ACAAG A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A U U G A A G A C G A G U G A A C U AAU U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U UUUGU A A AA A AACAA UGAAG AUA GAGGA A UAAAC CUUG C GA GAC UC AUCAG UGUU C UUCC C AU CUUUCC C U GAA G A GAC G A A G U G AUC1a 10a 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230 240 249a 10b 20b 30b 40b 50b 60b 70b 80b 90b 100b 110b 120b 130b 140b 150b 160b 170b 180b 190b 200b 210b 220b 230b 240b 249b 1b G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C 1a 10a 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230a 240a 249a 10b 20b 30b 40b 50b 60b 70b 80b 90b 100b 110b 120b 130b 140b 150b 160b 170b 180b 190b 200b 210b 220b 230b 240b 249b1b G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C 1a 10a 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230a 240a 249a 10b 20b 30b 40b 50b 60b 70b 80b 90b 100b 110b 120b 130b 140b 150b 160b 170b 180b 190b 200b 210b 220b 230b 240b 249b1b monomer 1 monomer 2 ASBVd(-): self-association 57
- 58. G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C 1a 10a 20a 30a 40a 50a 60a 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230a 240a 249a 10b 20b 30b 40b 50b 60b 70b 80b 90b 100b 110b 120b 130b 140b 150b 160b 170b 180b 190b 200b 210b 220b 230b 240b 249b 10c 20c 30c 40c 50c 60c 70c 80c 90c 100c 110c 120c 130c 140c 150c 160c 170c 180c 190c 200c 210c 220c 230c 240c 249c1b 1c GG A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U UA A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U CG G A A C AG A C C U G G U U U CG U C A AA C A A AG U U U A A U CA U A U C C UCA C U U C U U G U U C UA A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A G A G G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A UU G AA G AC G A G U G A A C U A A U U UUU U U A A U A A A A G U U C A C C A C G A C U C C U C C U U C U C U C A C A A G U CGA A ACUC A G A G U C G G A A A G U C G G A AC A G A C C U G G UU U C G U CA A A C A A A G U U U AA U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G A U U U U G U A A A A A A A C A A U G A A G A U A GAG G A A U A A A C C U U G C G A G A C U C A U C A G U G U U C U U C C C A U C U U U C C C U G A A G A G A C G A A G U G A U C G G A A G A G A U U G A A G A C G A G U G A A C U A A U U U U U U U A A U A A A A G U U C A C C A C G A CU C CU C C UU C U C U C A C A A G U C G A A A C U C A G A G U C G G A A A G U C G G A A C A G A C C U G G U U U C G U C A A A C A A A G U U U A A U C A U A U C C U C A C U U C U U G U U C U A A U A A A C A A G AU U U U G U A A A A A A A C A AUG A A G A U A G A G G A A UAAACC UUG CG A GAC UC AUCAGU G U UC UUCC CA U C U U U C C C UGA A G A G A C G A A G U G A UC 1a 10a 20a 30a 40a 50a 60 70a 80a 90a 100a 110a 120a 130a 140a 150a 160a 170a 180a 190a 200a 210a 220a 230a 240a 249a 10b 20b 30b 40b 50b 60b 70b 80b 90b 100b 110b 120b 130b 140b 150b 160b 170b 180b 190b 200b 210b 220b 230b 240b 249b 10c 20c 30c 40c 50c 60c 70c 80c 90c 100c 110c 120c 130c 140c 150c 160c 170c 180c 190c 200c 210c 220c 230c 240c 249c intra-molecular base-pairs inter-molecular contacts HHR motif nucleotide in tertiary contact cleavage site ASBVd(-): self-association 58
- 59. What did we learn ? • for RNA and ribozymes: « too short » may be bad • don’t forget about dynamics • SANS & modeling approaches to infer self- association modes • theoretical approaches to explore reaction mechanisms and pathways
- 60. Viroids: Plant Parasites Genus Pospiviroids: PSTVd (potato spindle tuber) Genus Hostuviroids: HSVd (hop stunt) Genus Cocadviroids: CCCVd (coconut cadang-cadang) Genus Apscaviroids: ASSVd (apple scar skin) Genus Coleviroids: CbVd 1 (coleus blumei 1) Genus Avsunviroids: ASBVd (avocado sunblotch) Genus Pelamoviroids: PLMVd (peach latent mosaic) 60
- 61. Viroid & HDV: Hepatitis D 61
- 62. Acknowledgments •Zdenek Chval (University of South Bohemia, CK) •Daniela Chvalová (University of South Bohemia, CK) •Xavier Lopez (Euskal Herriko Unibertsitatea, SP) •Annick Dejaegere (ESBS Strasbourg) •Darrin M. York (Rutgers University, USA) •Martin Karplus (Harvard University, USA) •Giuseppe Zaccai (IBS, Grenoble) •Jacques Vergne (MNHN, Paris) •Anne Martel (ILL, Grenoble) •Martina Rihova (Institute of Physics, Prague, CK) •Marie-Christine Maurel (MNHN, Paris) •William G. Scott (UCSC, Santa-Cruz, USA) 62 ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! !
- 63. NITA SAHAI and HUSSEIN KADDOUR, Guest Editors 63 Fabrice Leclerc, Ph.D. I2BC /Dept. de Biologie des Génomes, « Séquence Structure Fonction des ARN » SSFA (D. Gautheret) fabrice.leclerc@u-psud.fr
