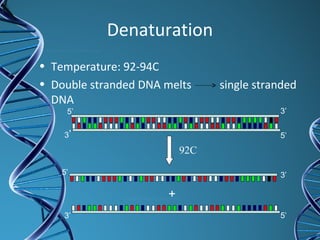
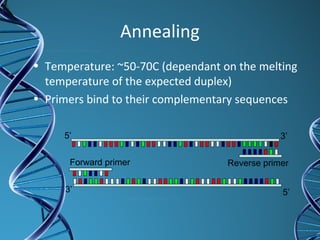
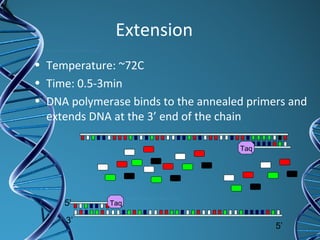
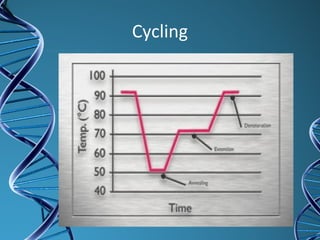
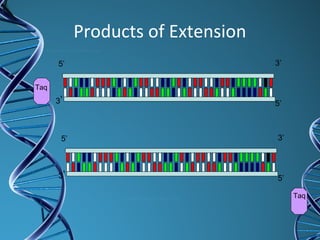
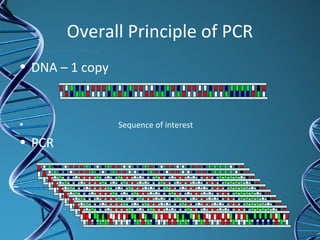
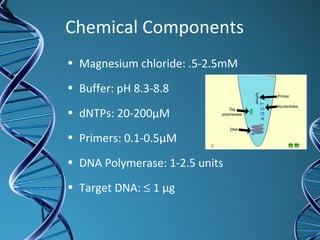
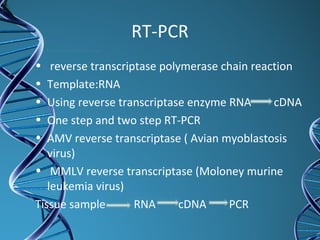
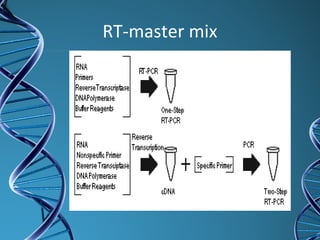
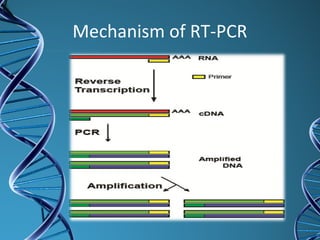
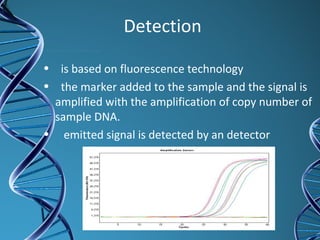
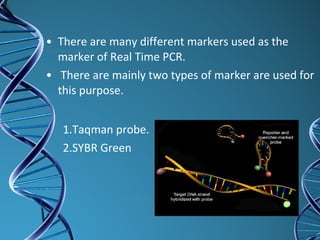
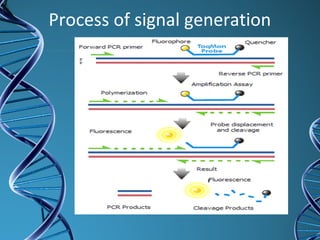
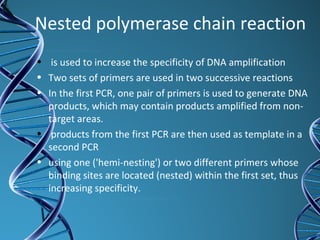
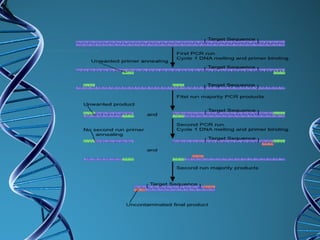
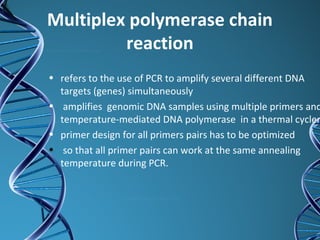
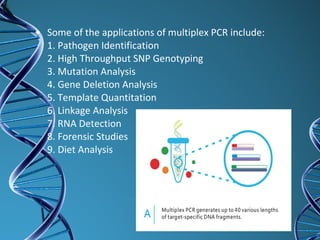
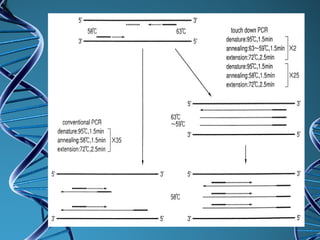
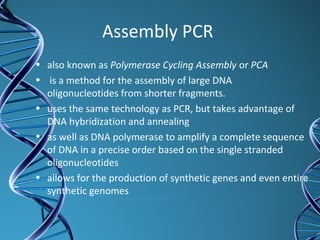
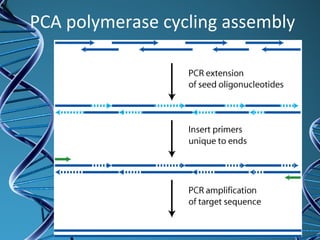
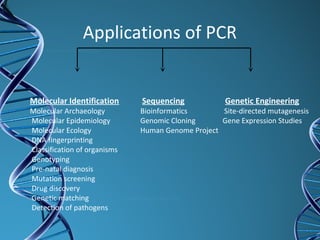
PCR is a technique that amplifies a specific DNA sequence. It works by repeated cycles of heating and cooling of the DNA sample to separate, copy, and recombine strands. Each cycle approximately doubles the number of target sequences. This allows a very small initial sample to generate millions of copies of the target sequence. PCR is used in various applications including DNA sequencing, genetic disease diagnosis, cloning, and forensic analysis. It has become essential to many areas of biological research and medical diagnostics.