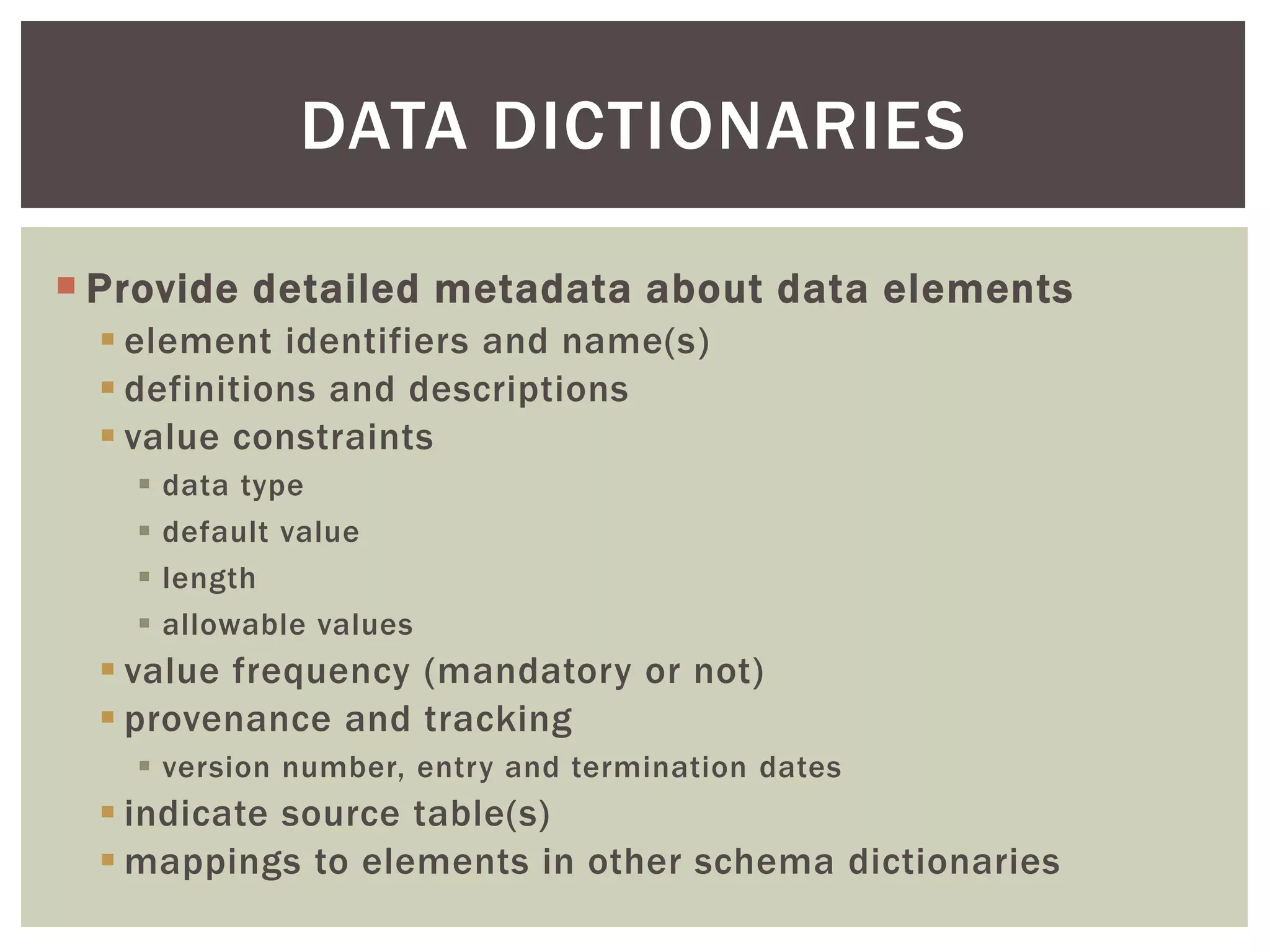
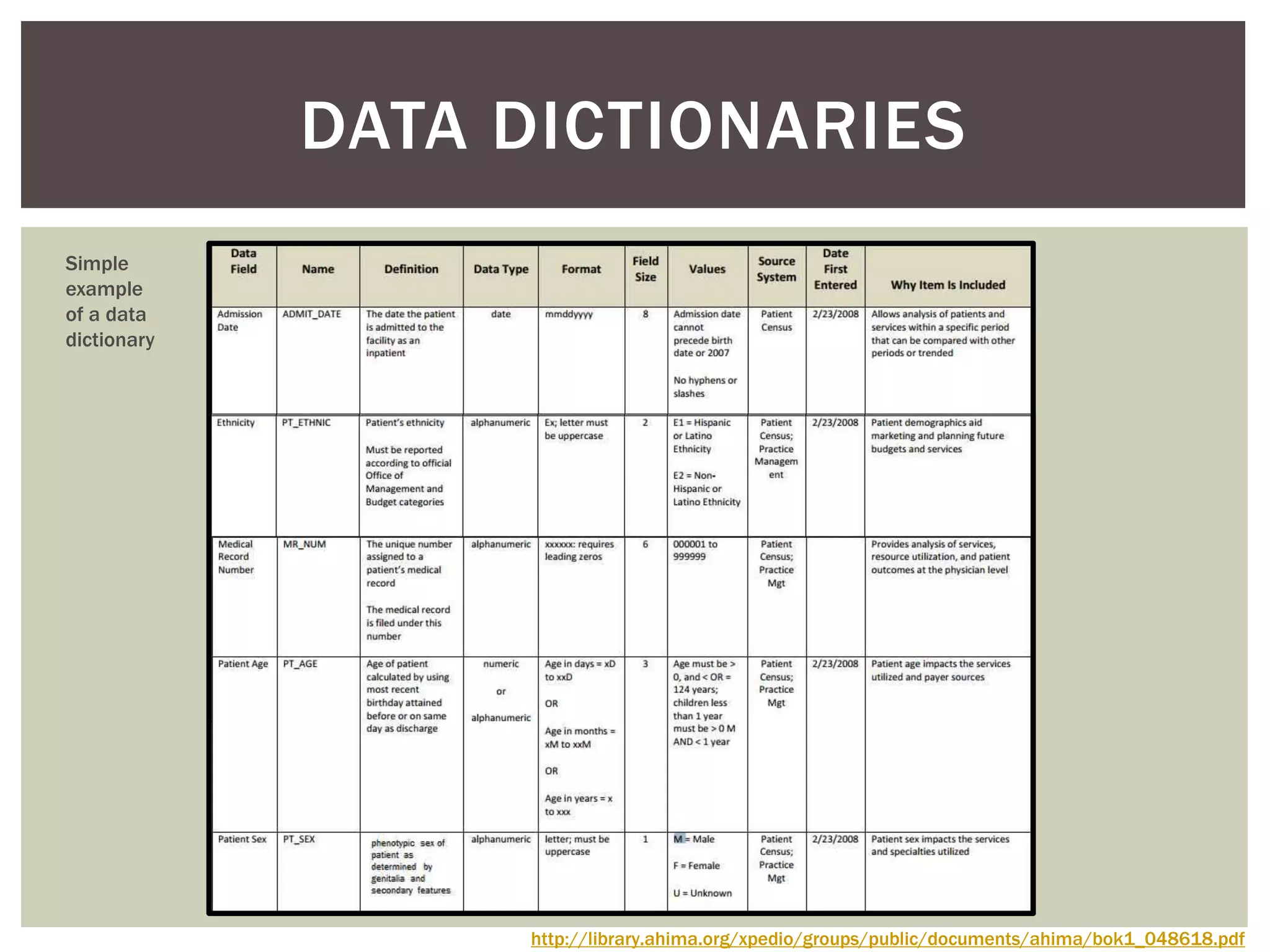
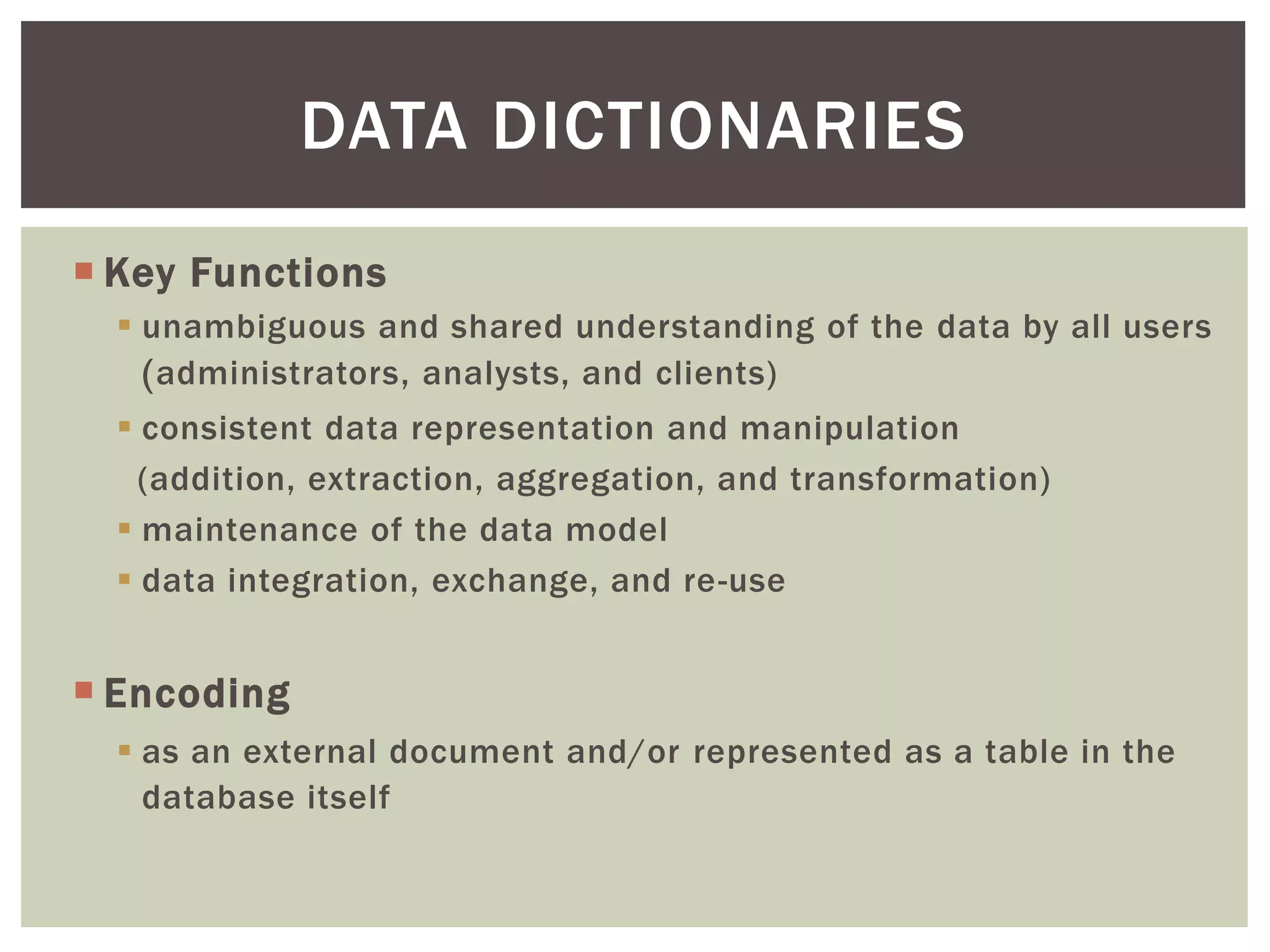
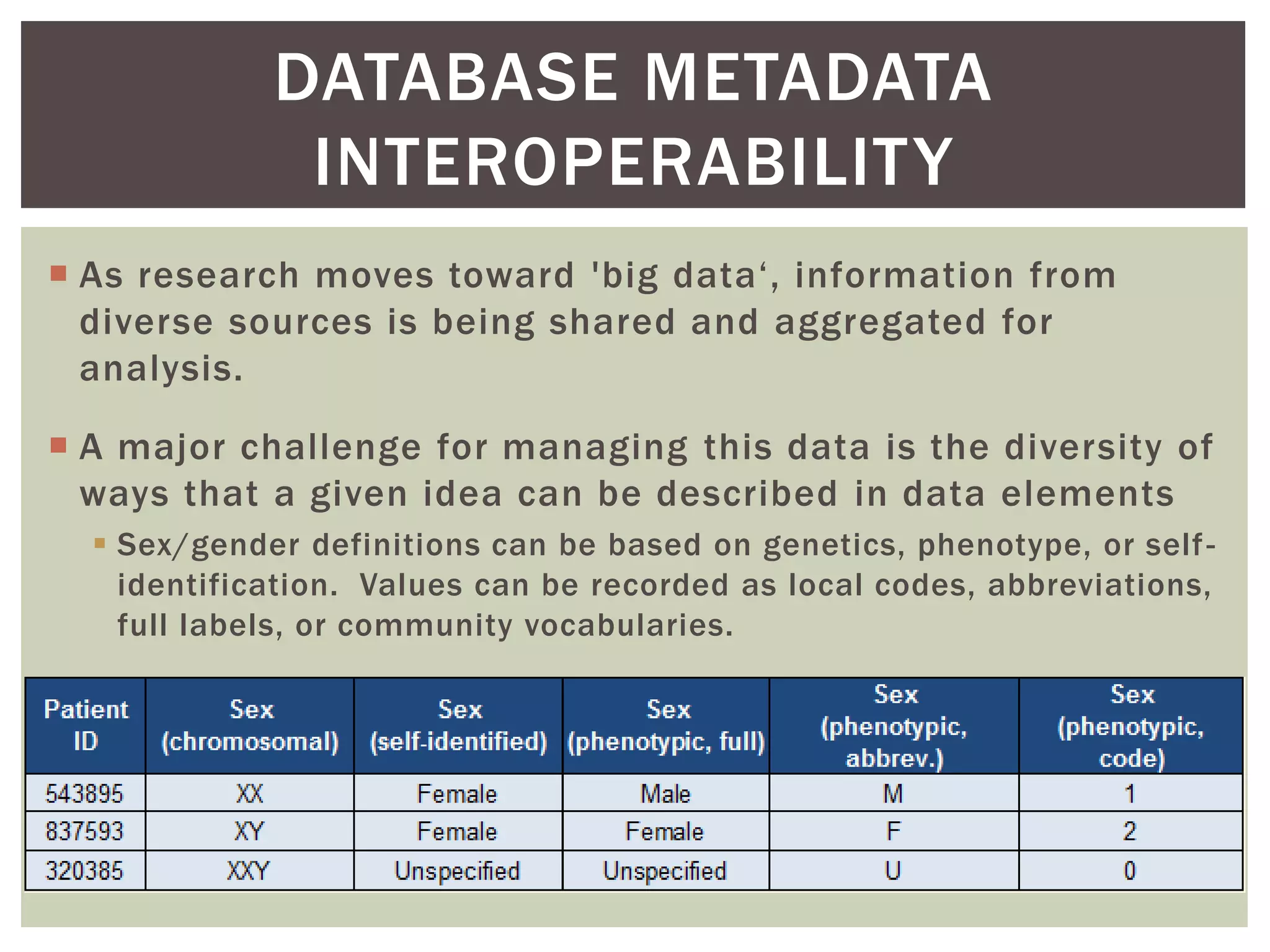
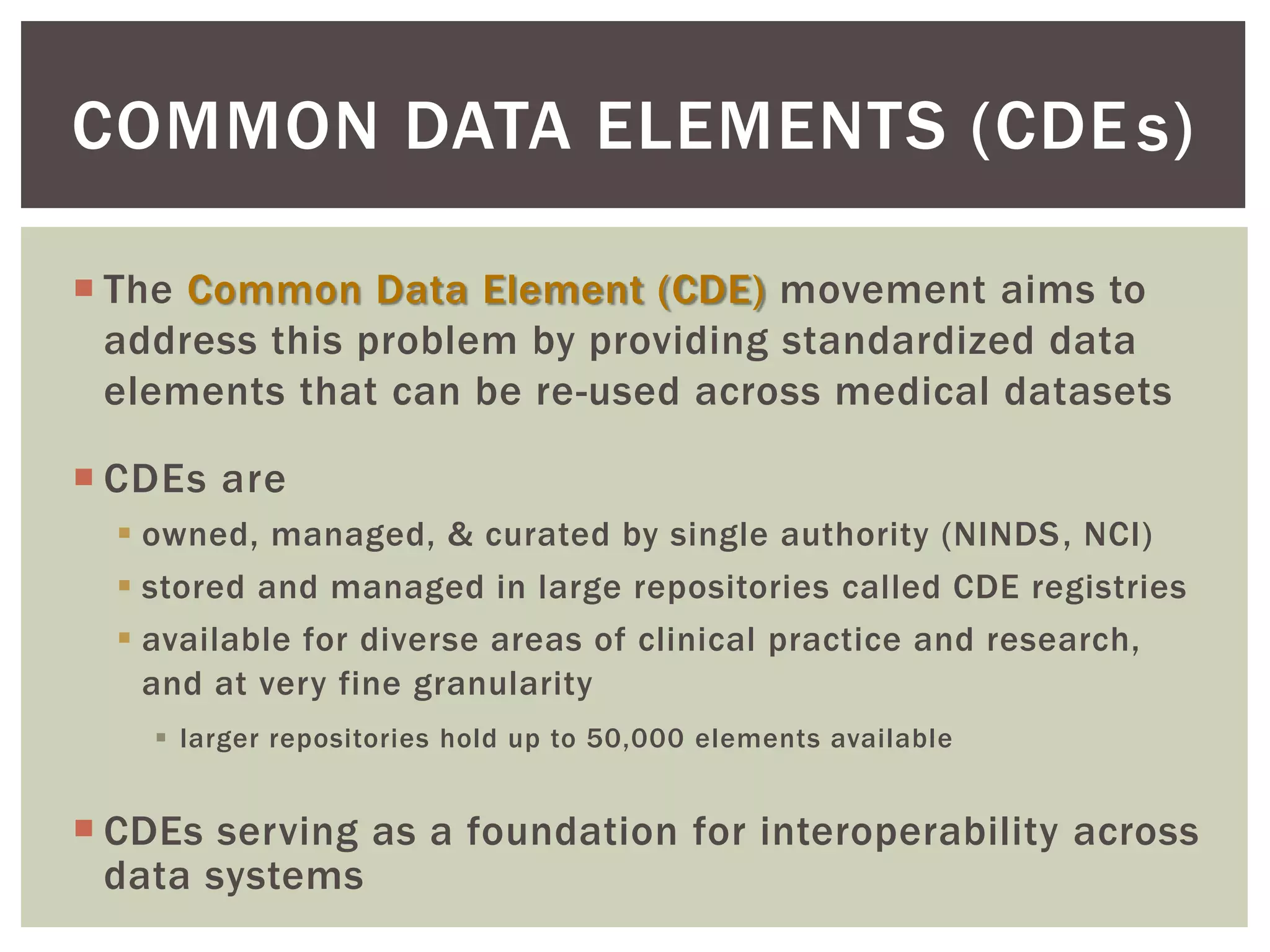
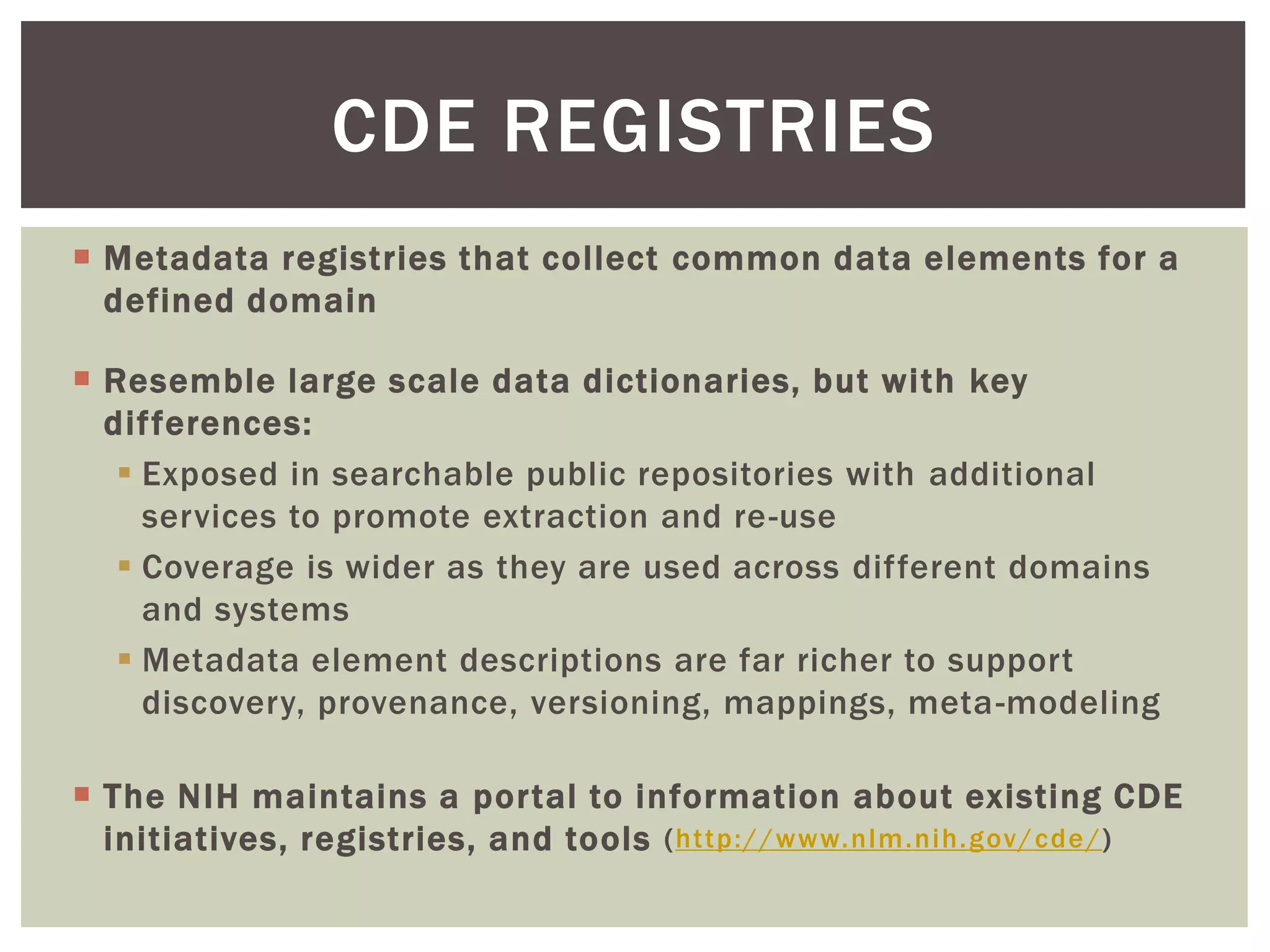
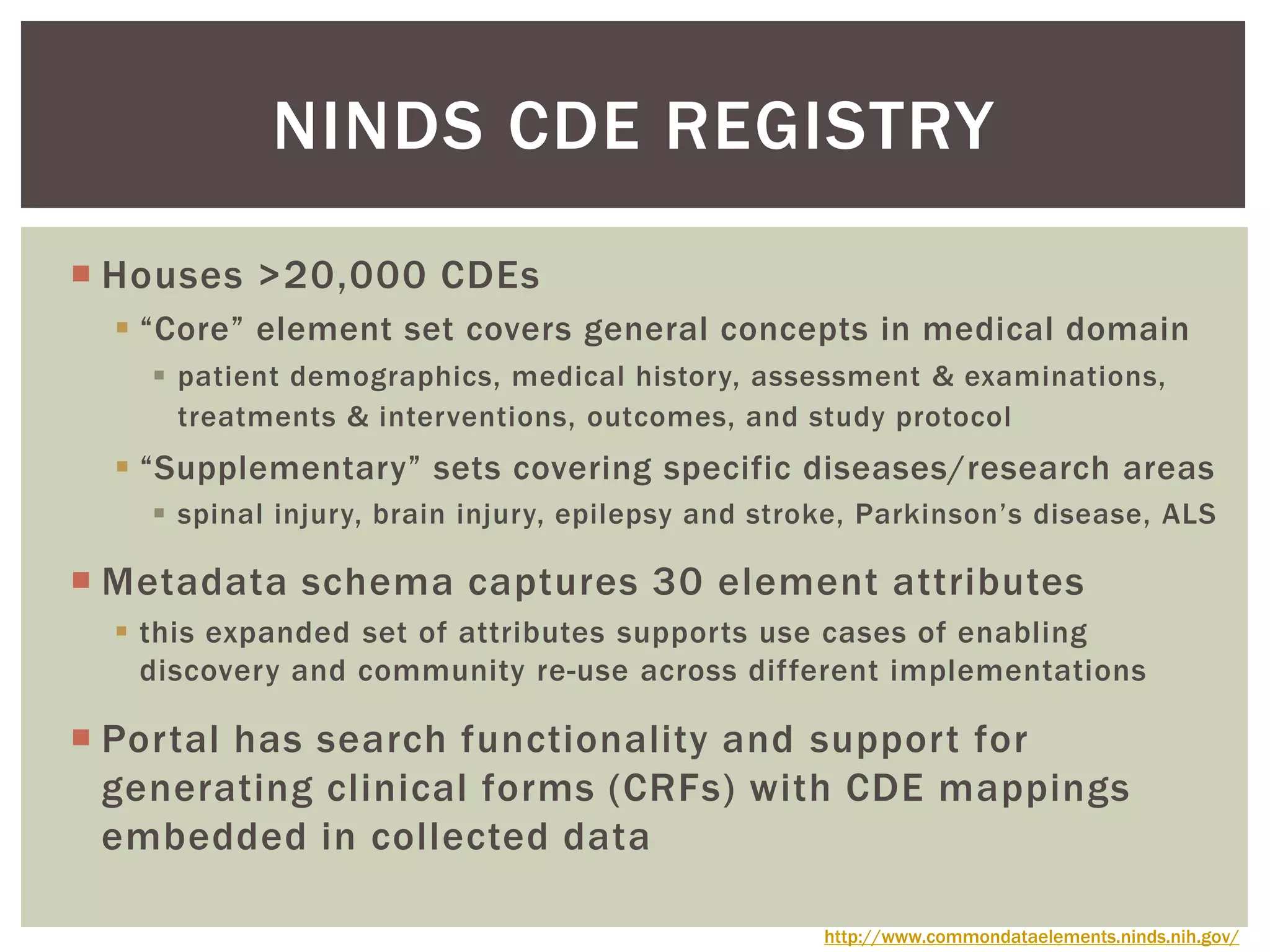
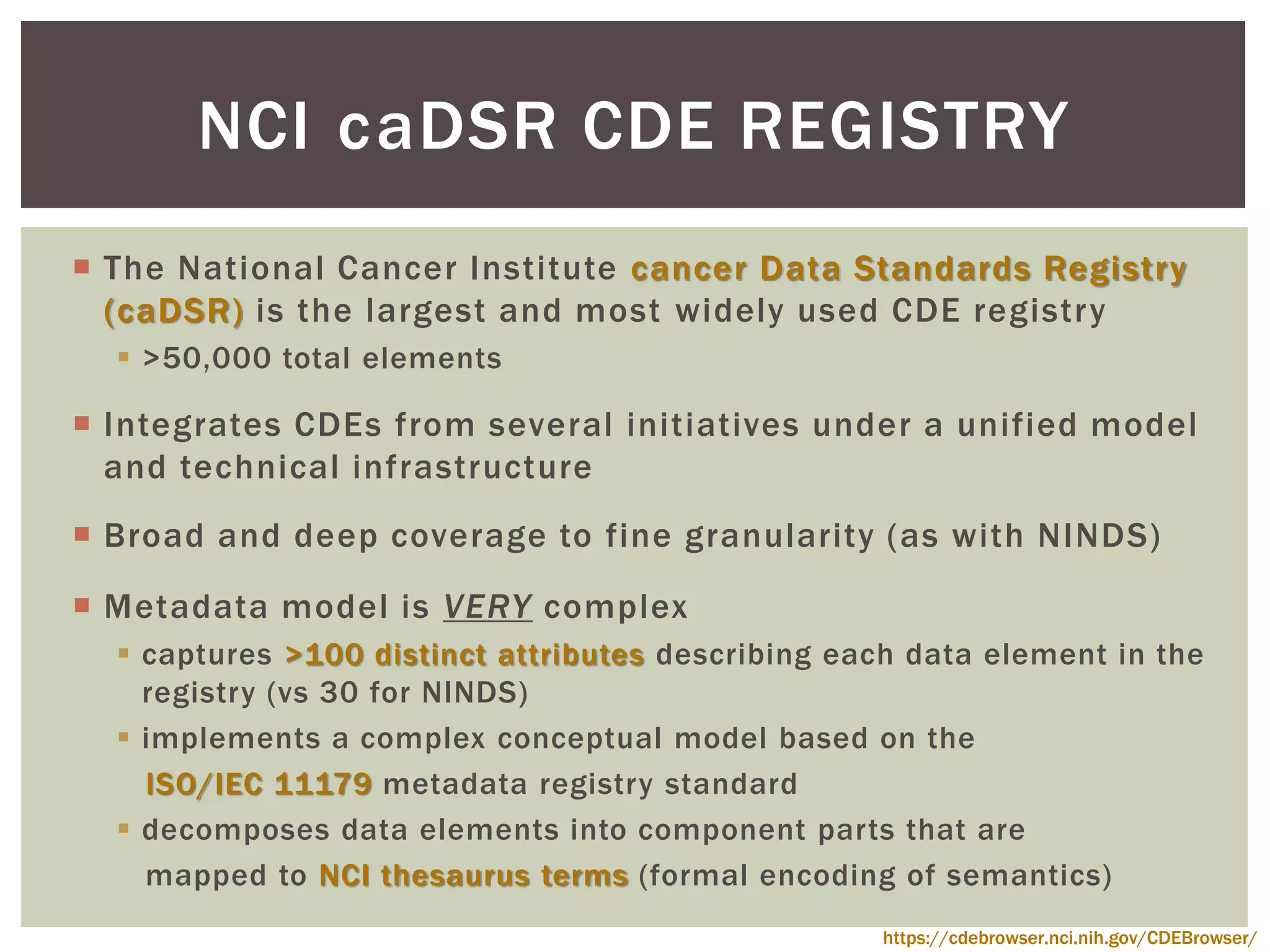
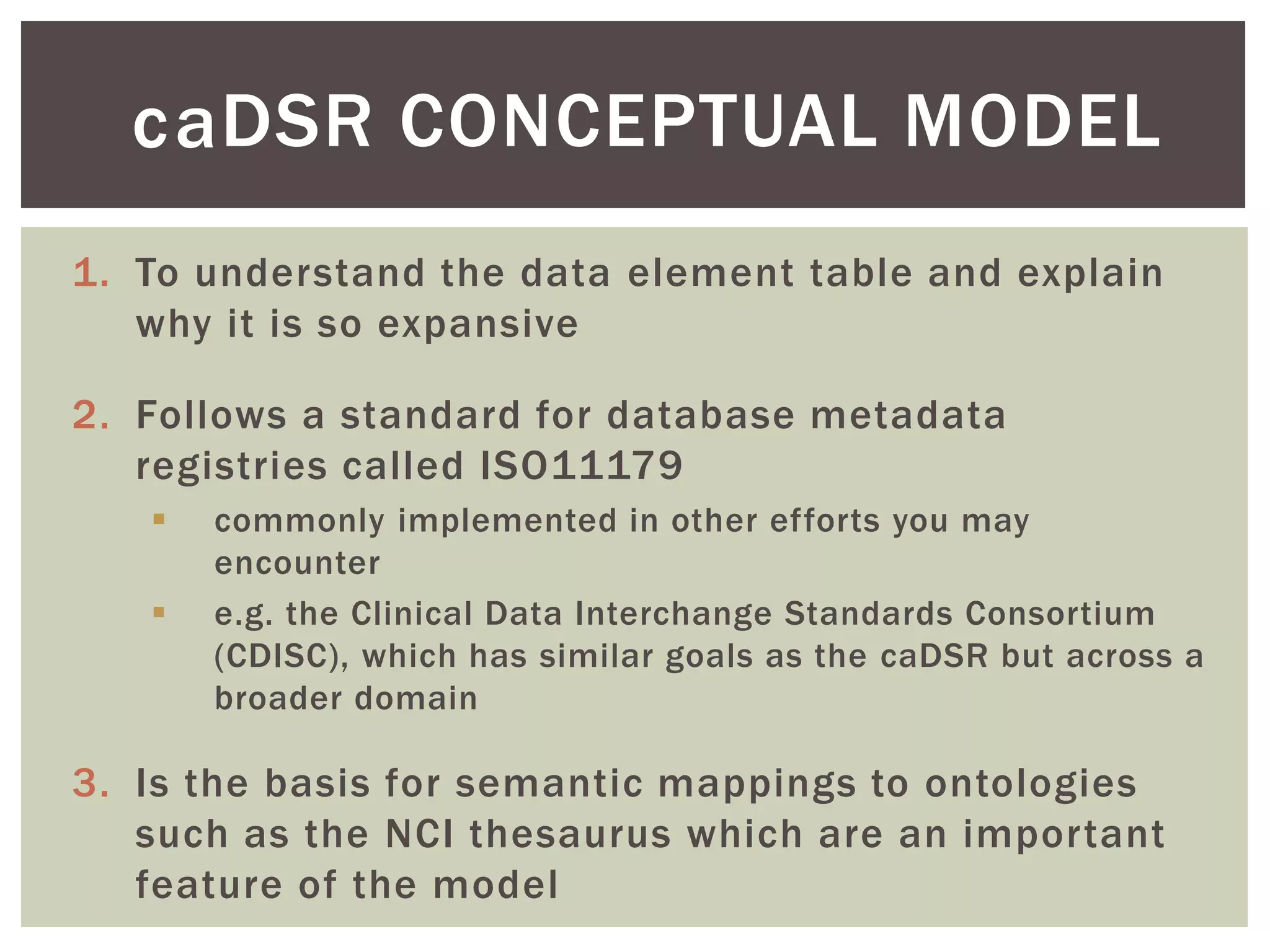
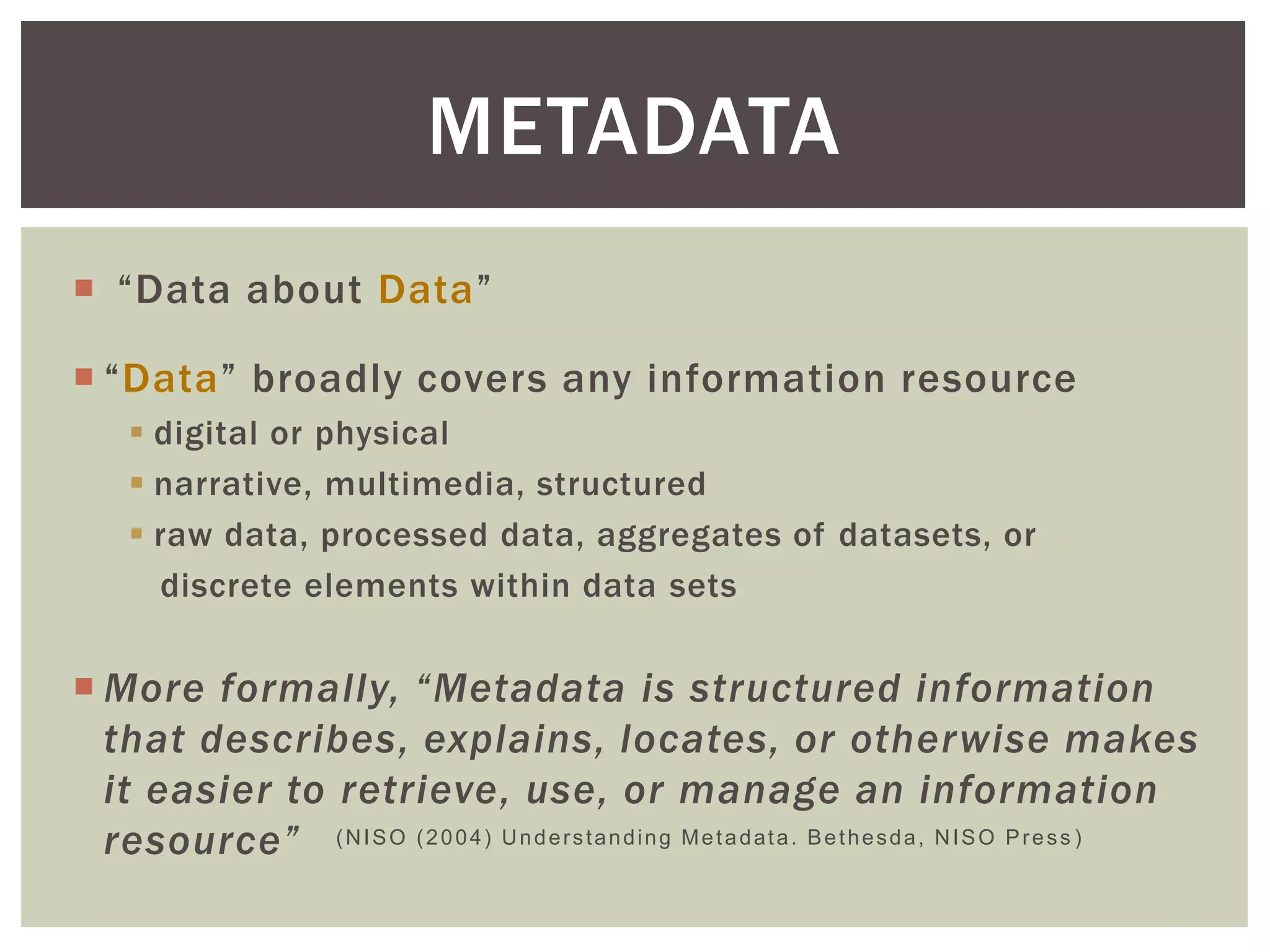
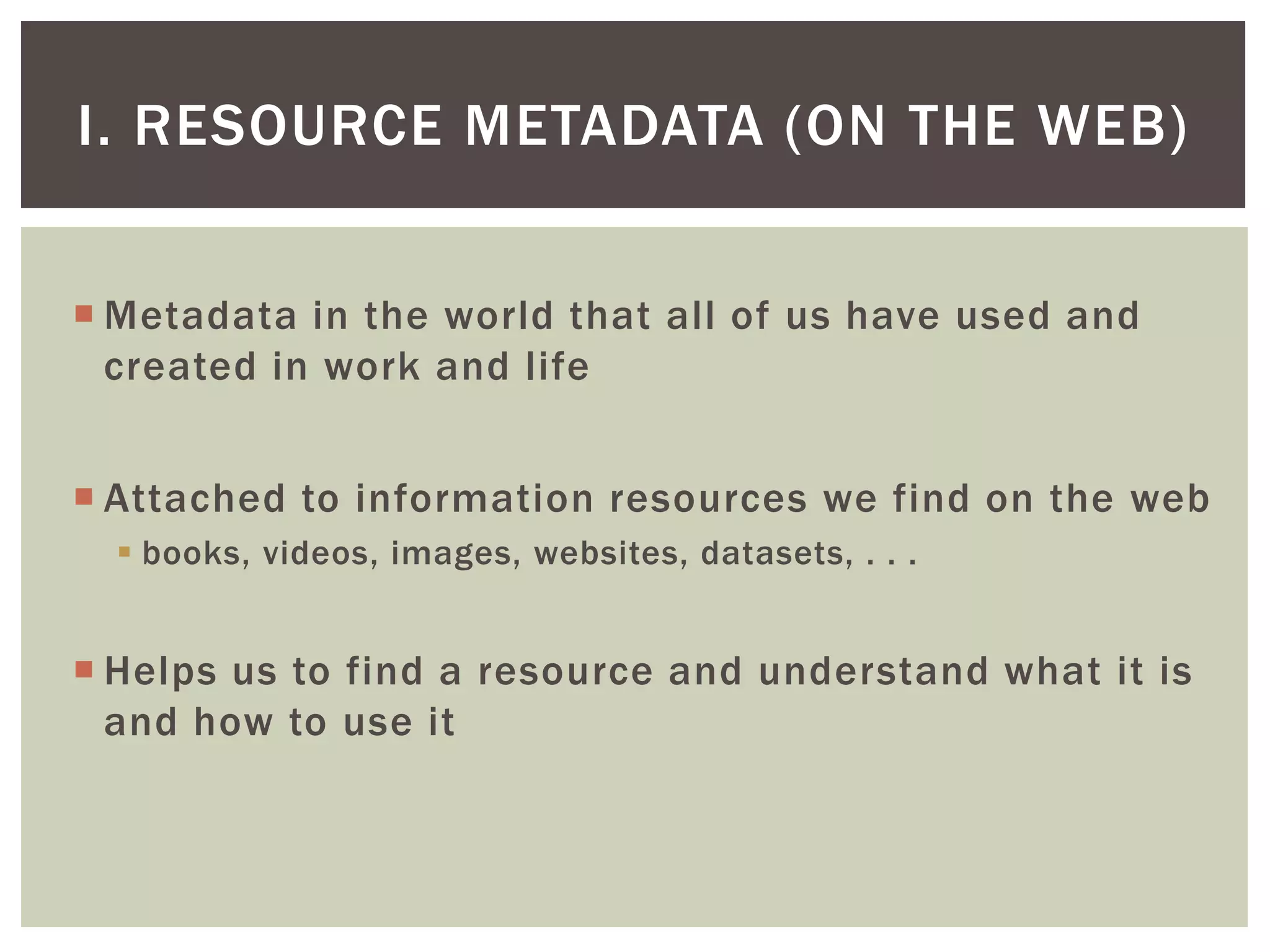
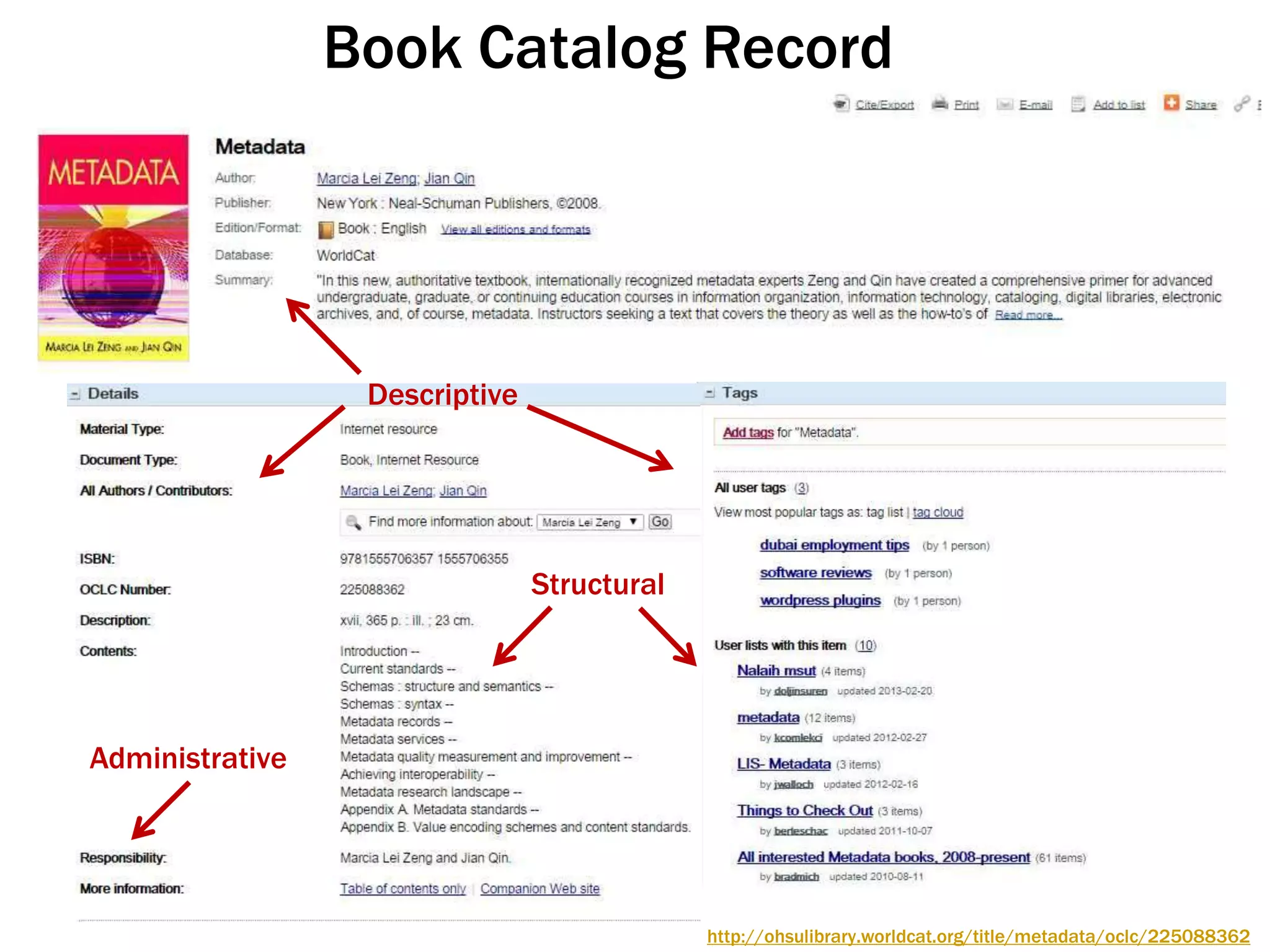
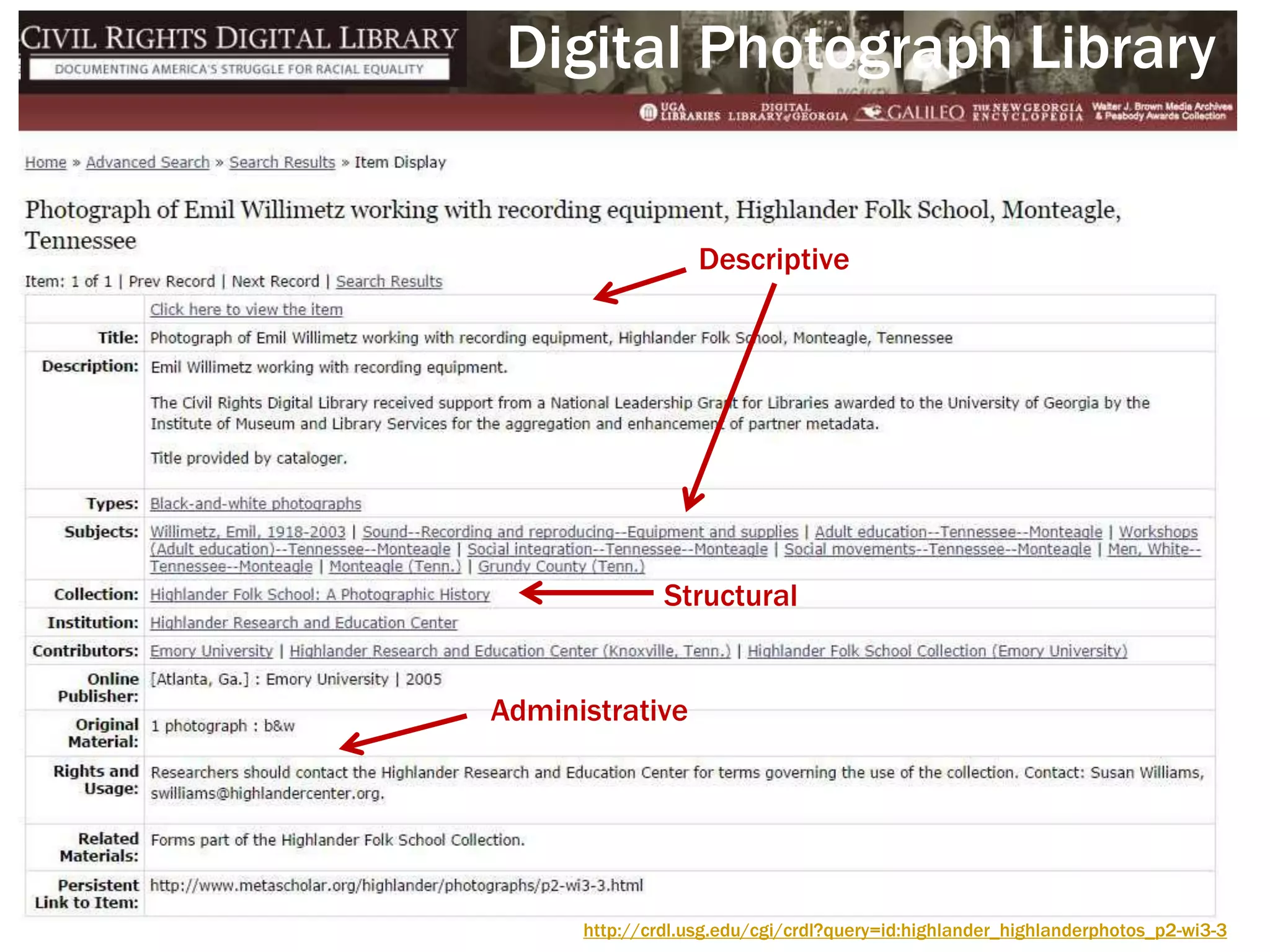
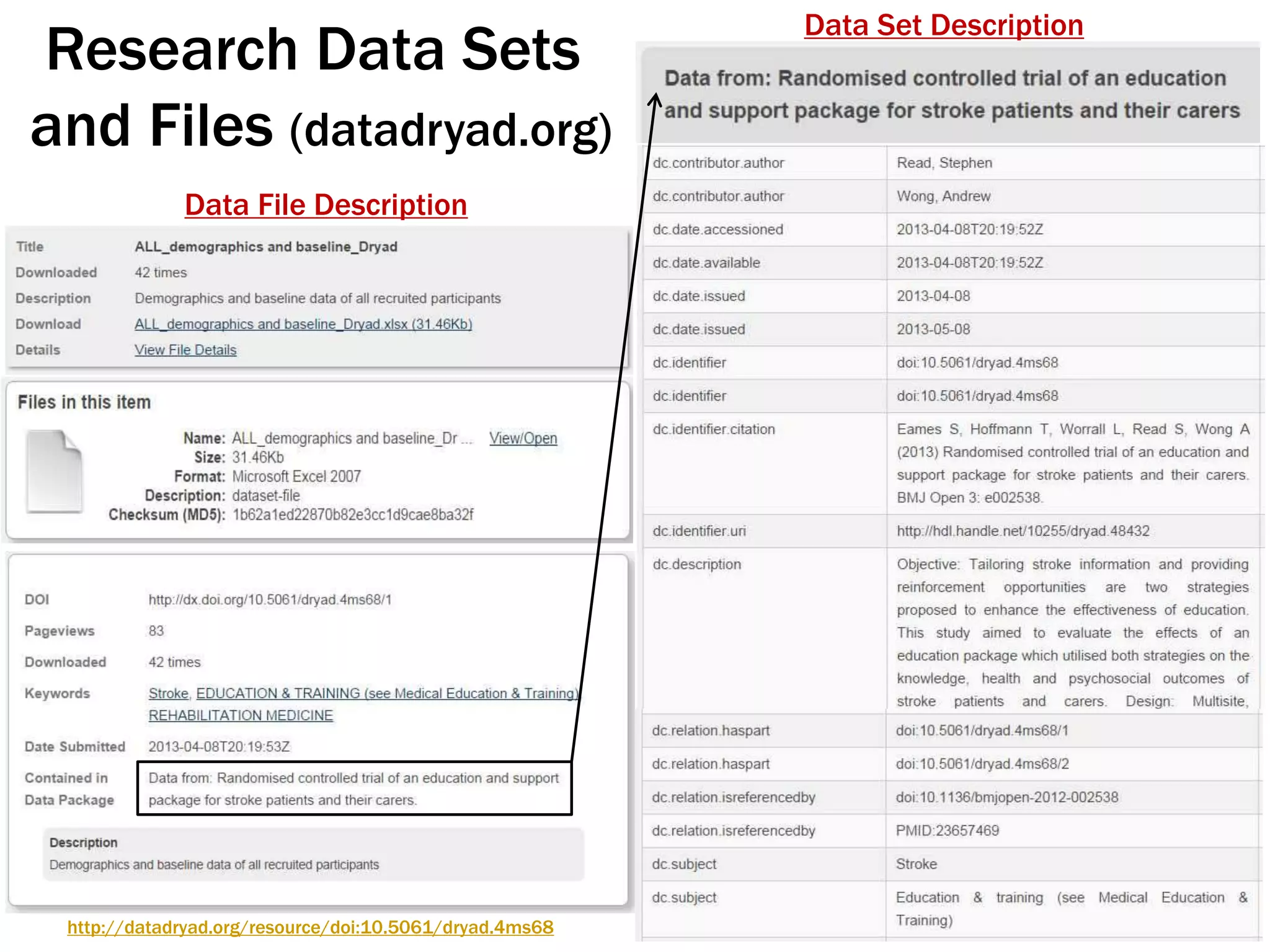
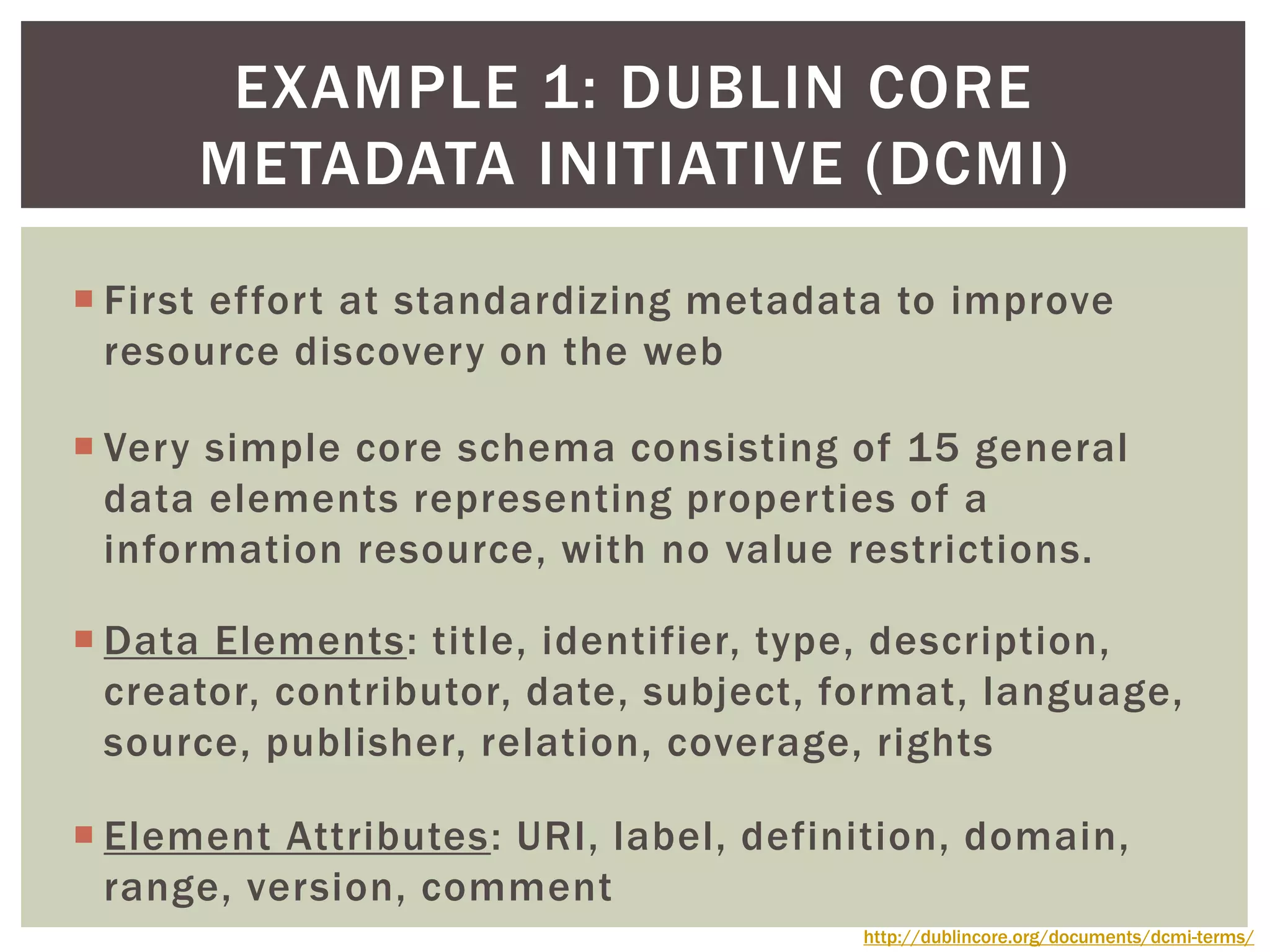
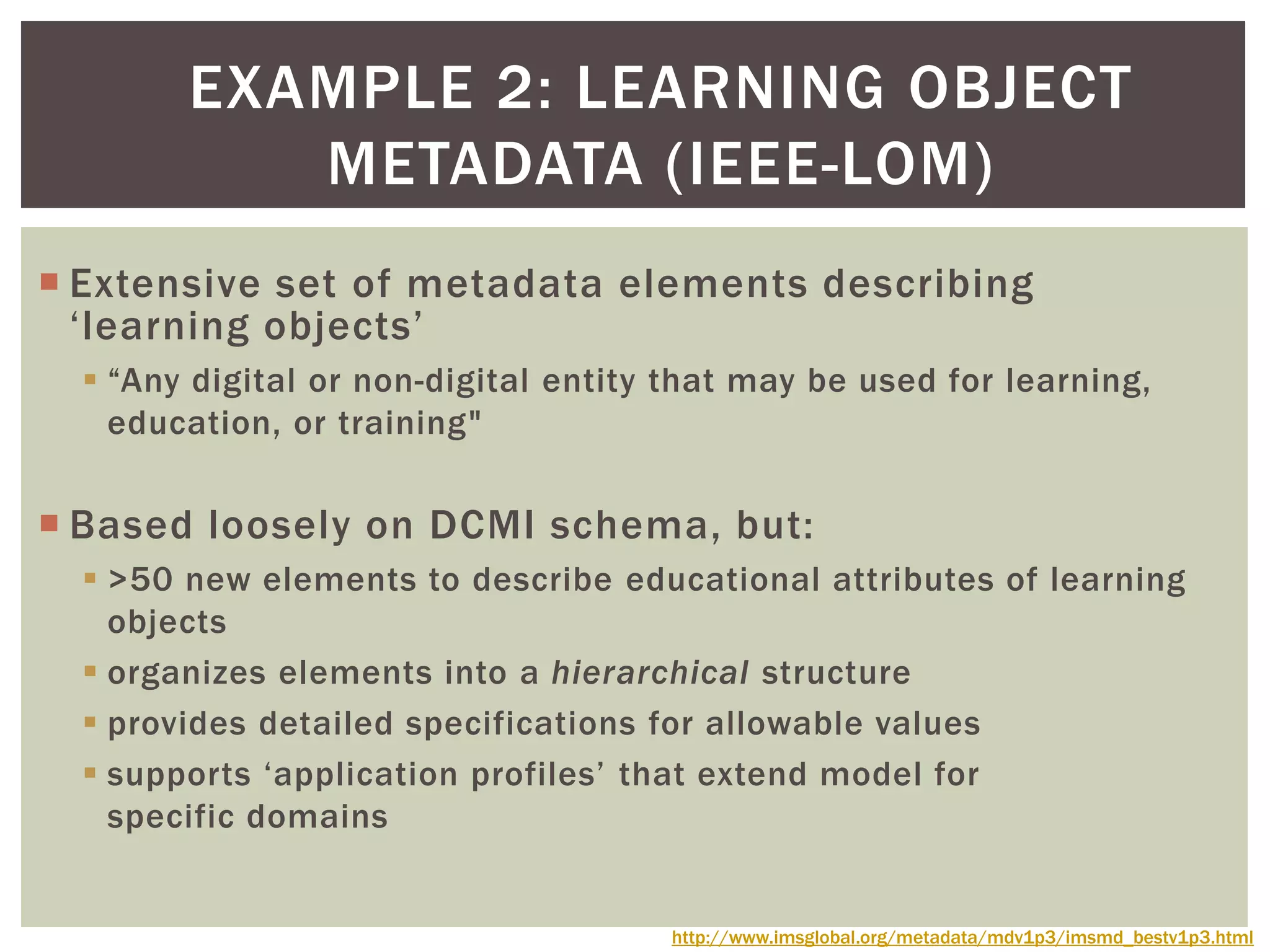
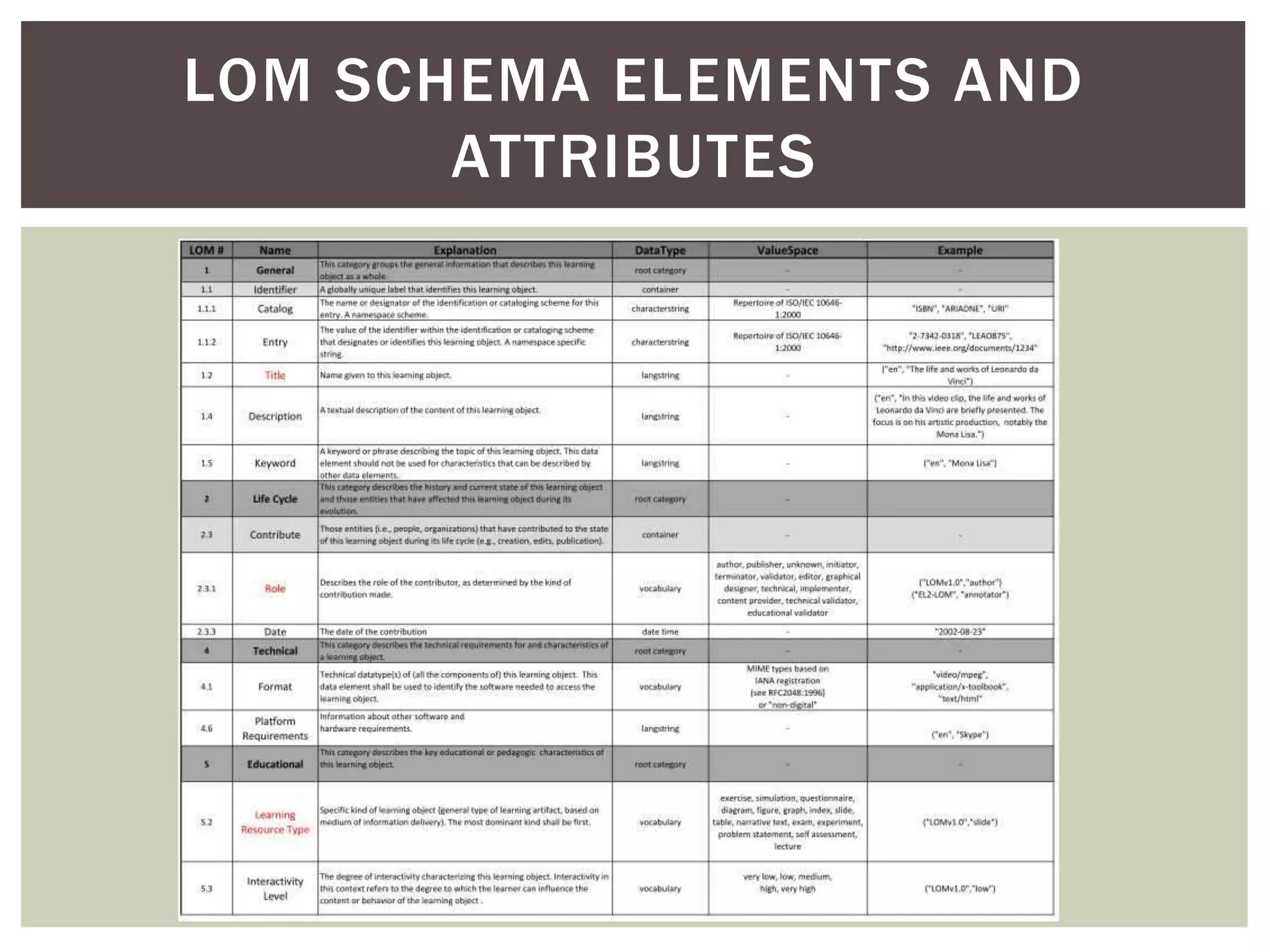
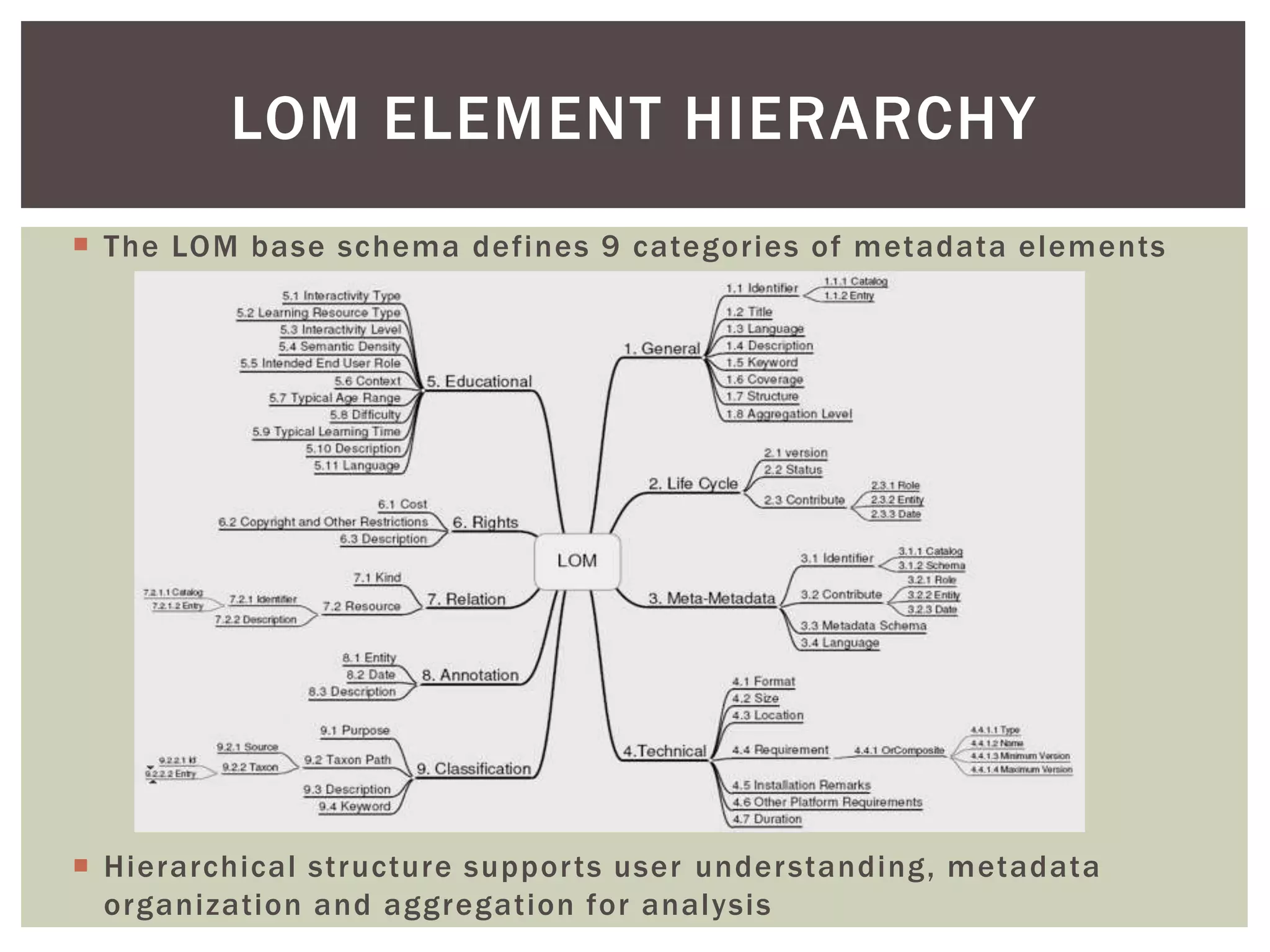
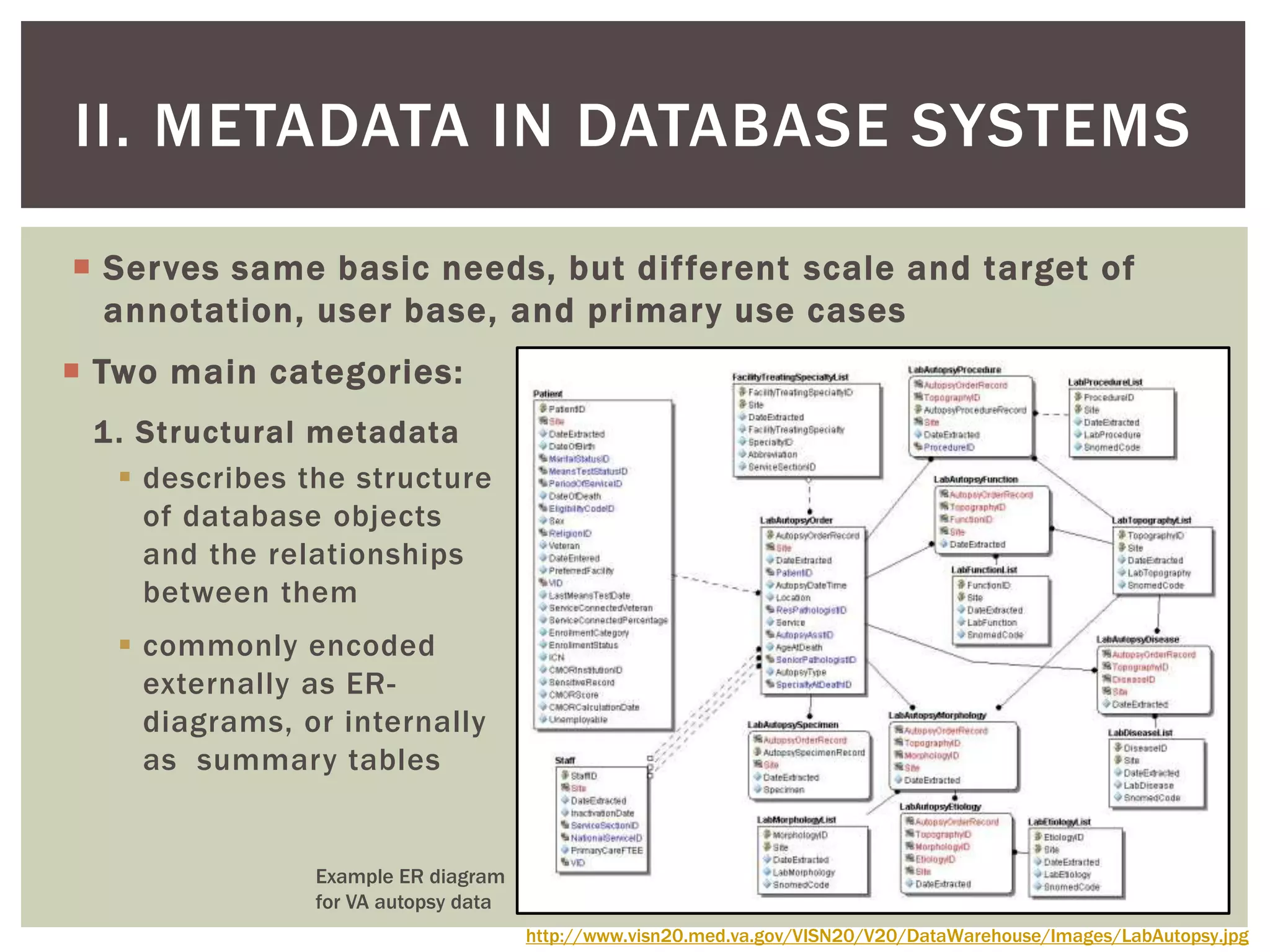
The document discusses perspectives on metadata from web resources and database systems. It describes how metadata comes in many forms and serves various purposes, such as supporting discovery and identification of information resources on the web (resource metadata), and ensuring consistency and analysis of structured data in databases (metadata in database systems). Resource metadata commonly follows standards and is stored separately from the resources it describes, while database metadata includes both structural metadata describing data organization and content metadata in the form of data dictionaries.






![ The notion of a ‘data element’ obtains a more precise meaning
and specification in the context of a database.
elements can be specified at finer granularity in a databases holding
structured data in a controlled operational system
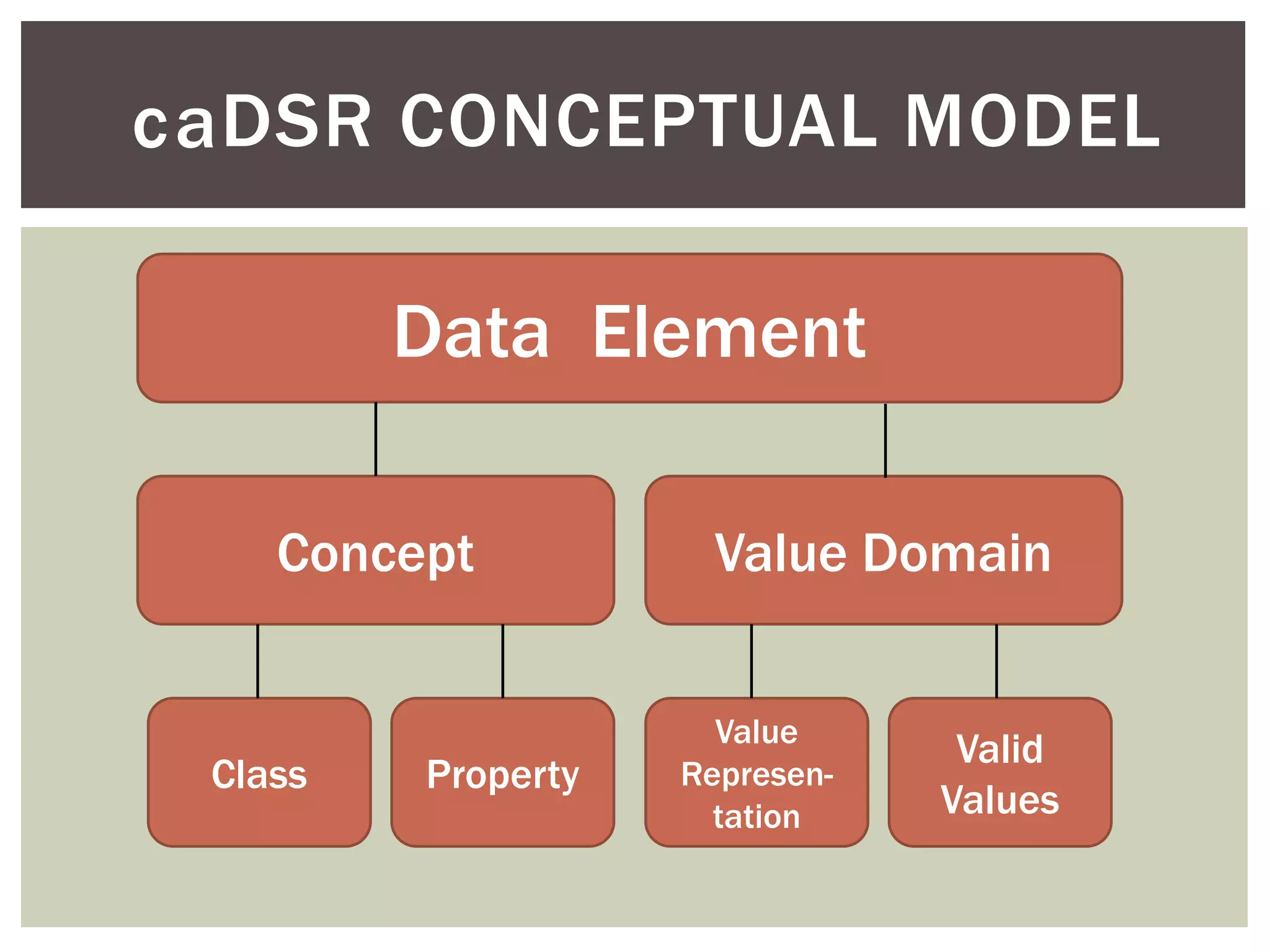
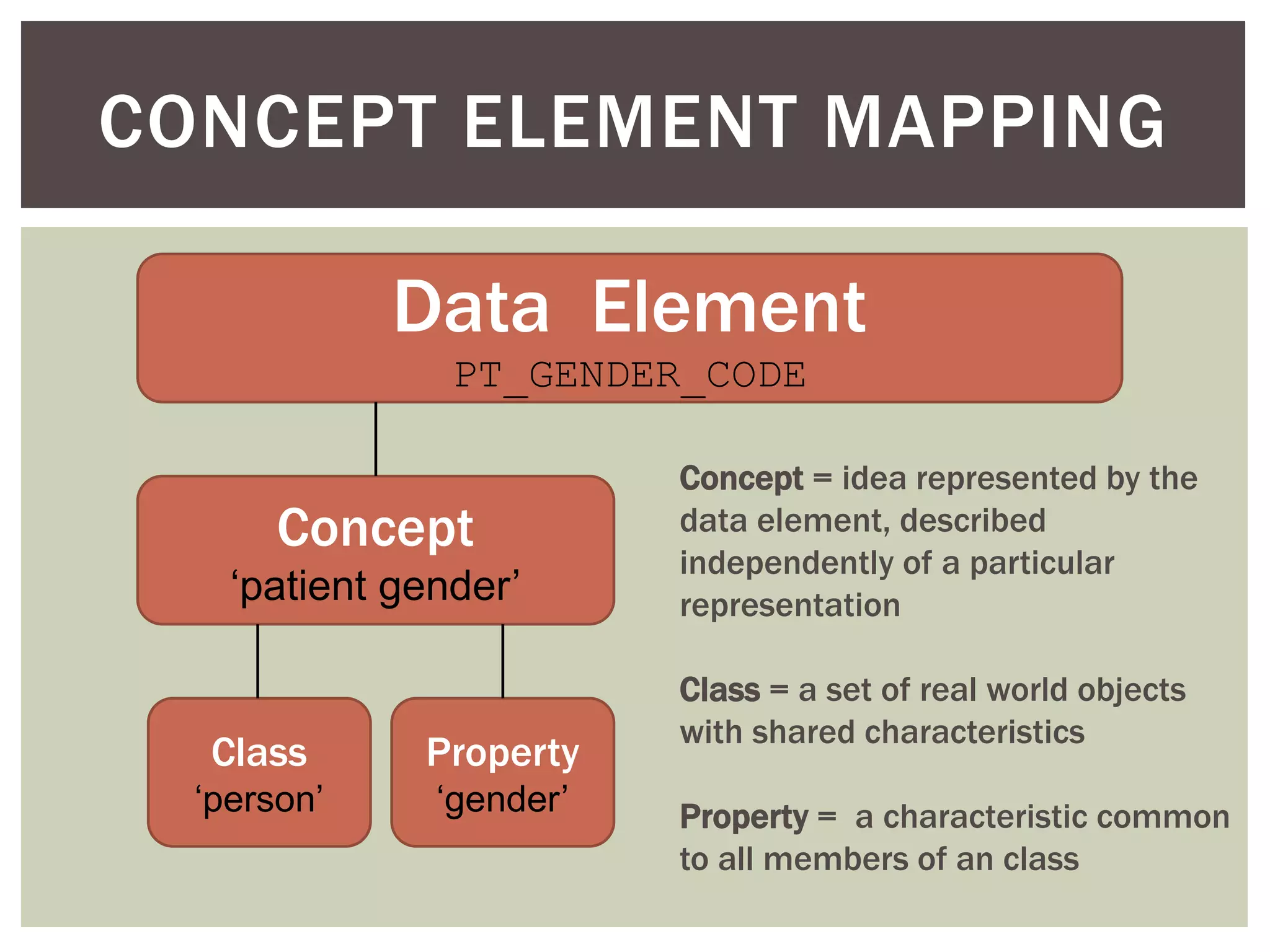
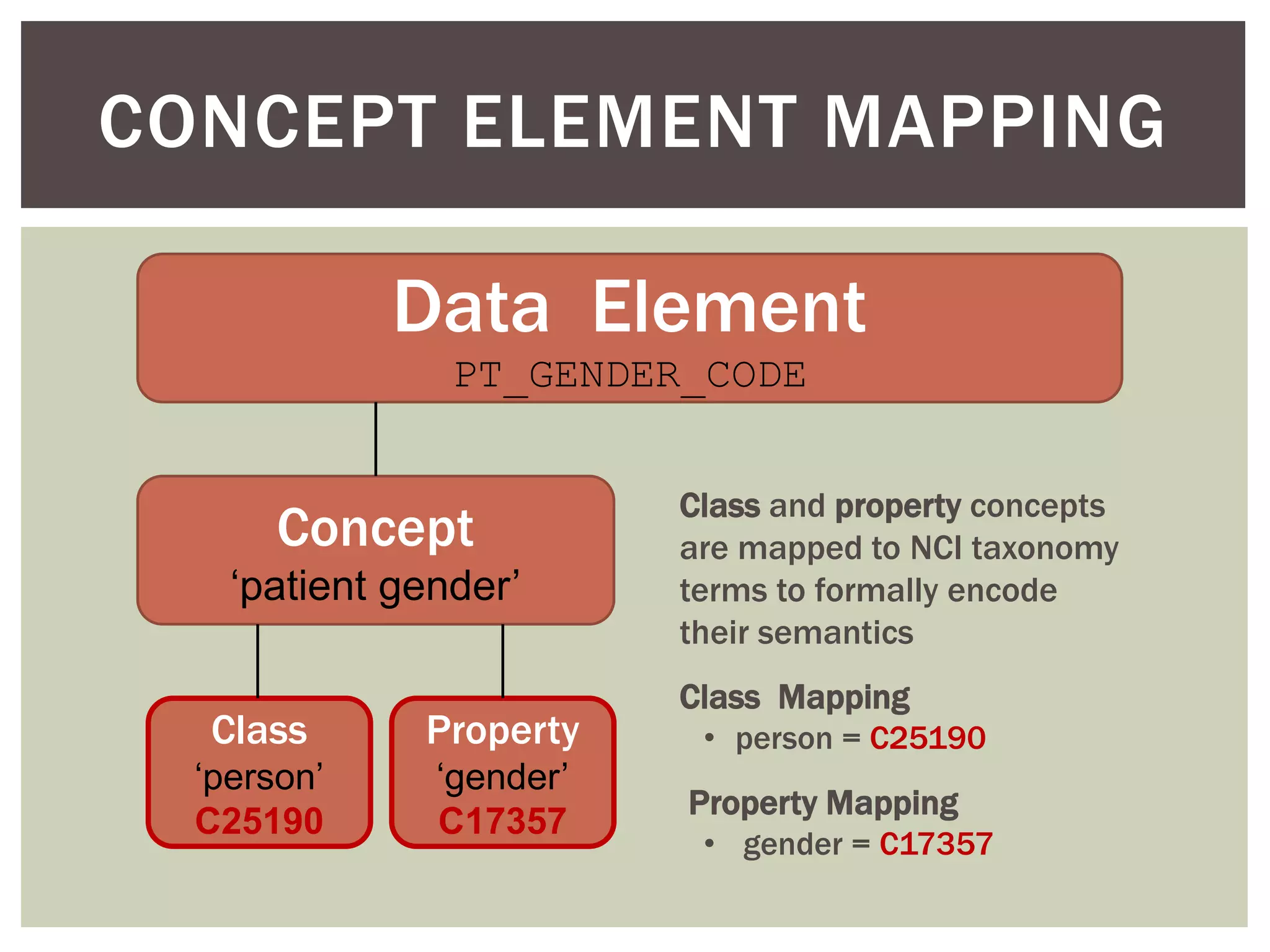
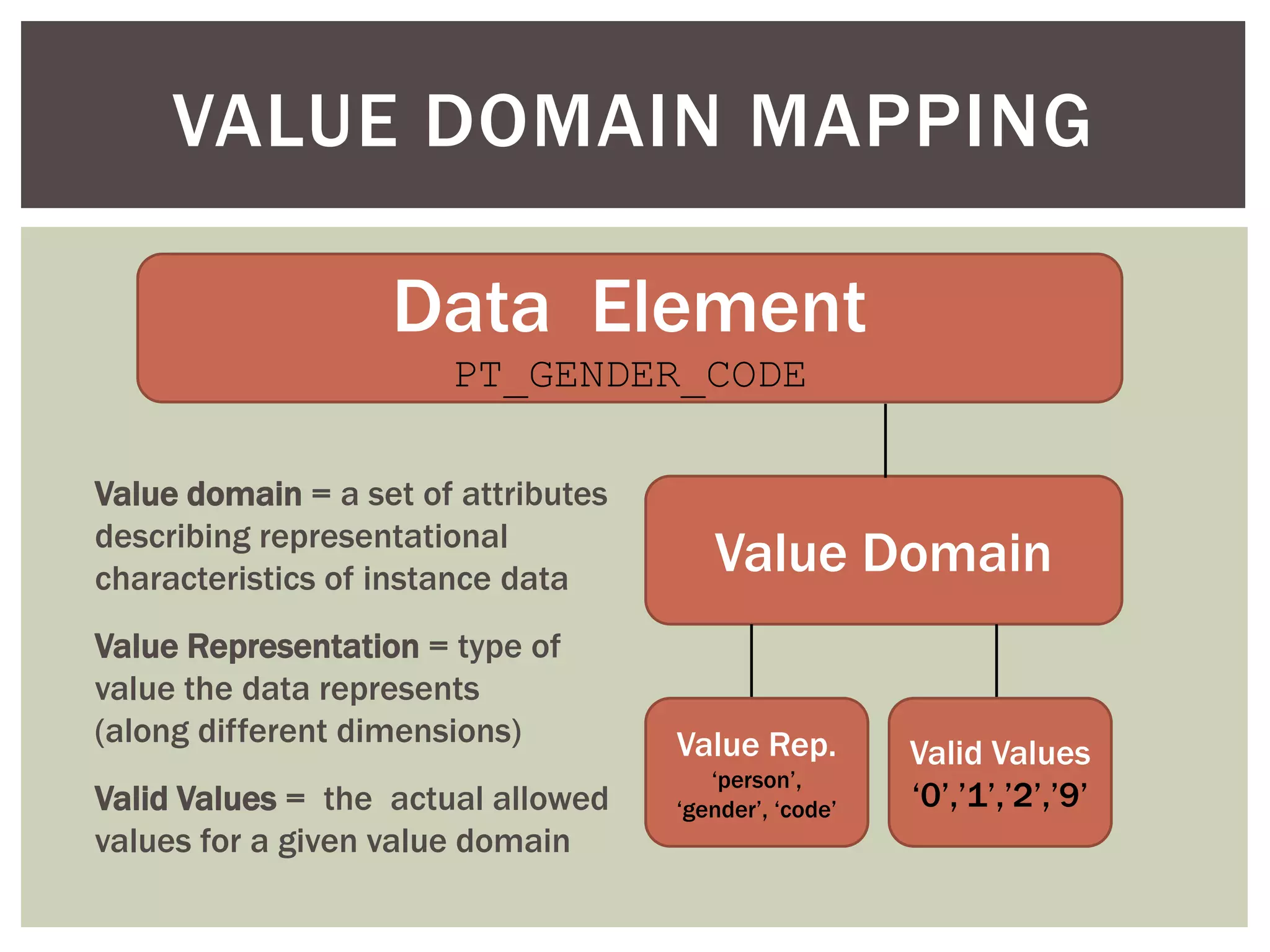
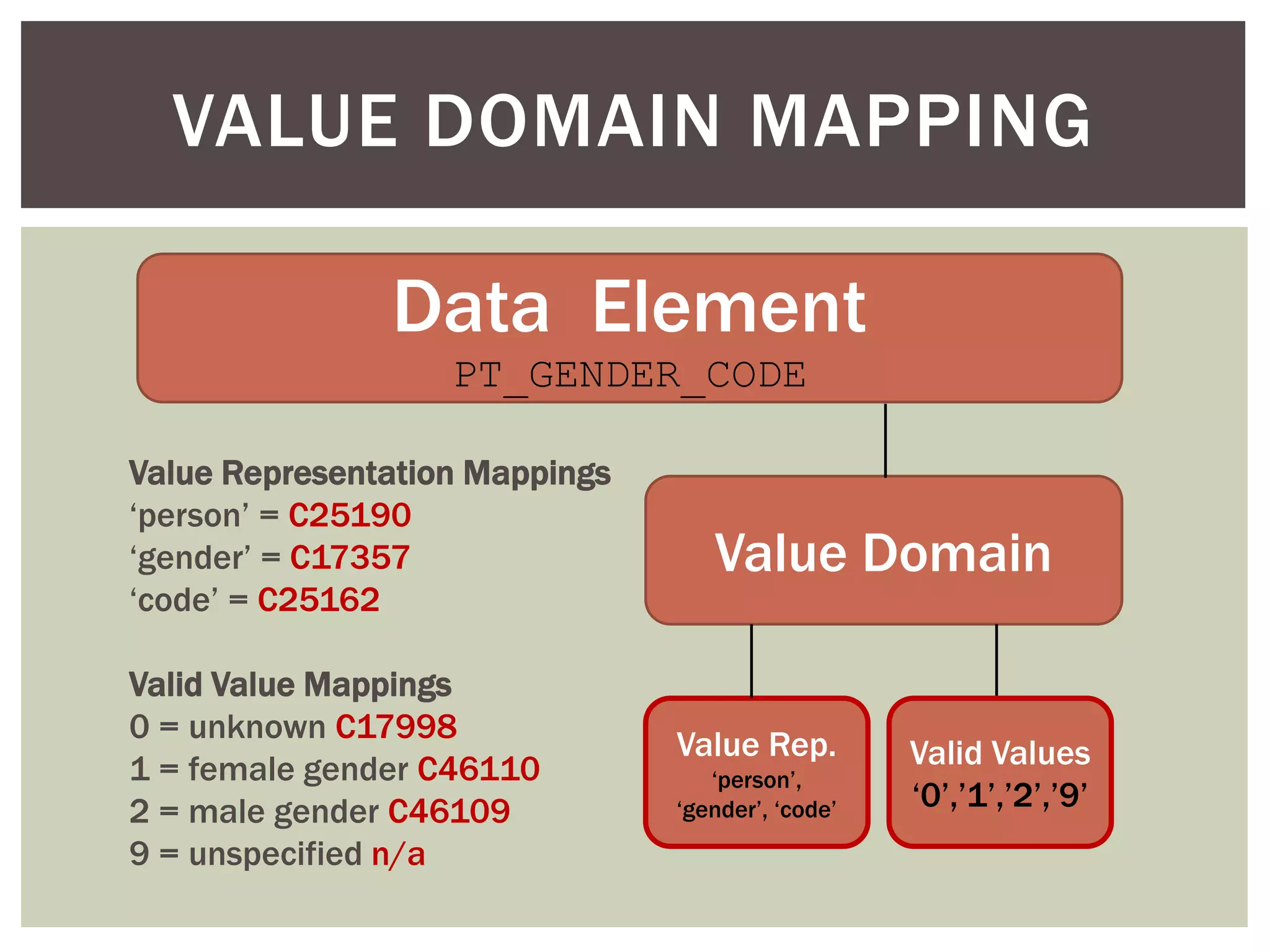
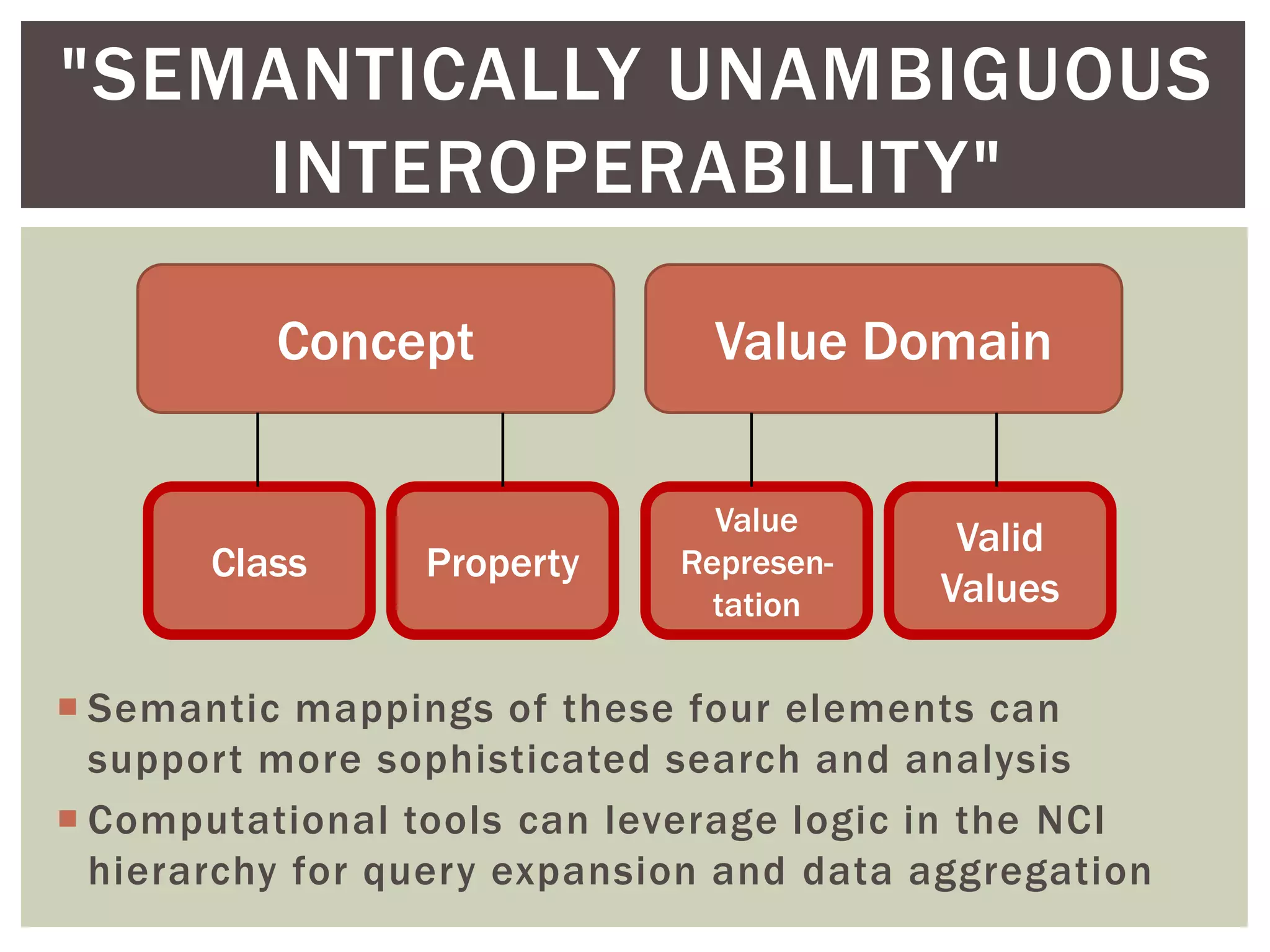
Conceptually, a data element is comprised of a concept and a
value domain
concept = the subject of the data recorded for a given element
value domain = the defined value set for how that data is recorded
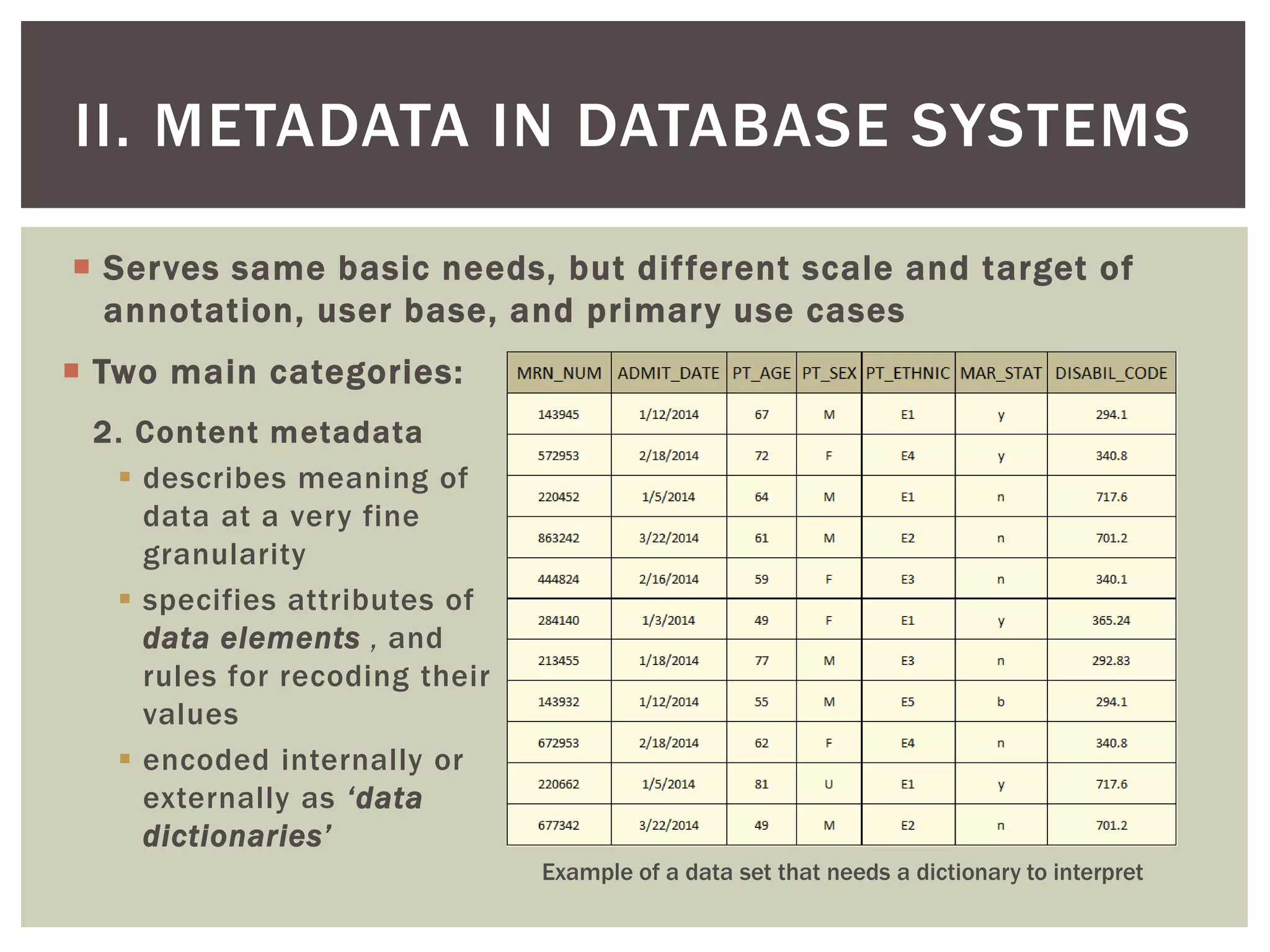
Example: PT_ETHNIC
concept = patient ethnicity
value domain = [ E1 (caucasian), E2 (hispanic/latino), E3 (african),
E4 (asian), E5 (mixed) ]
DATA ELEMENTS](https://image.slidesharecdn.com/metadatalecture9-17-14-140926152131-phpapp02/75/Metadata-lecture-9-17-14-27-2048.jpg)