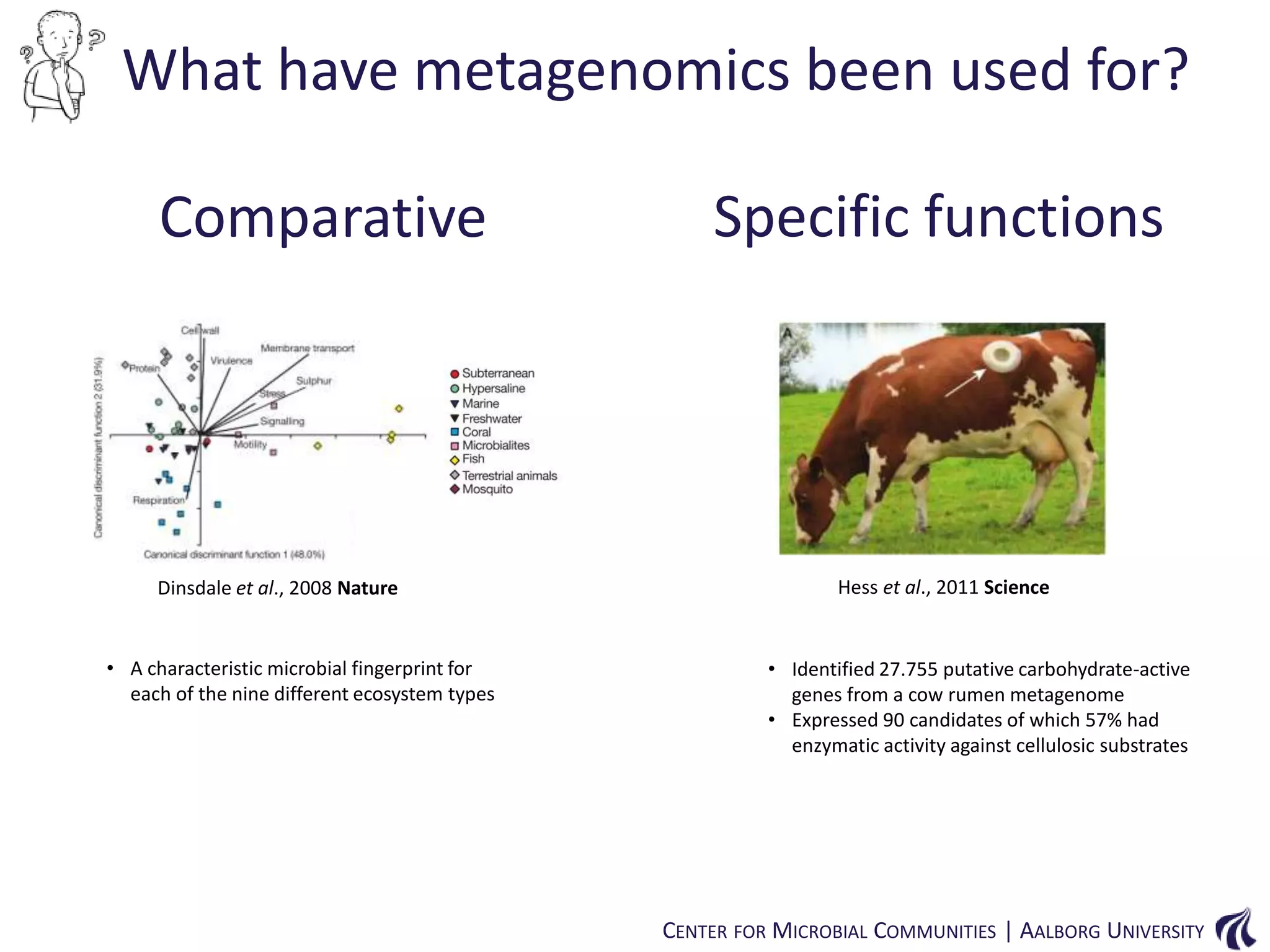
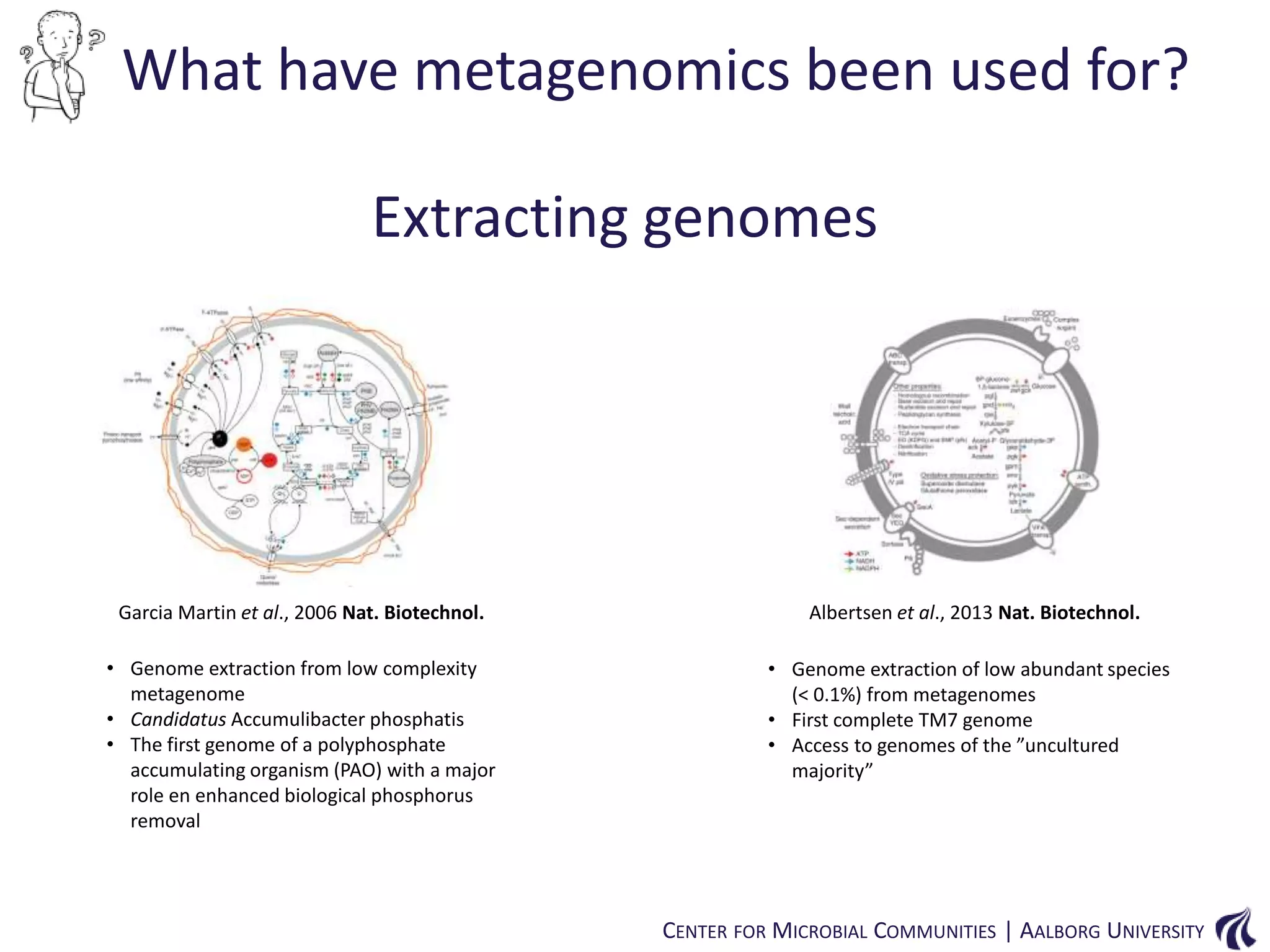
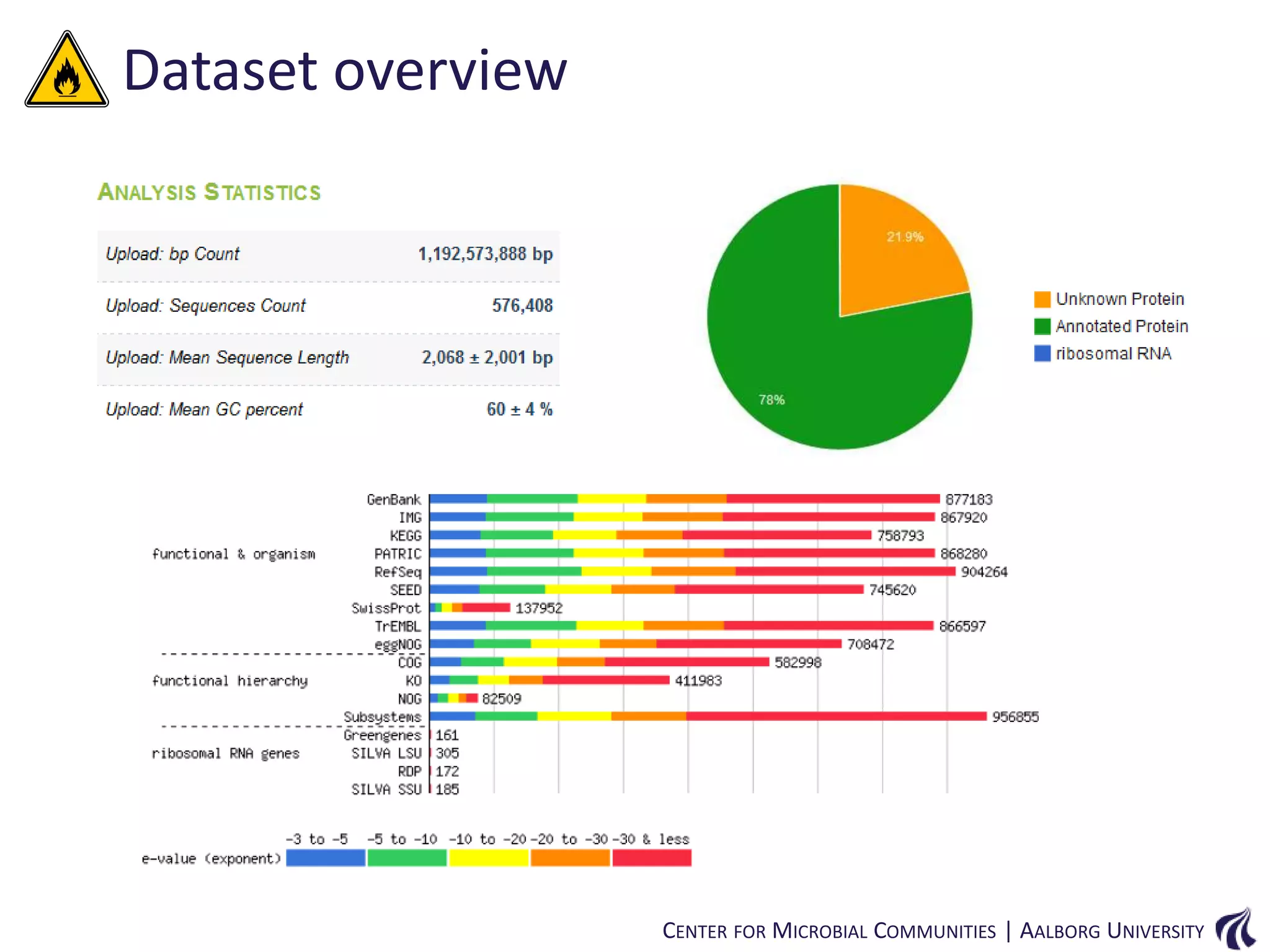
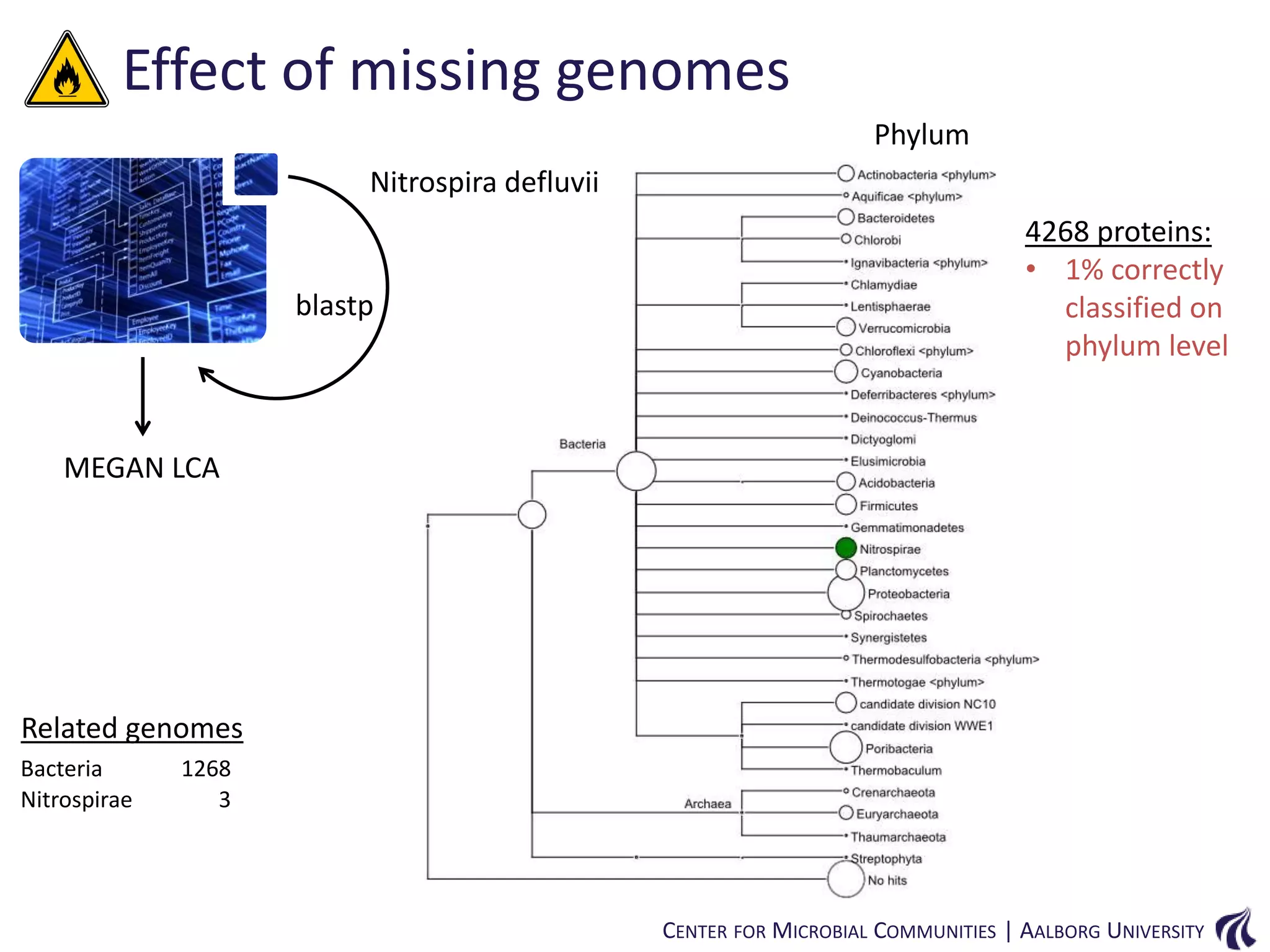
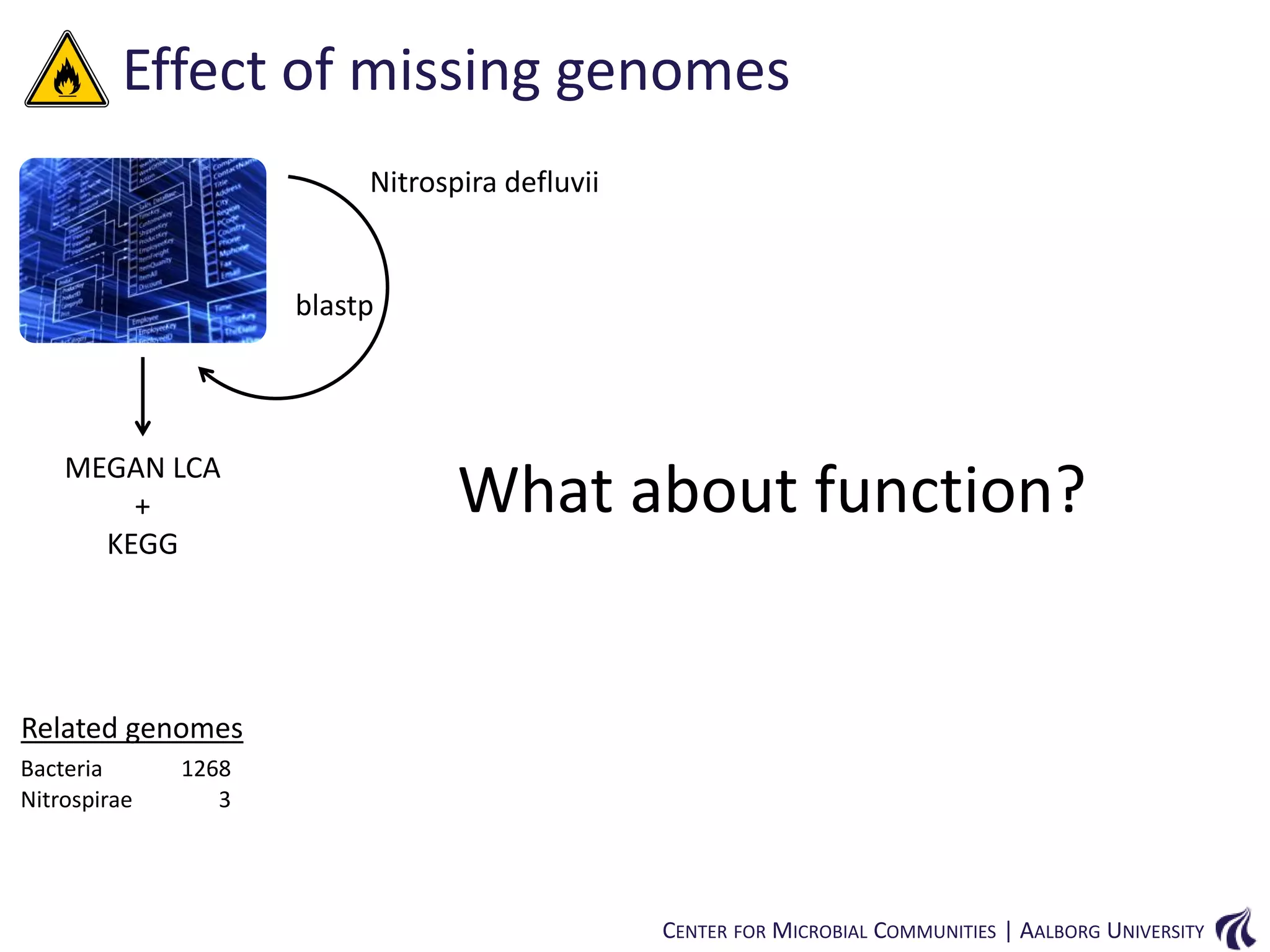
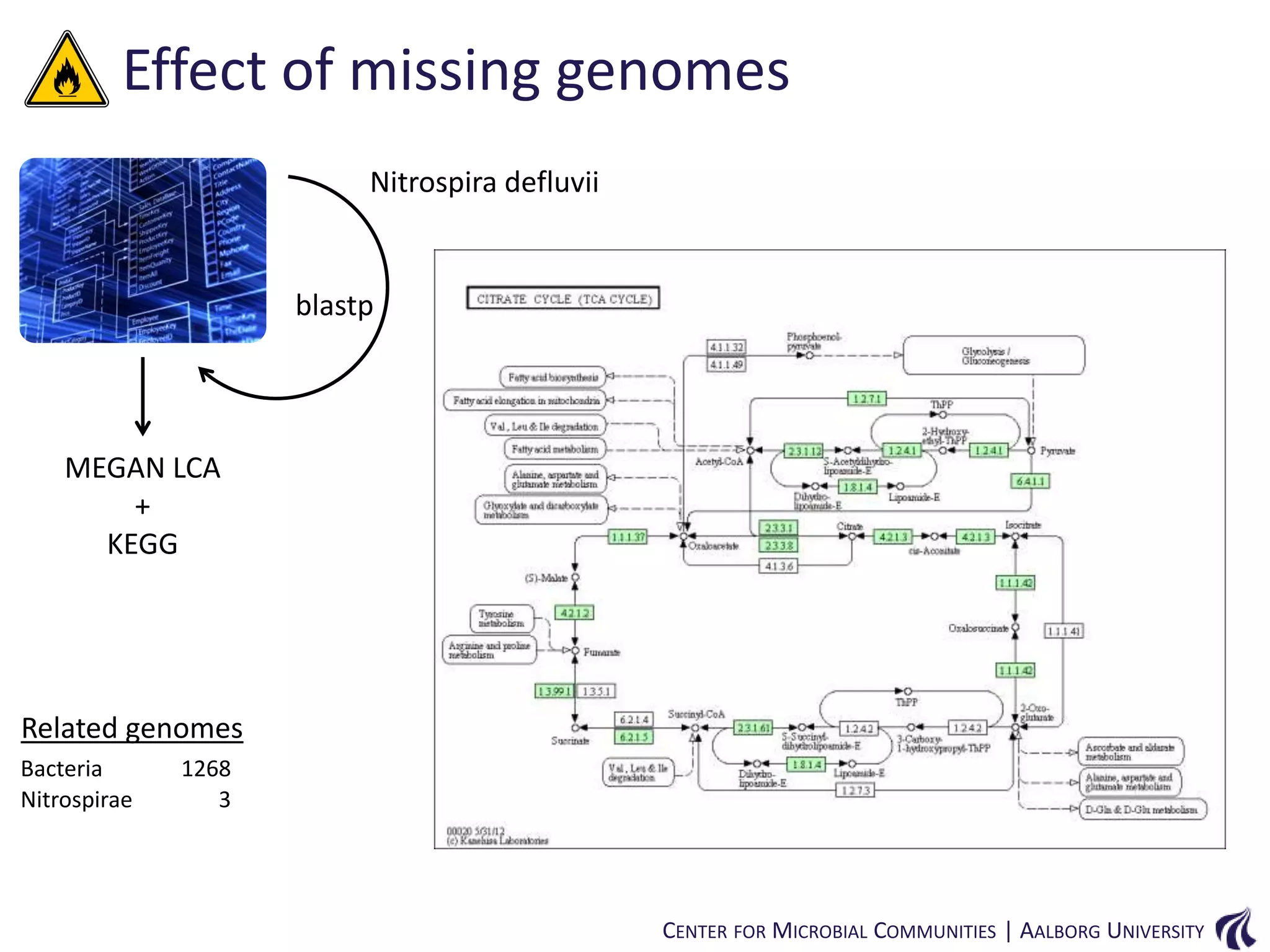
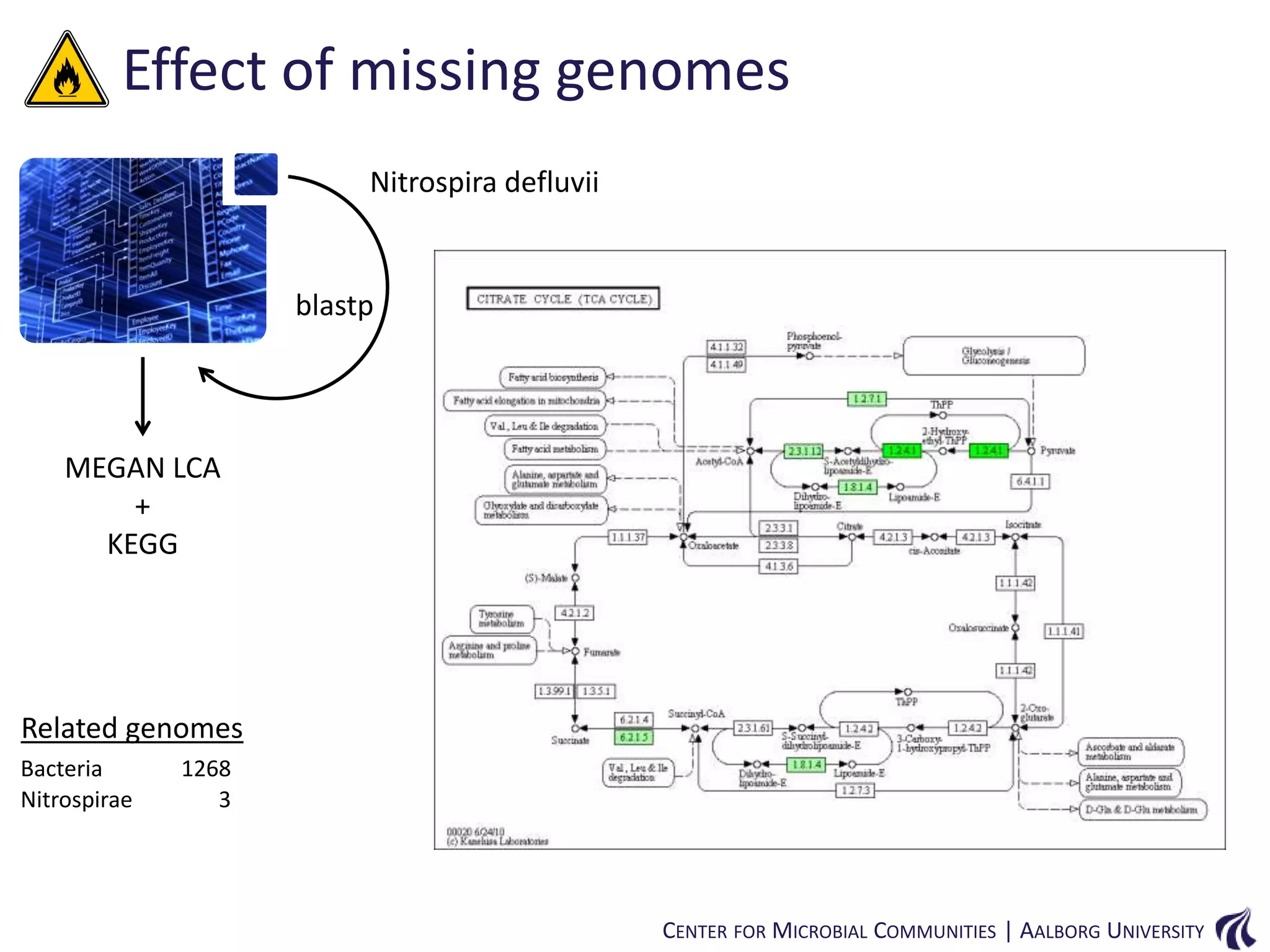
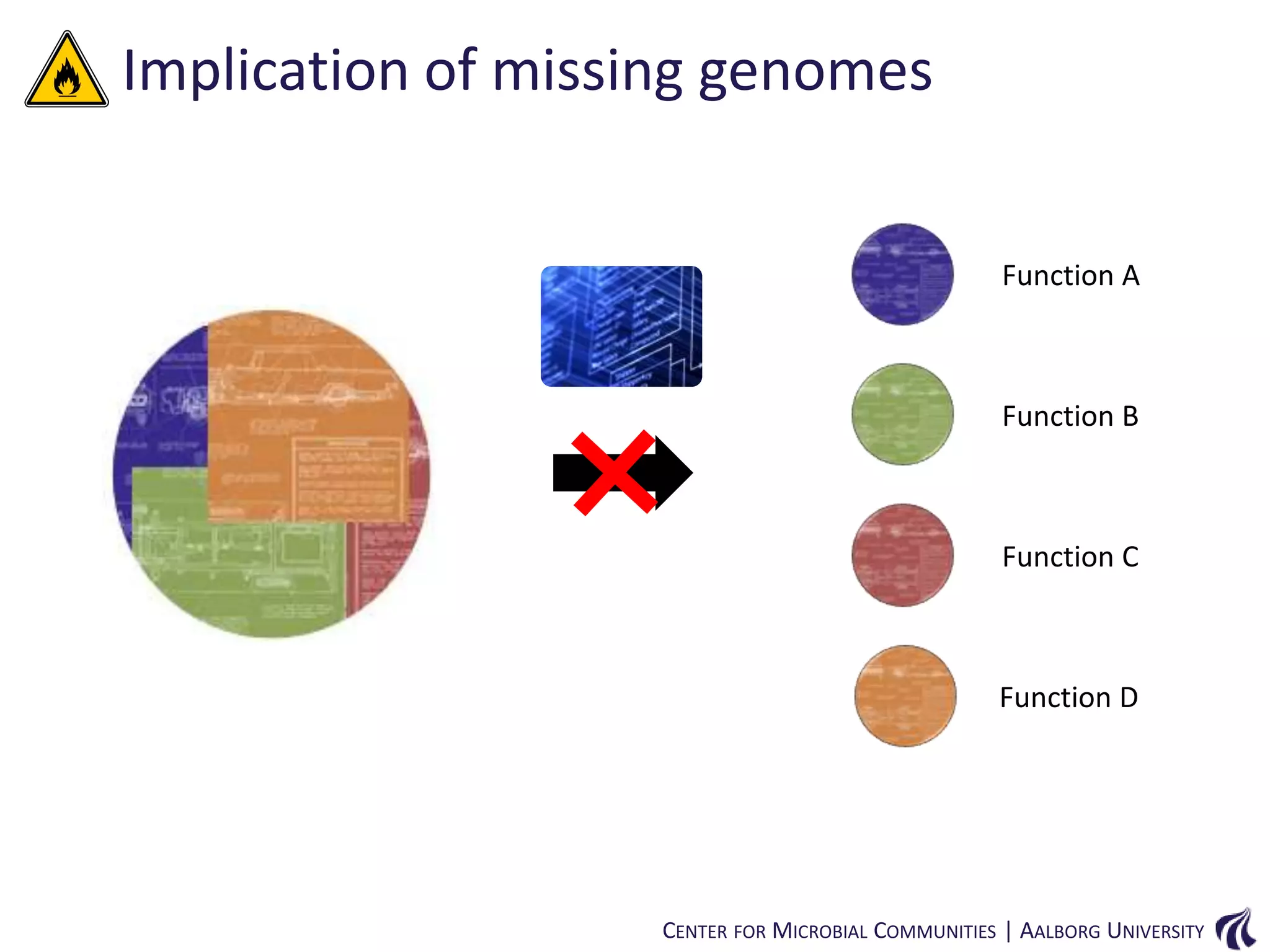
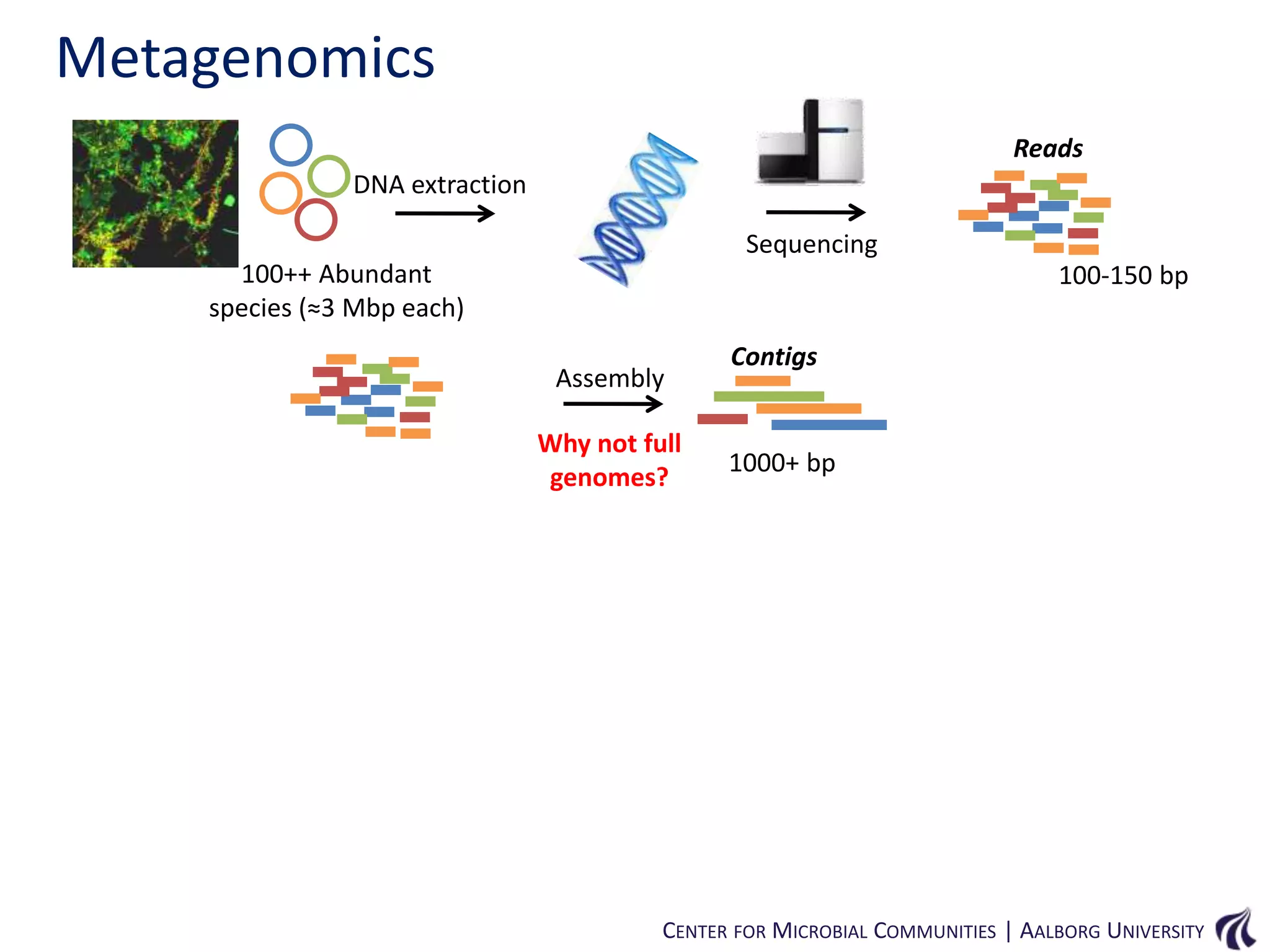
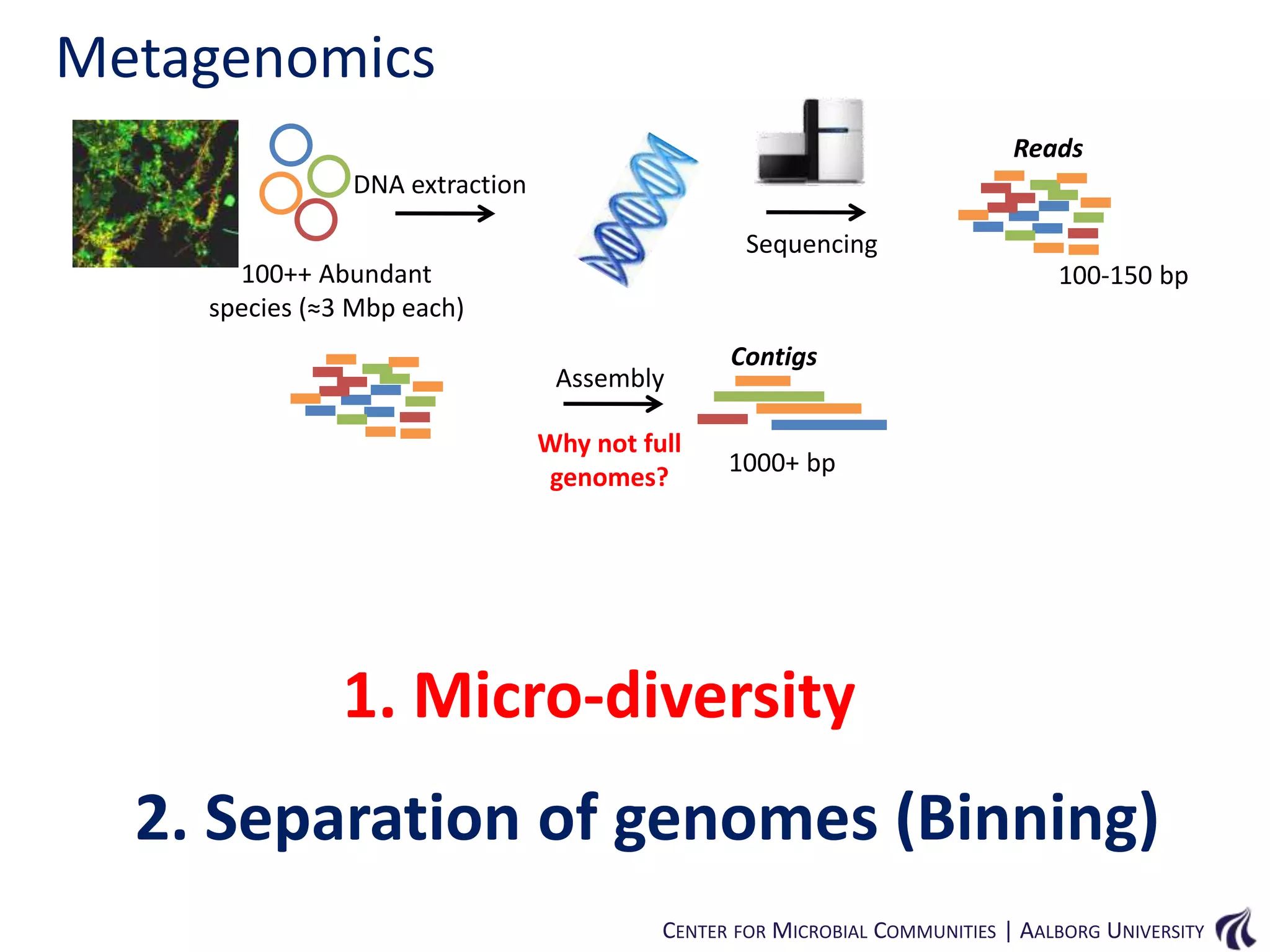
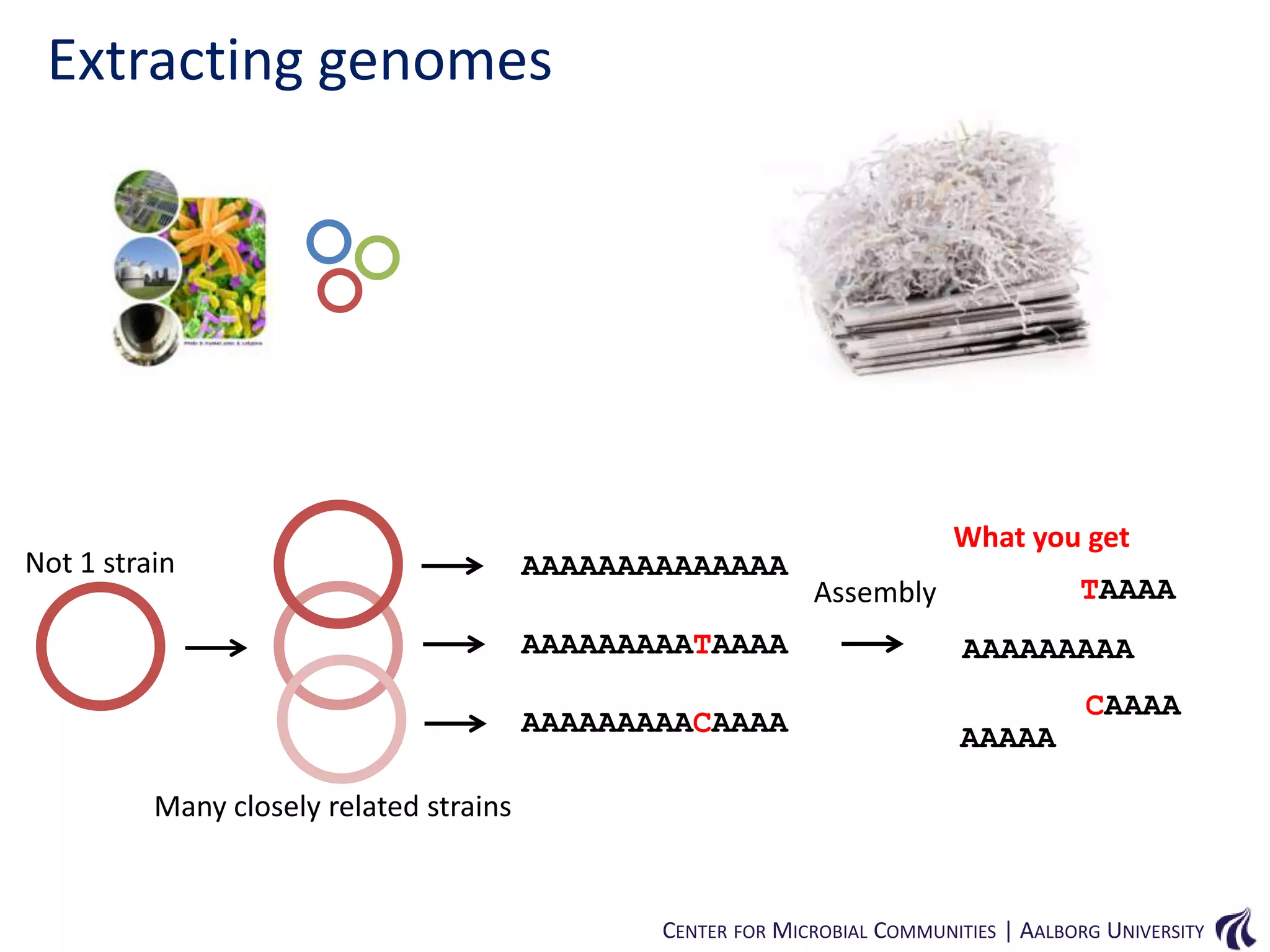
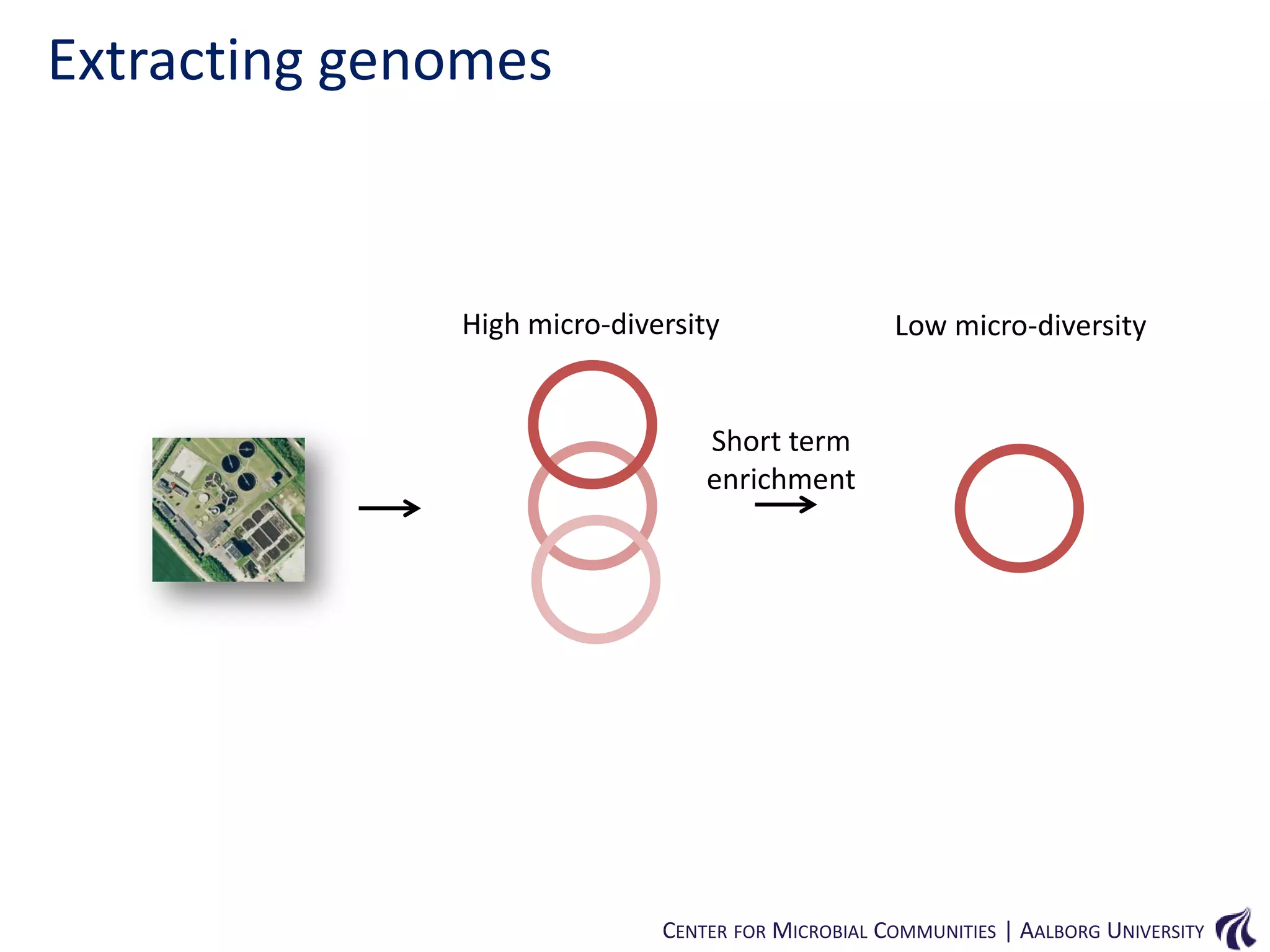
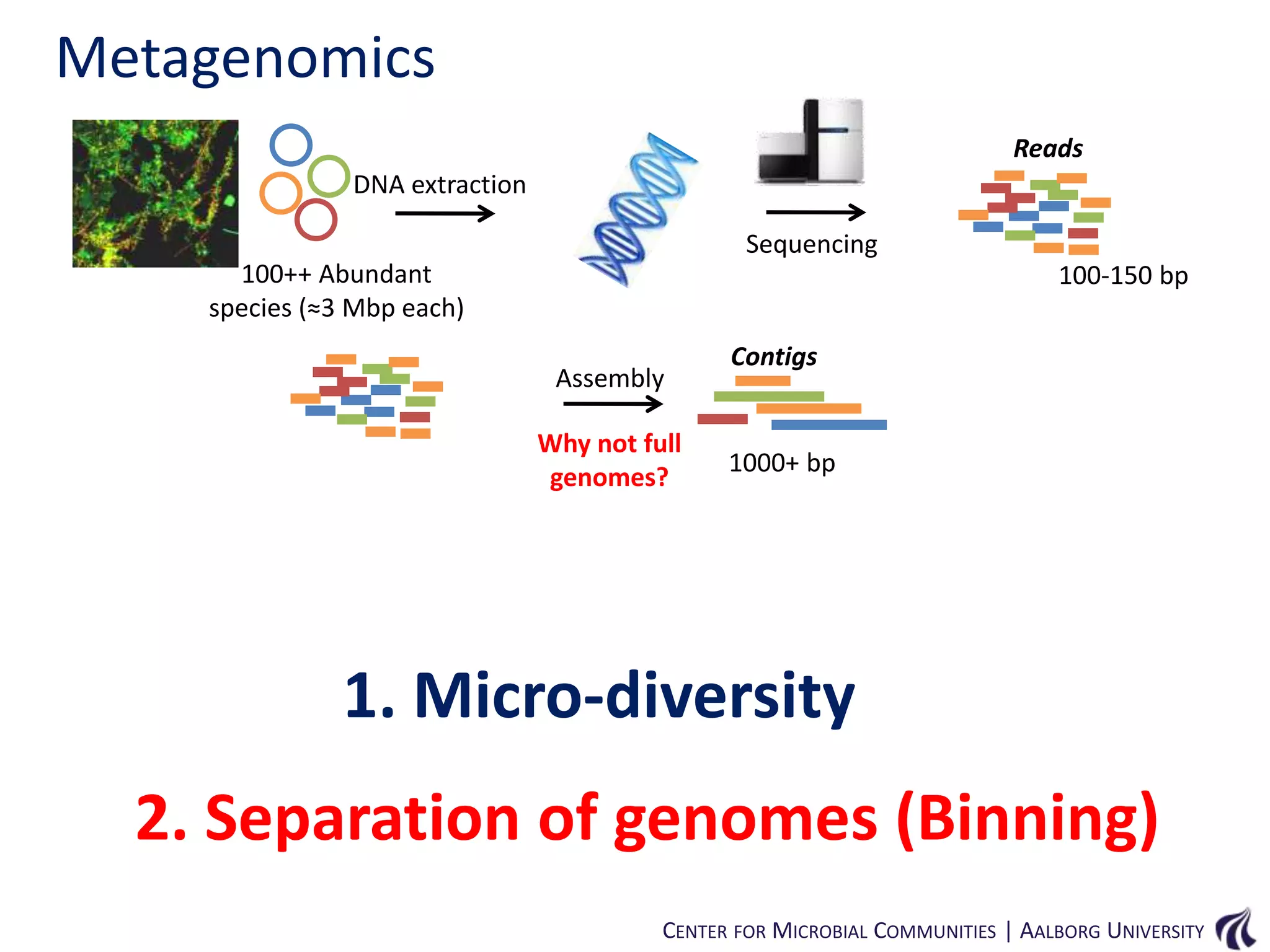
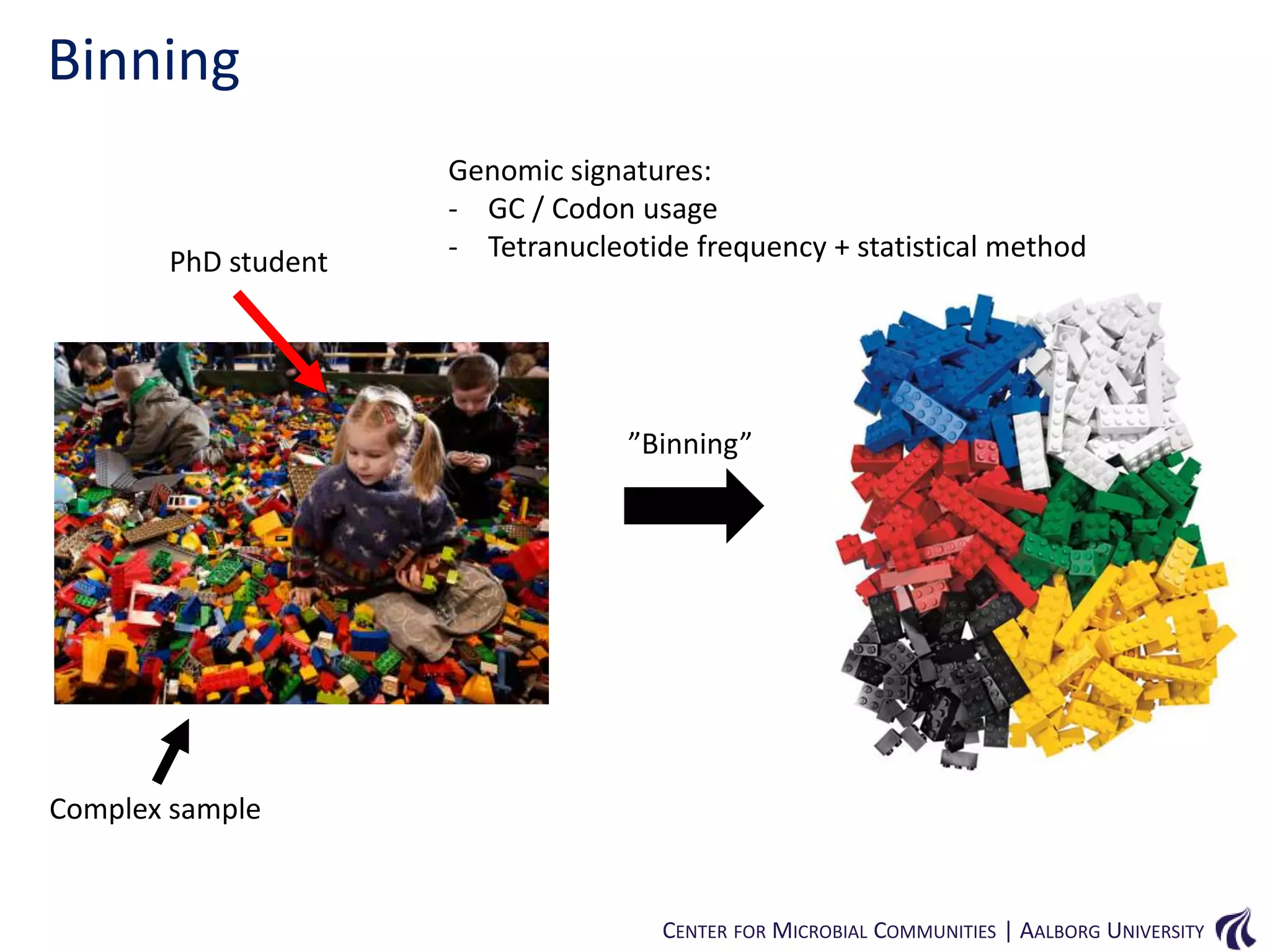
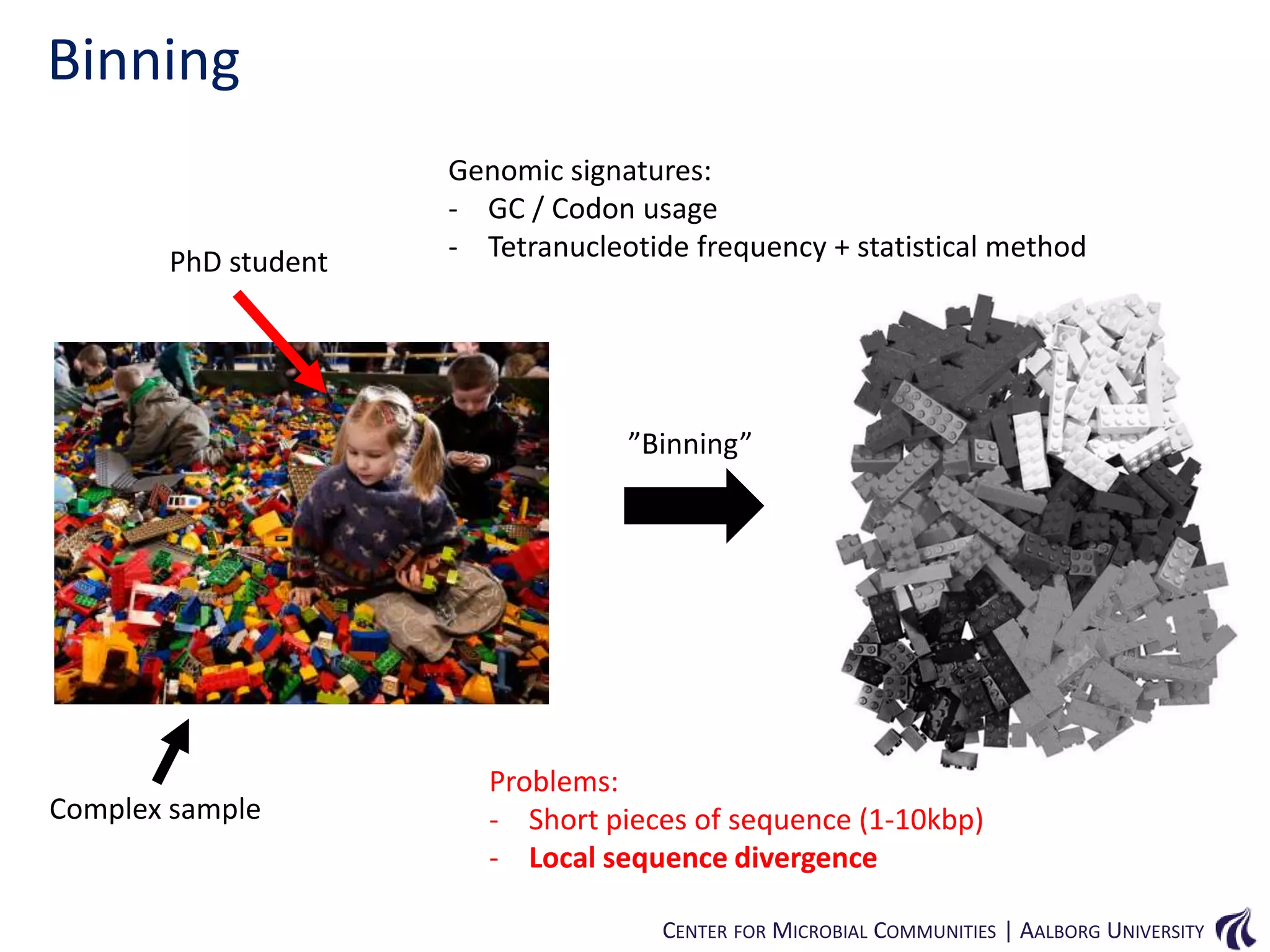
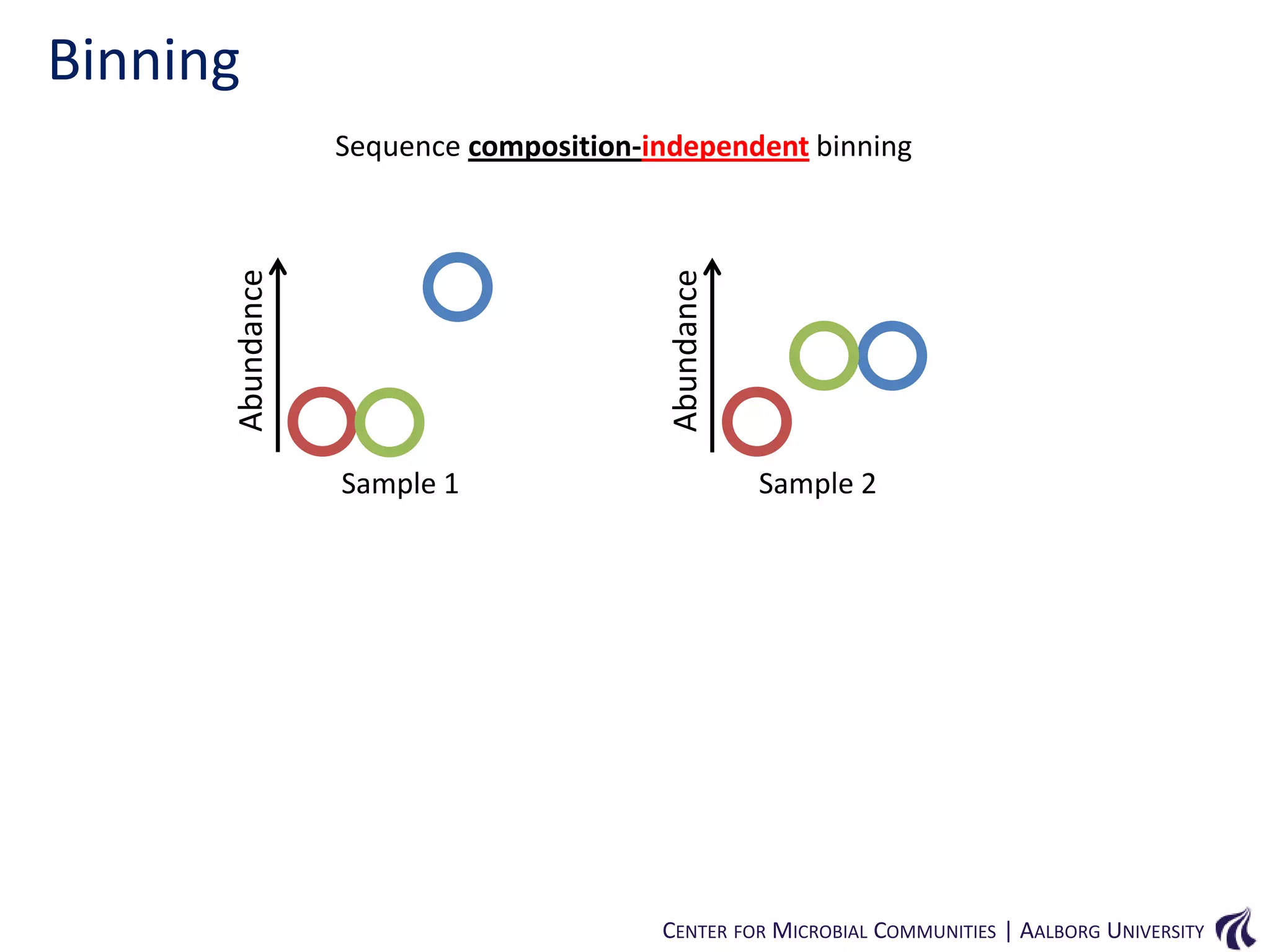
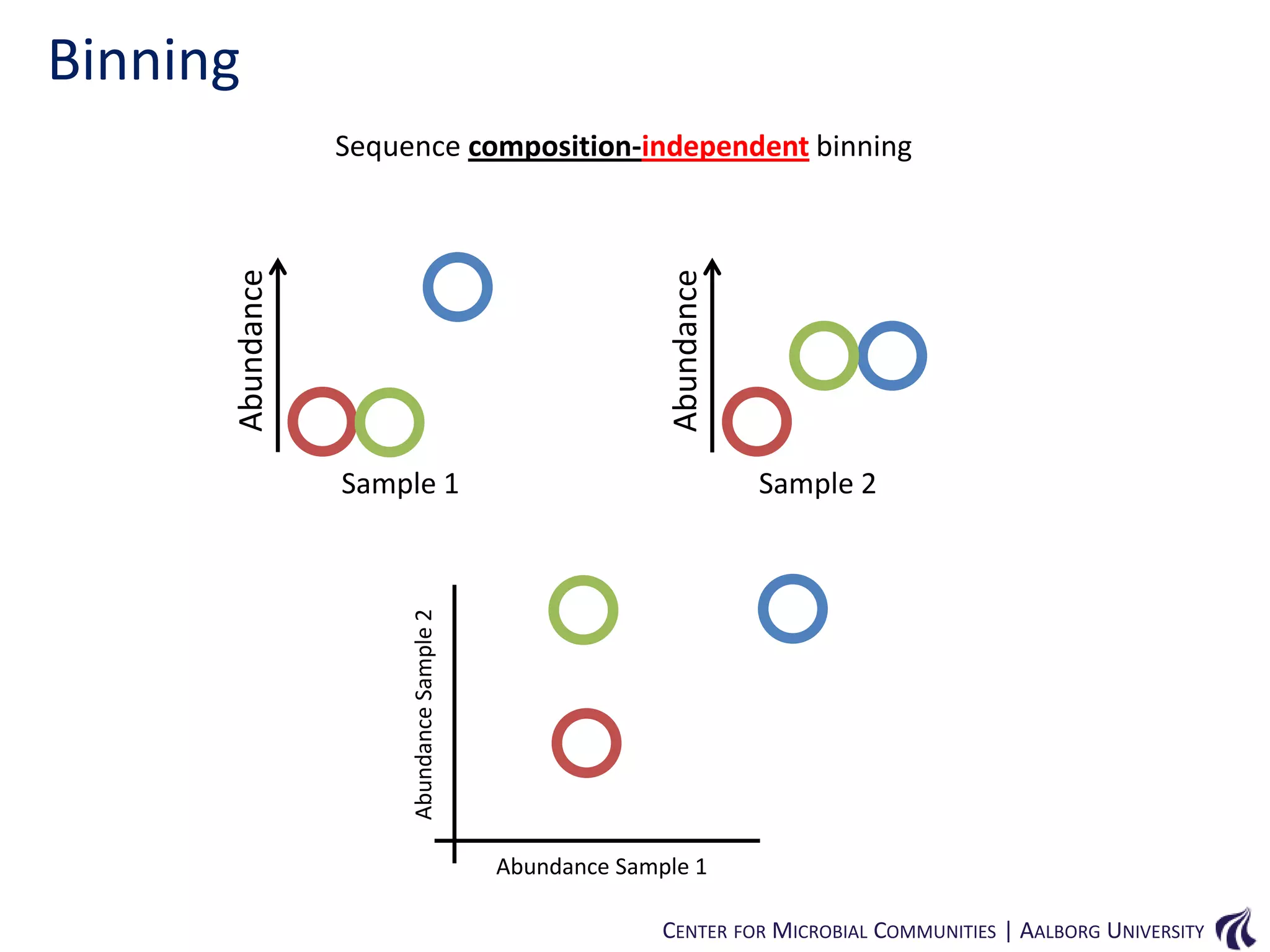
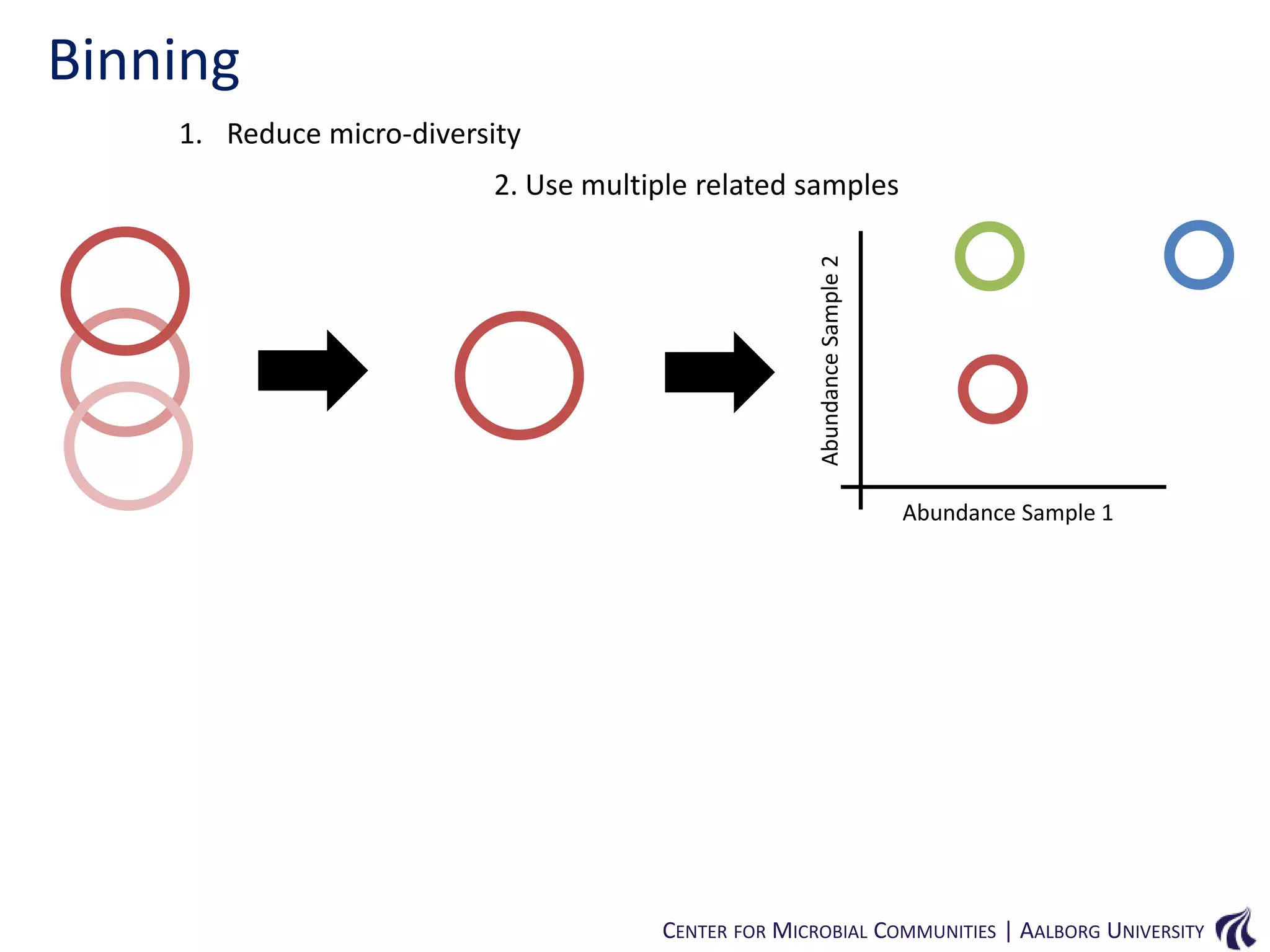
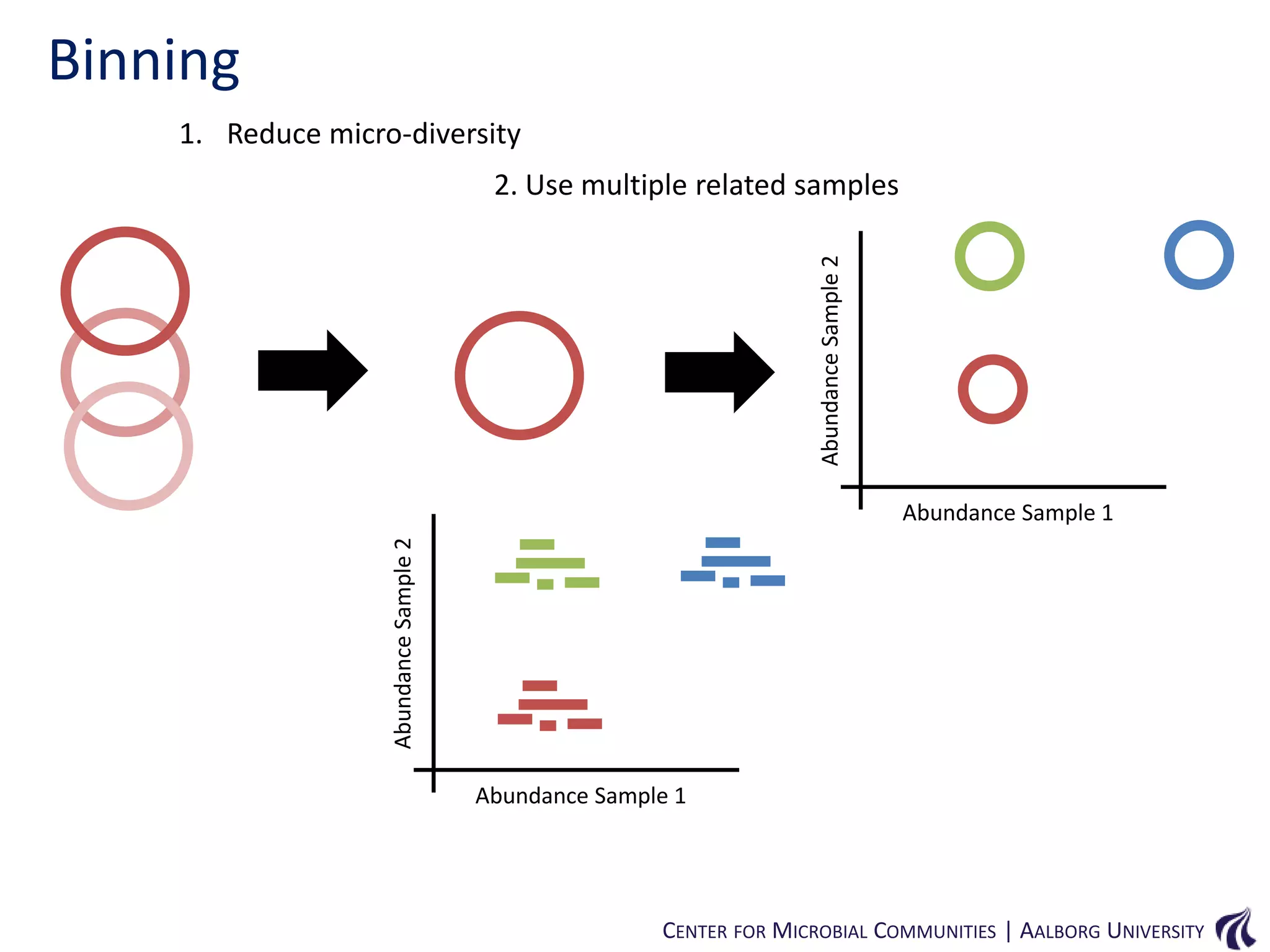
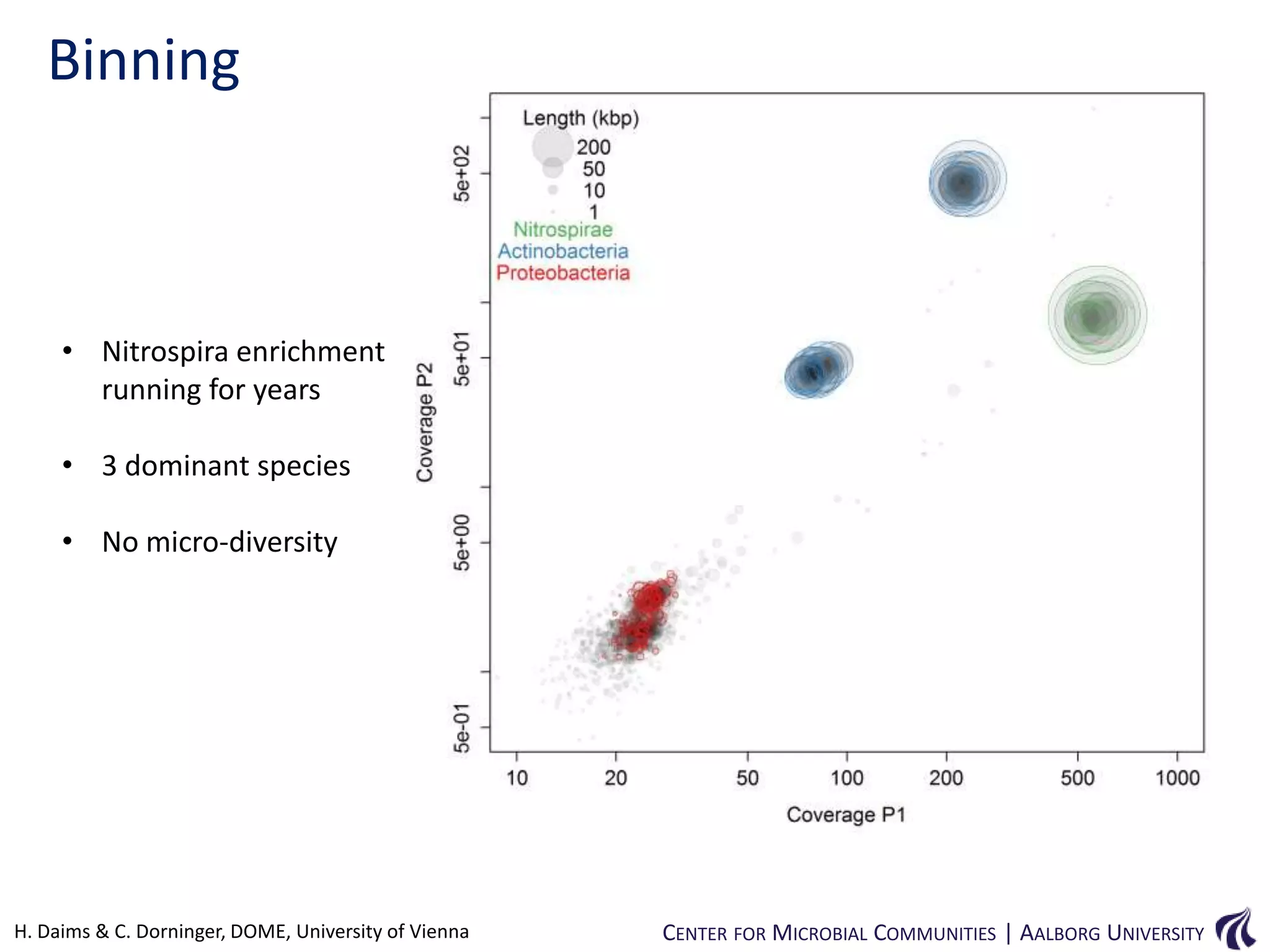
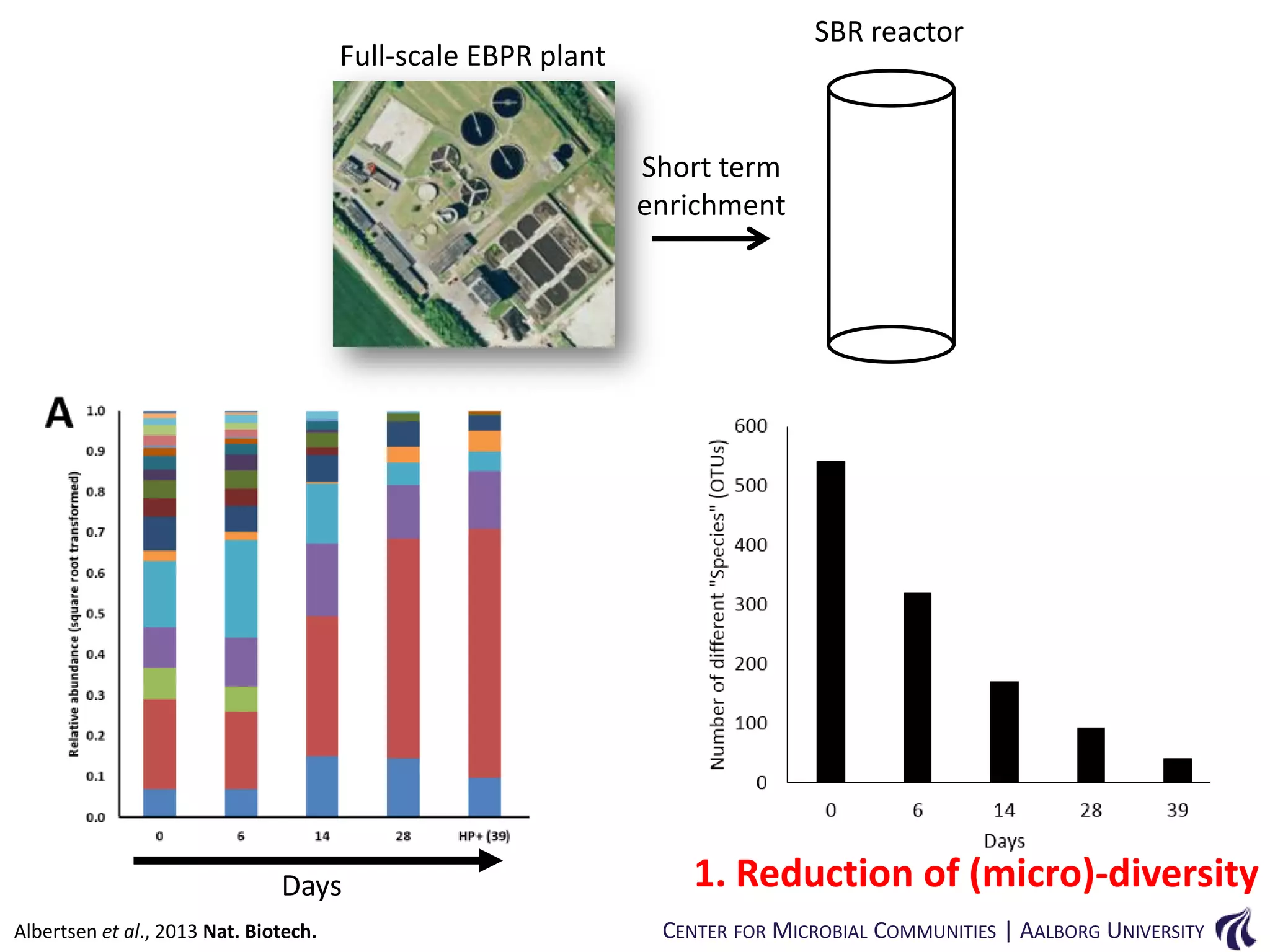
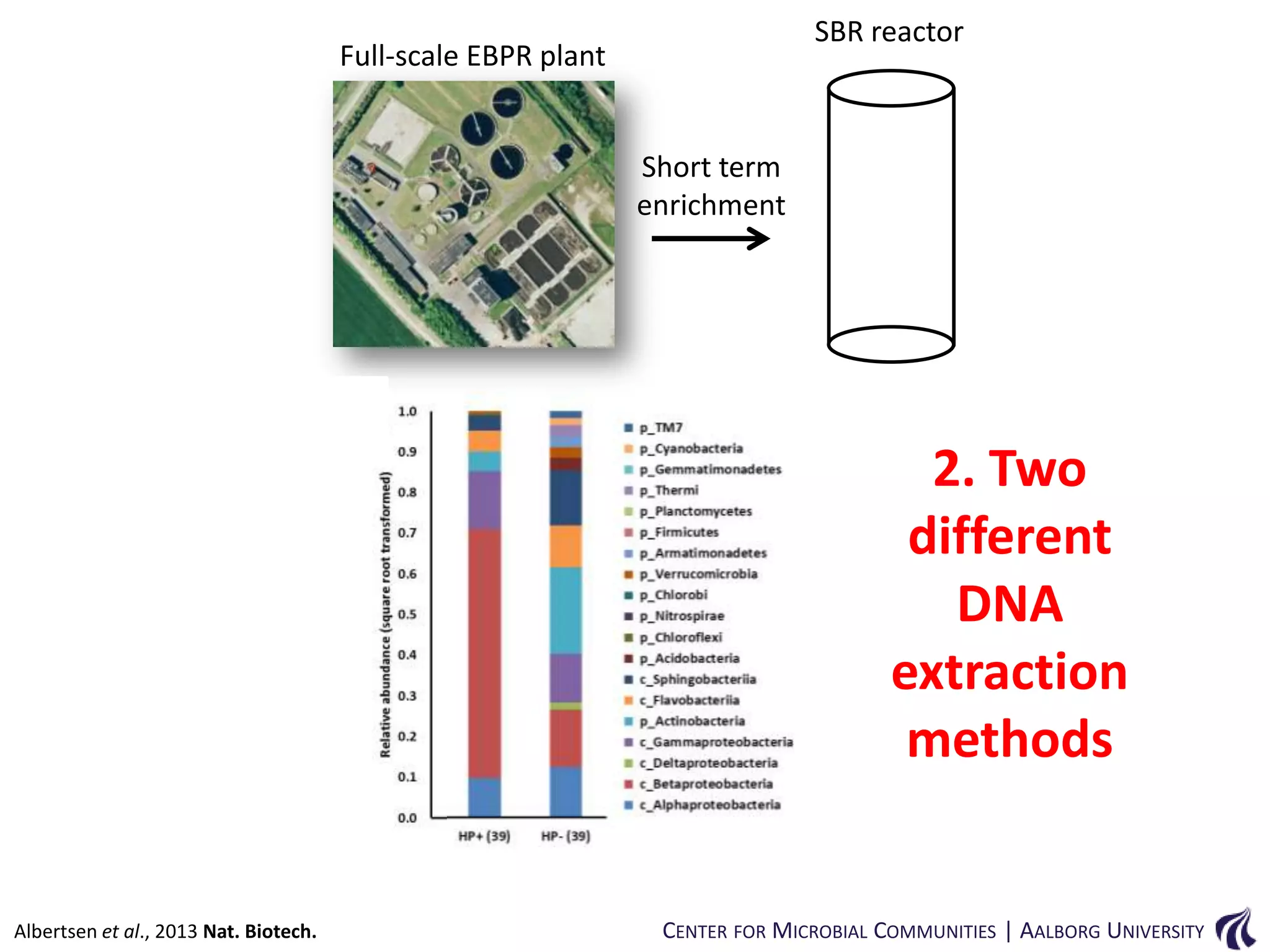
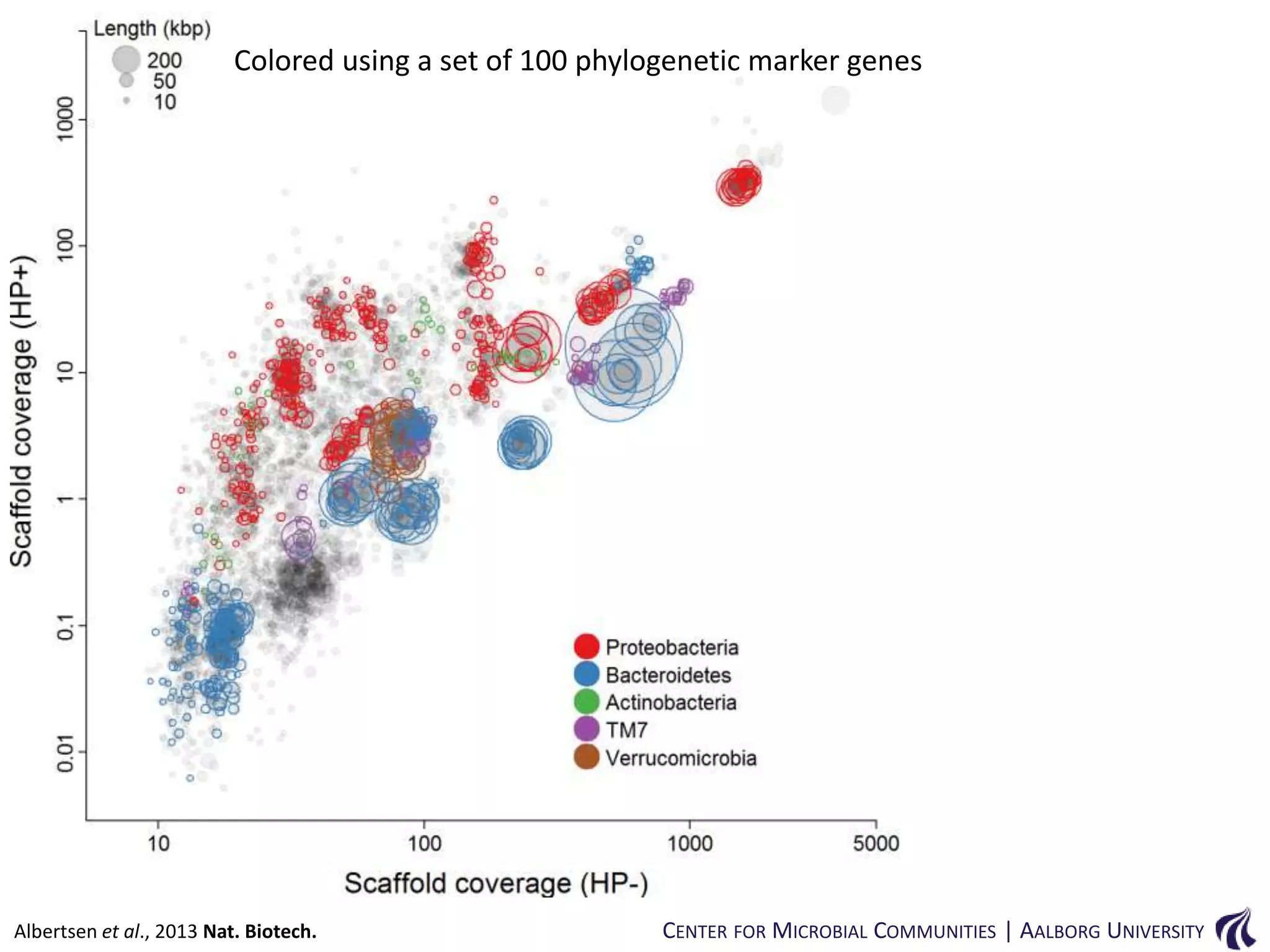
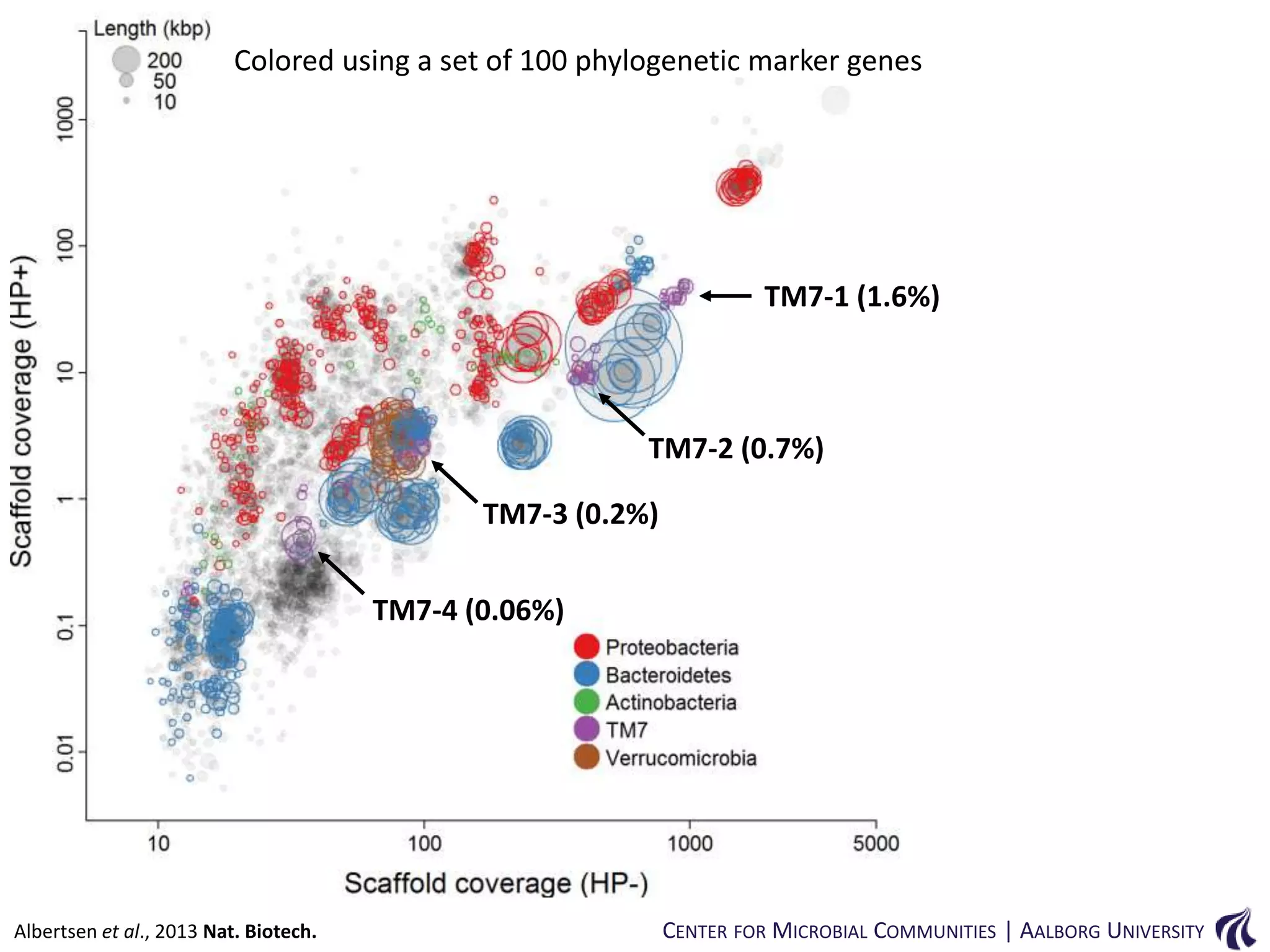
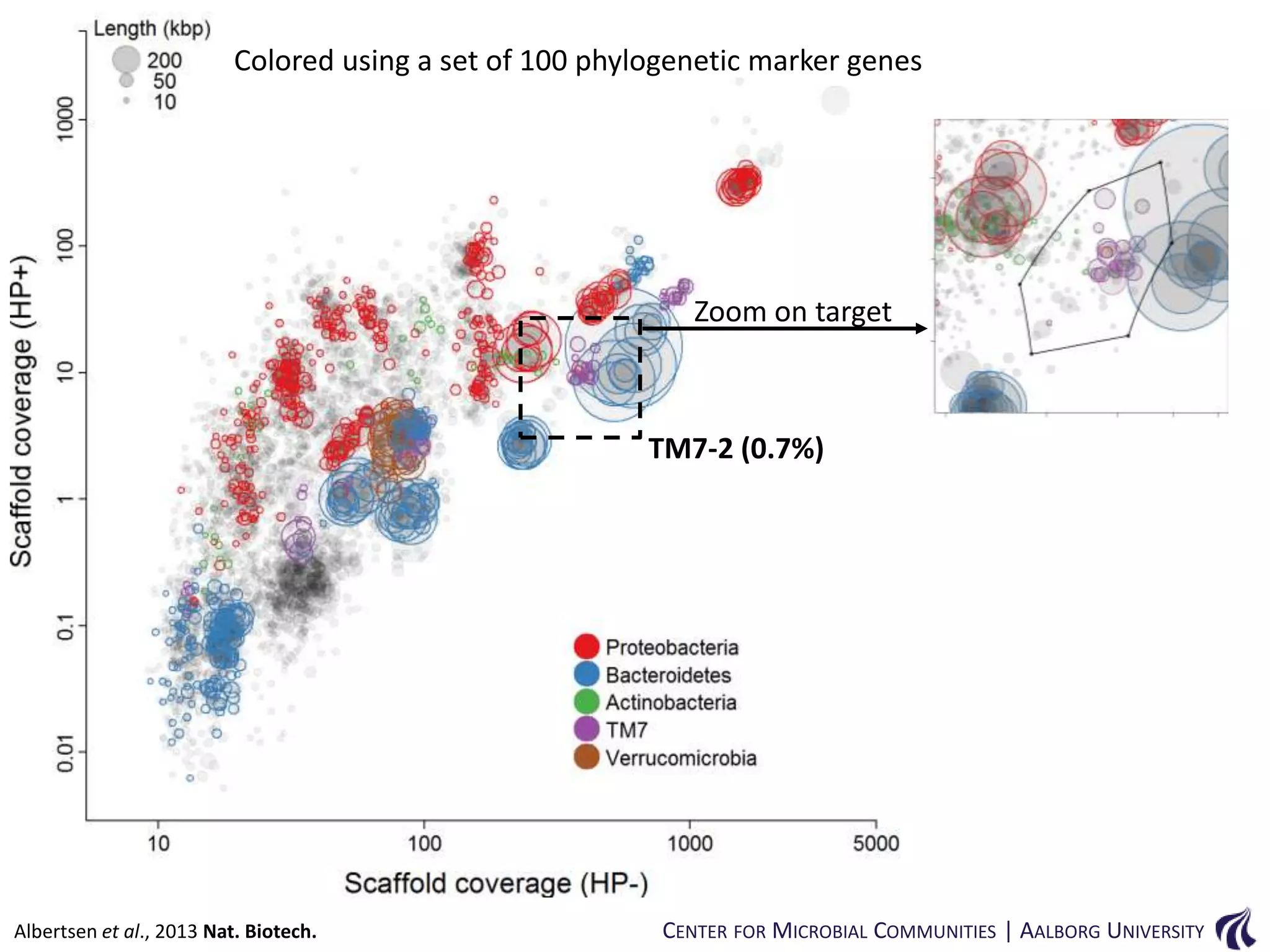
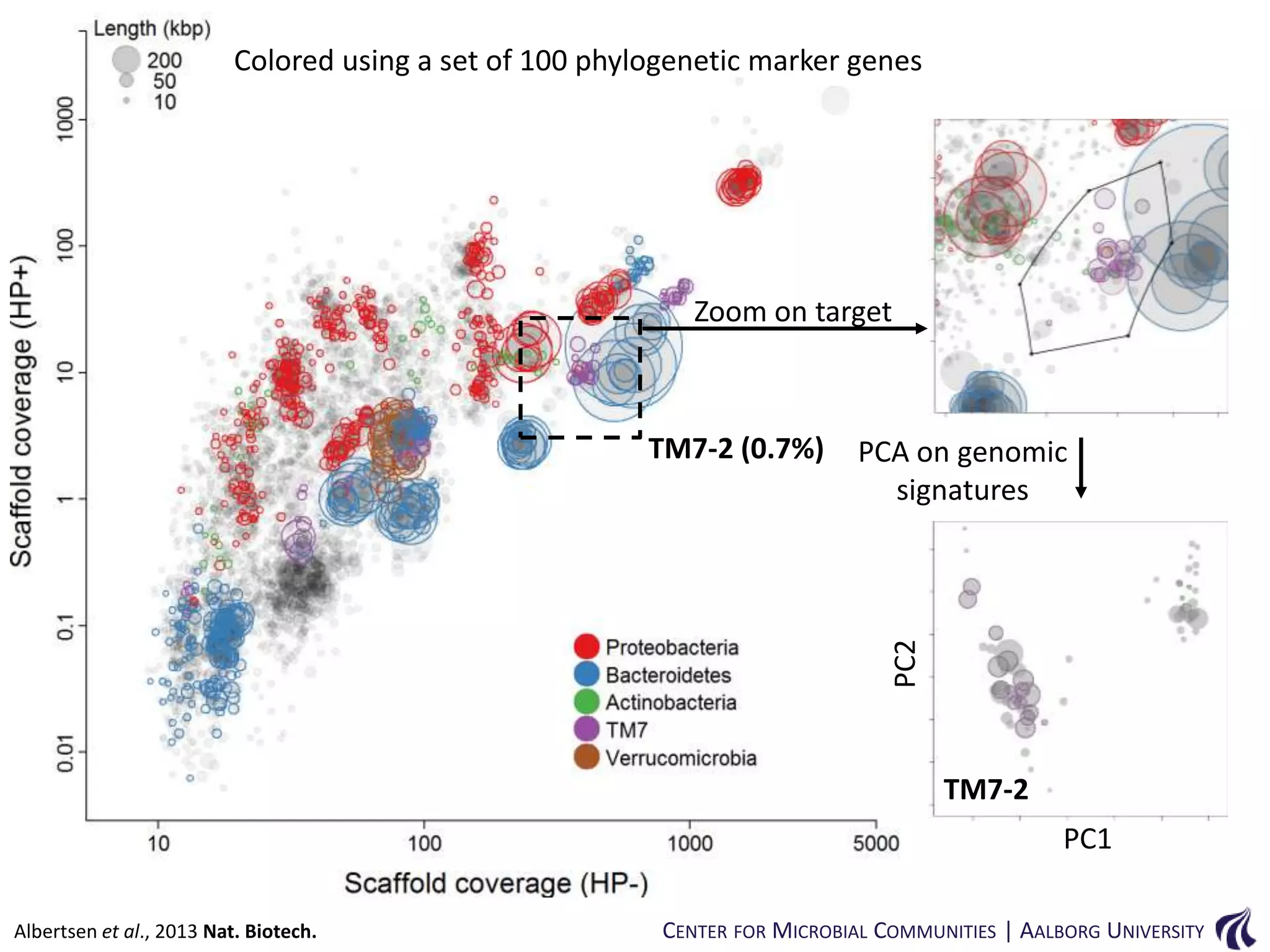
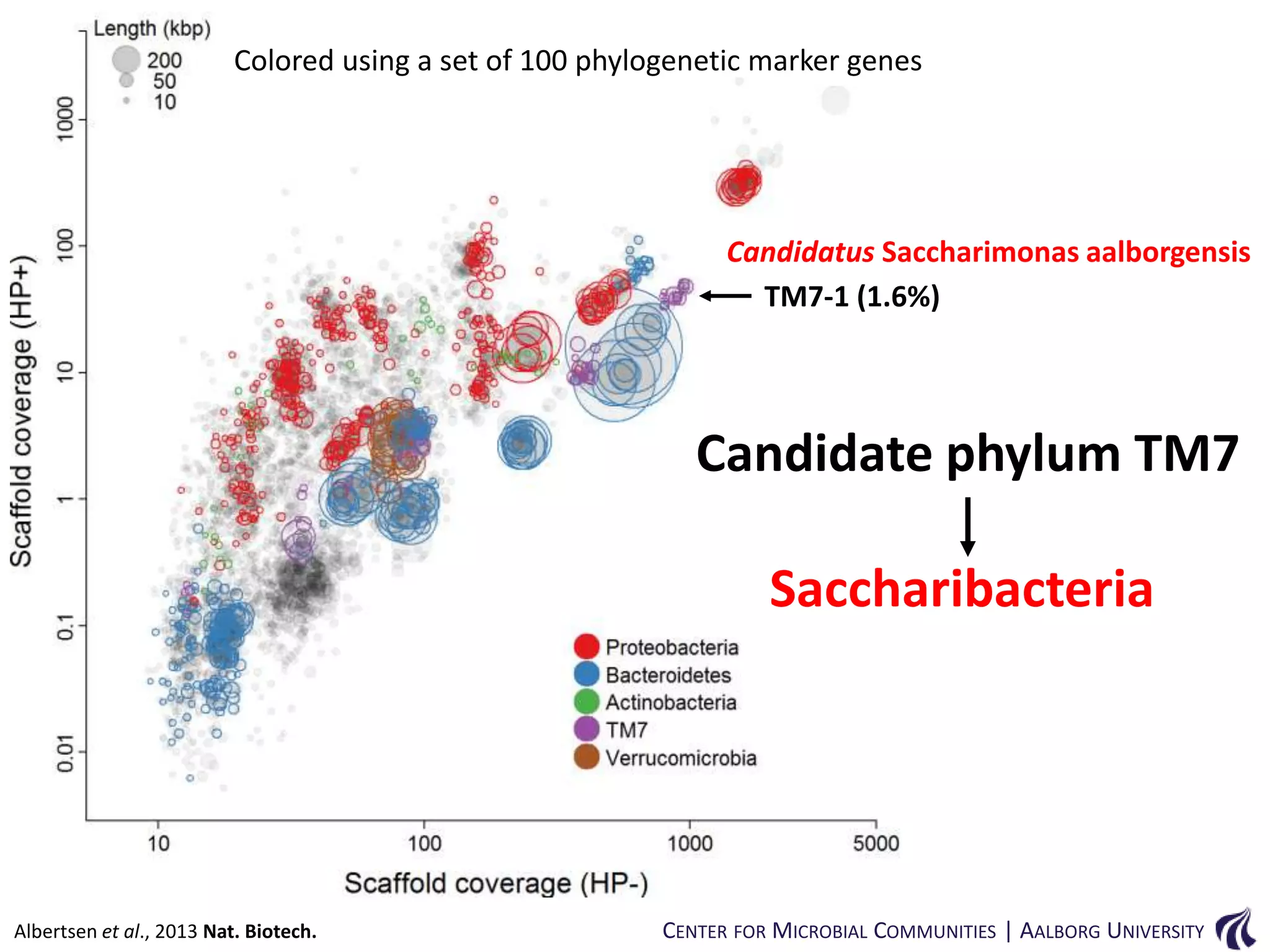
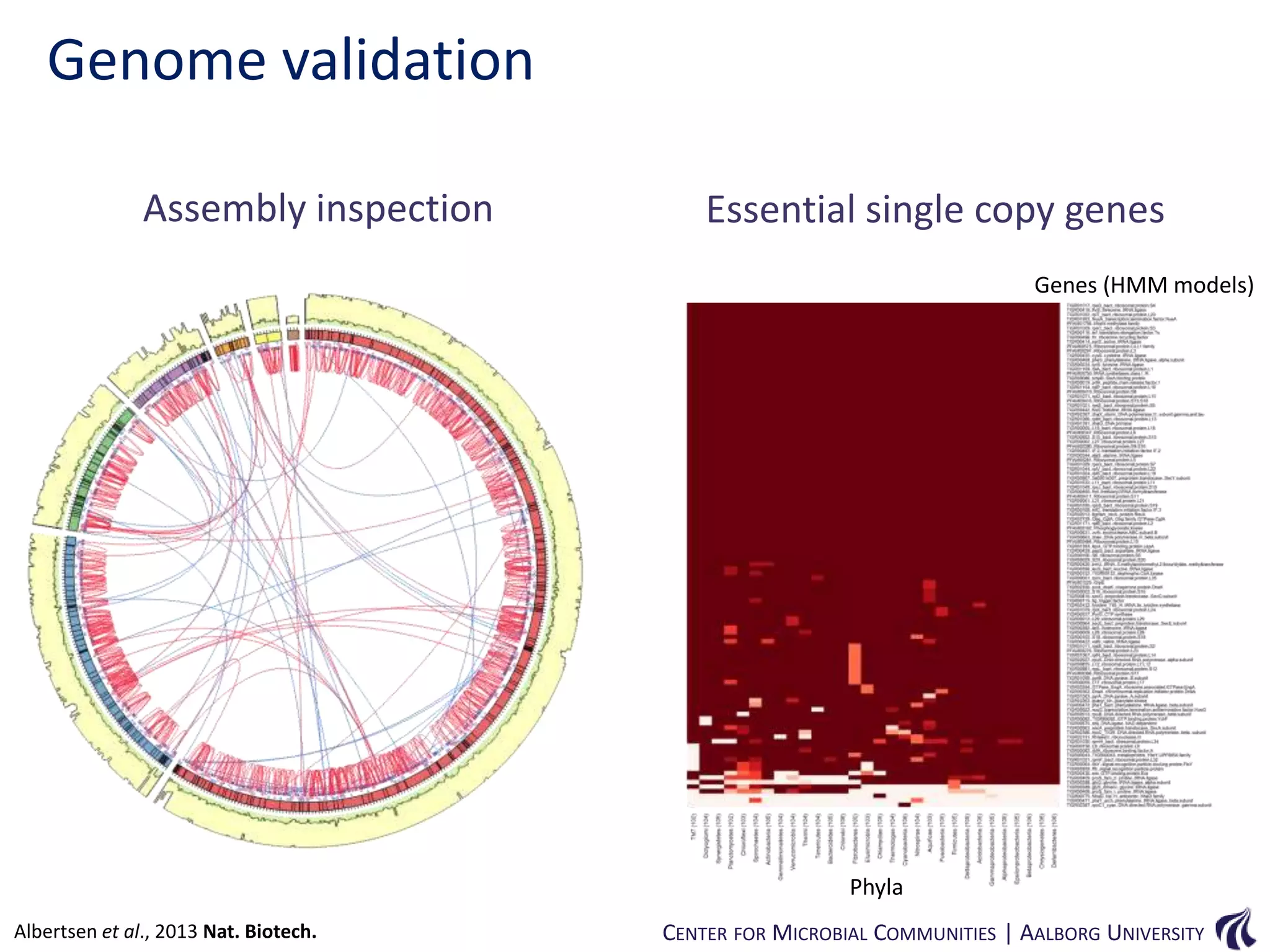
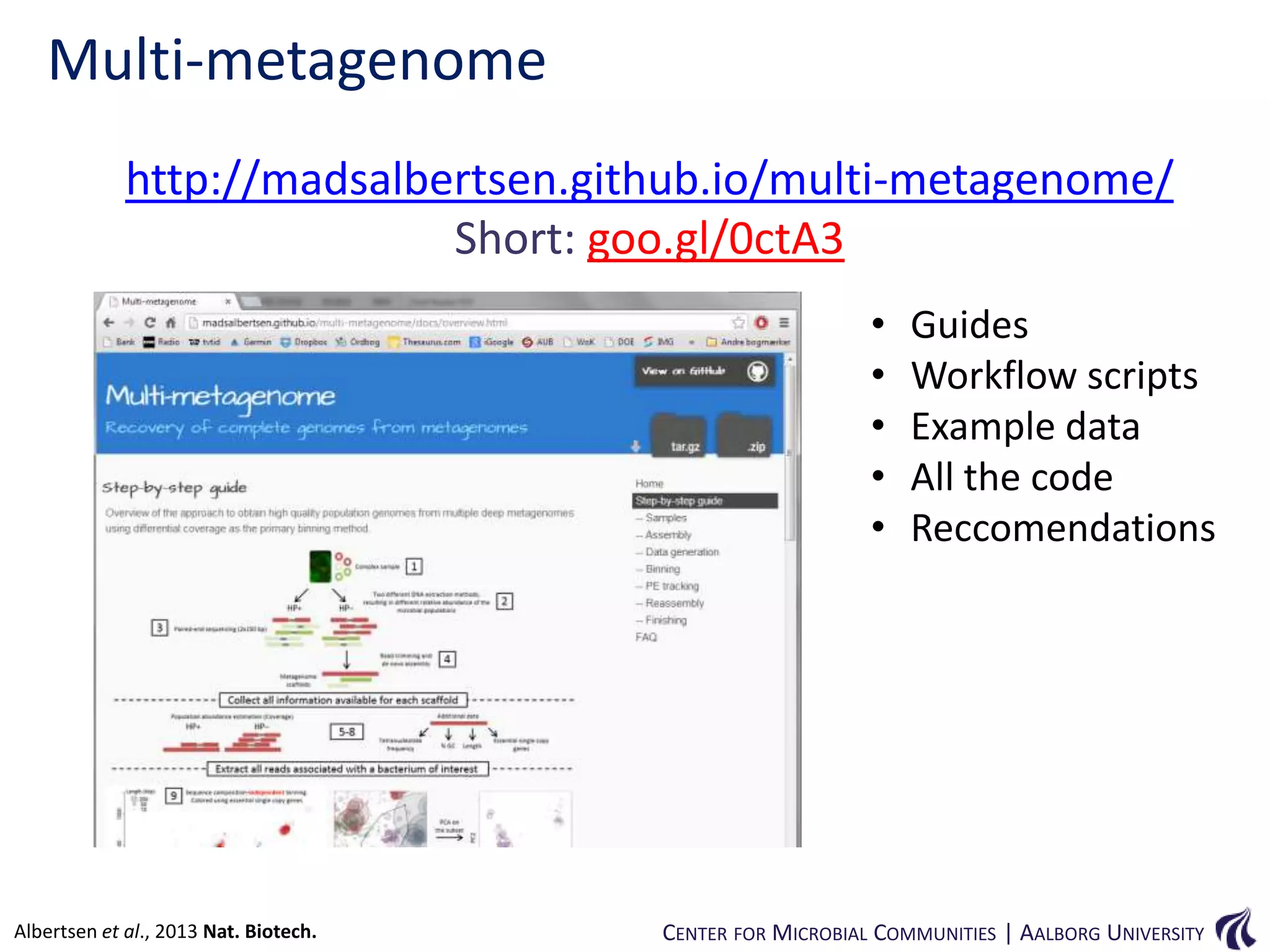
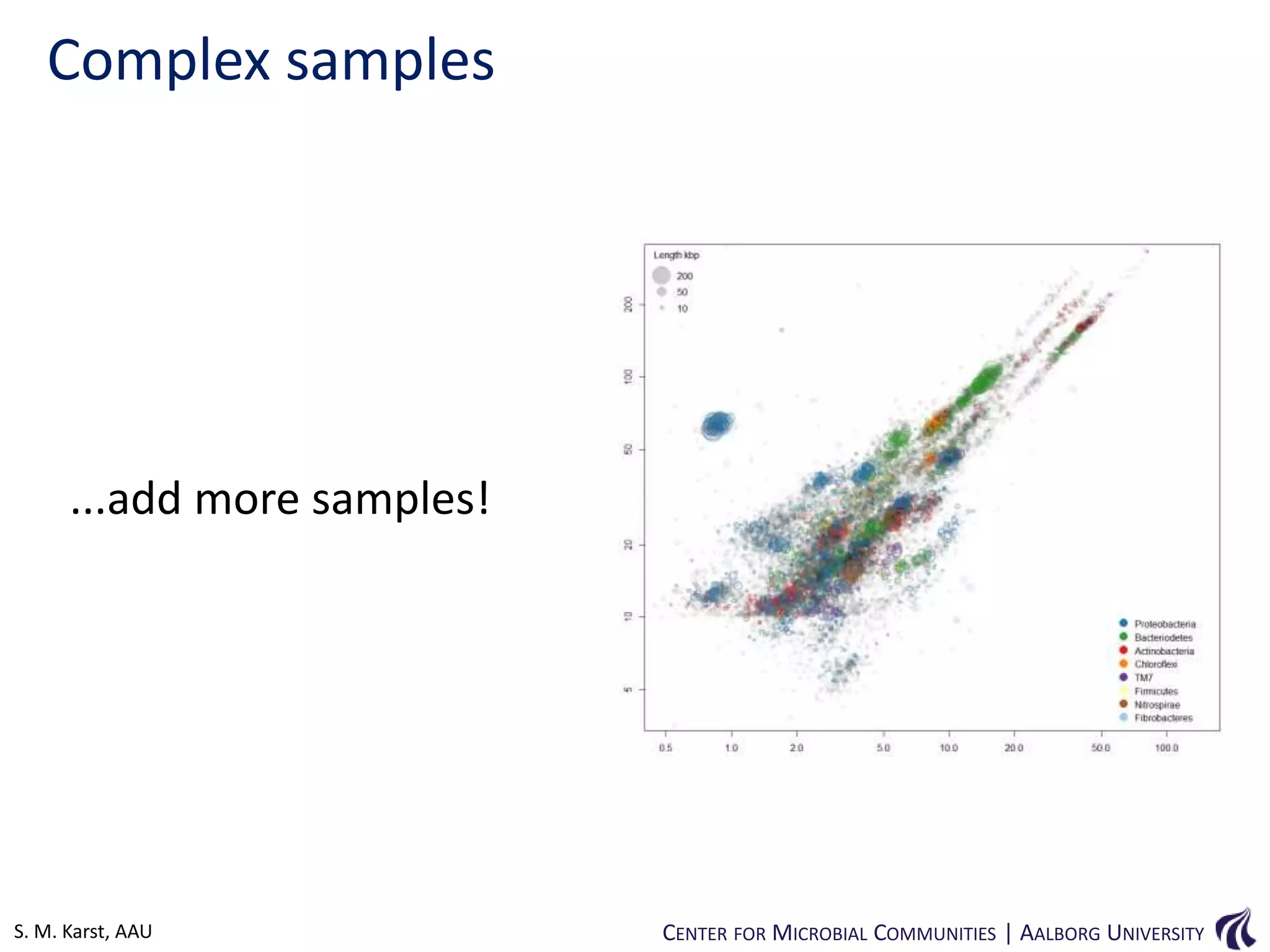
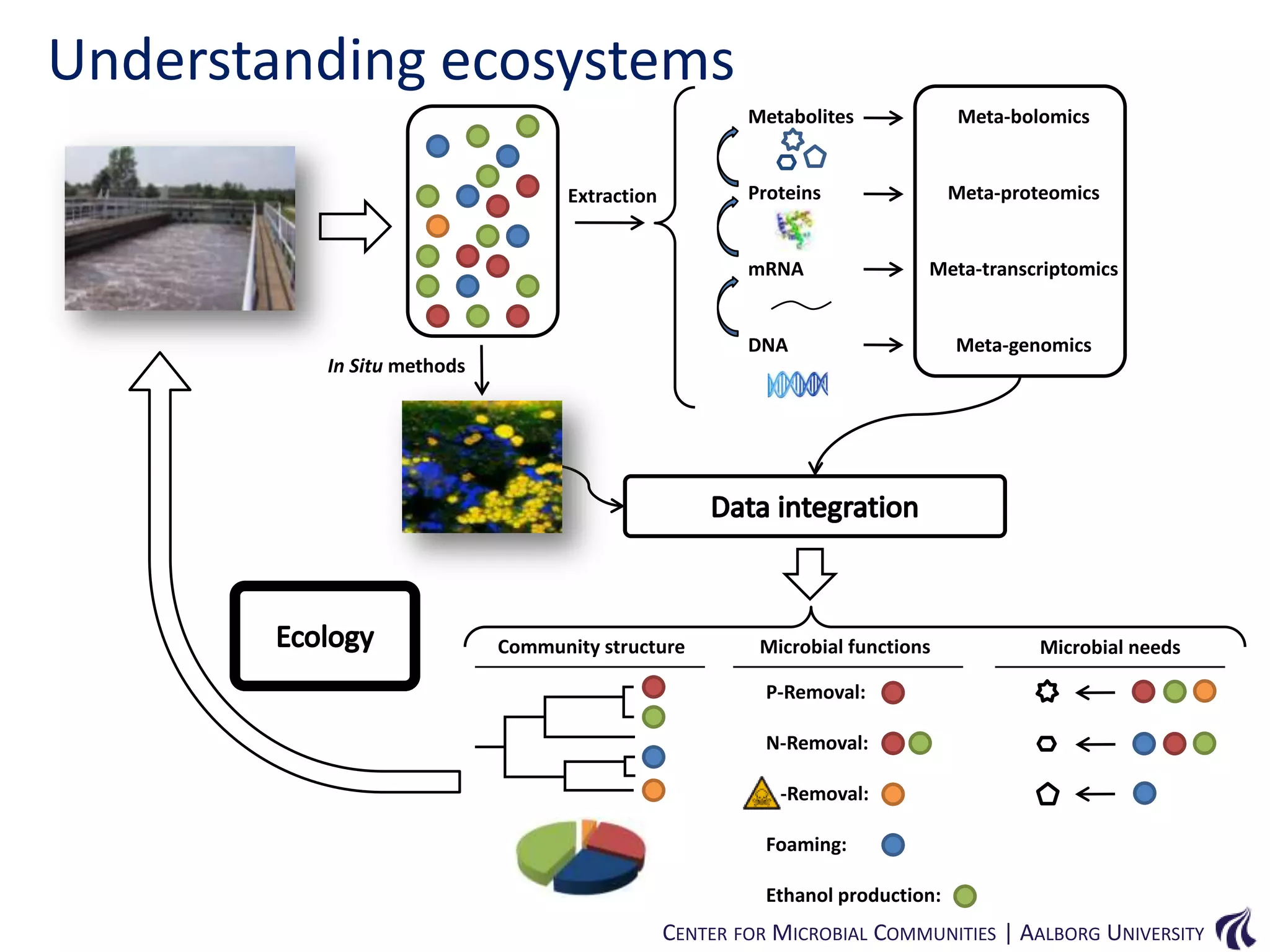
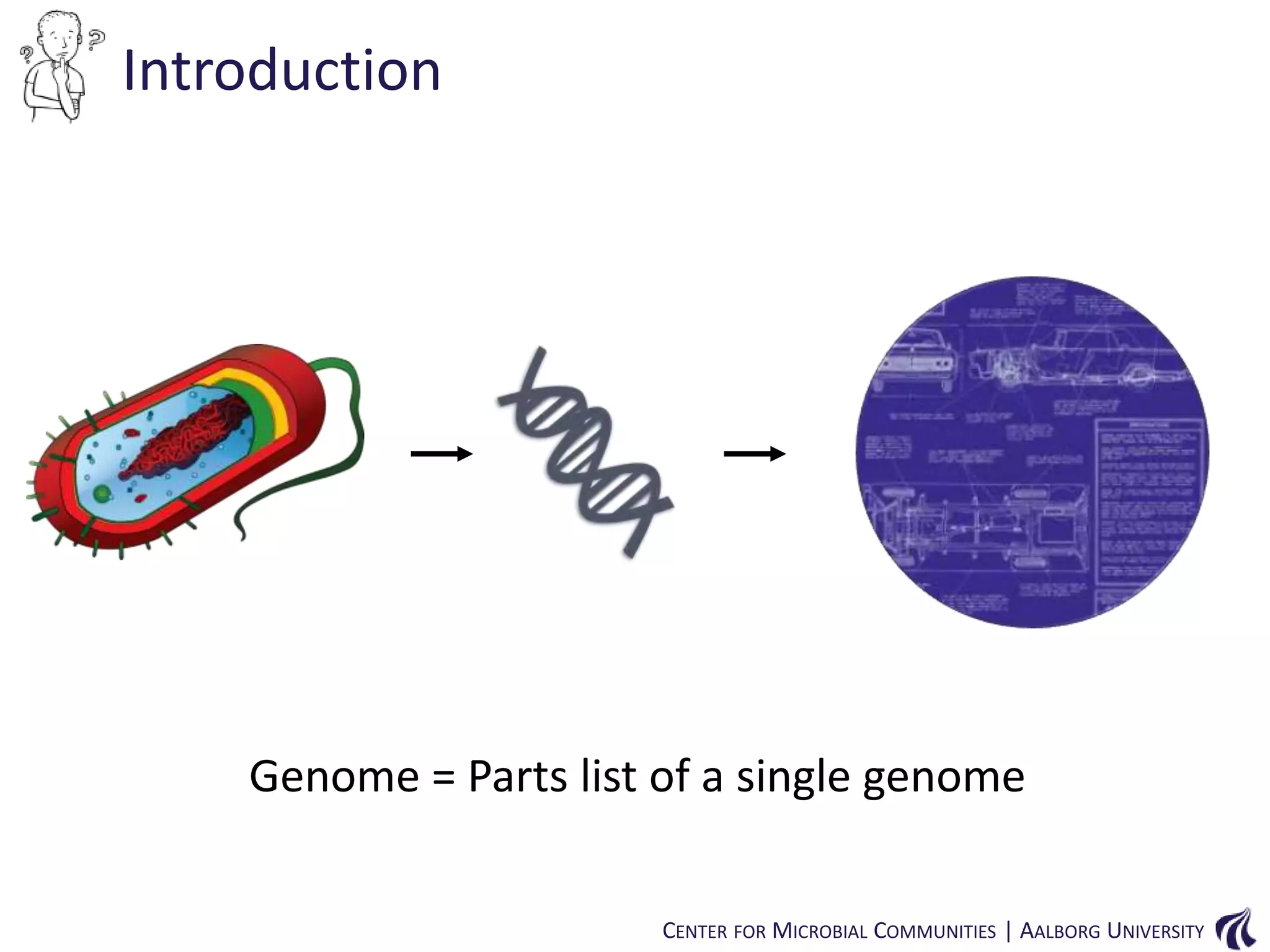
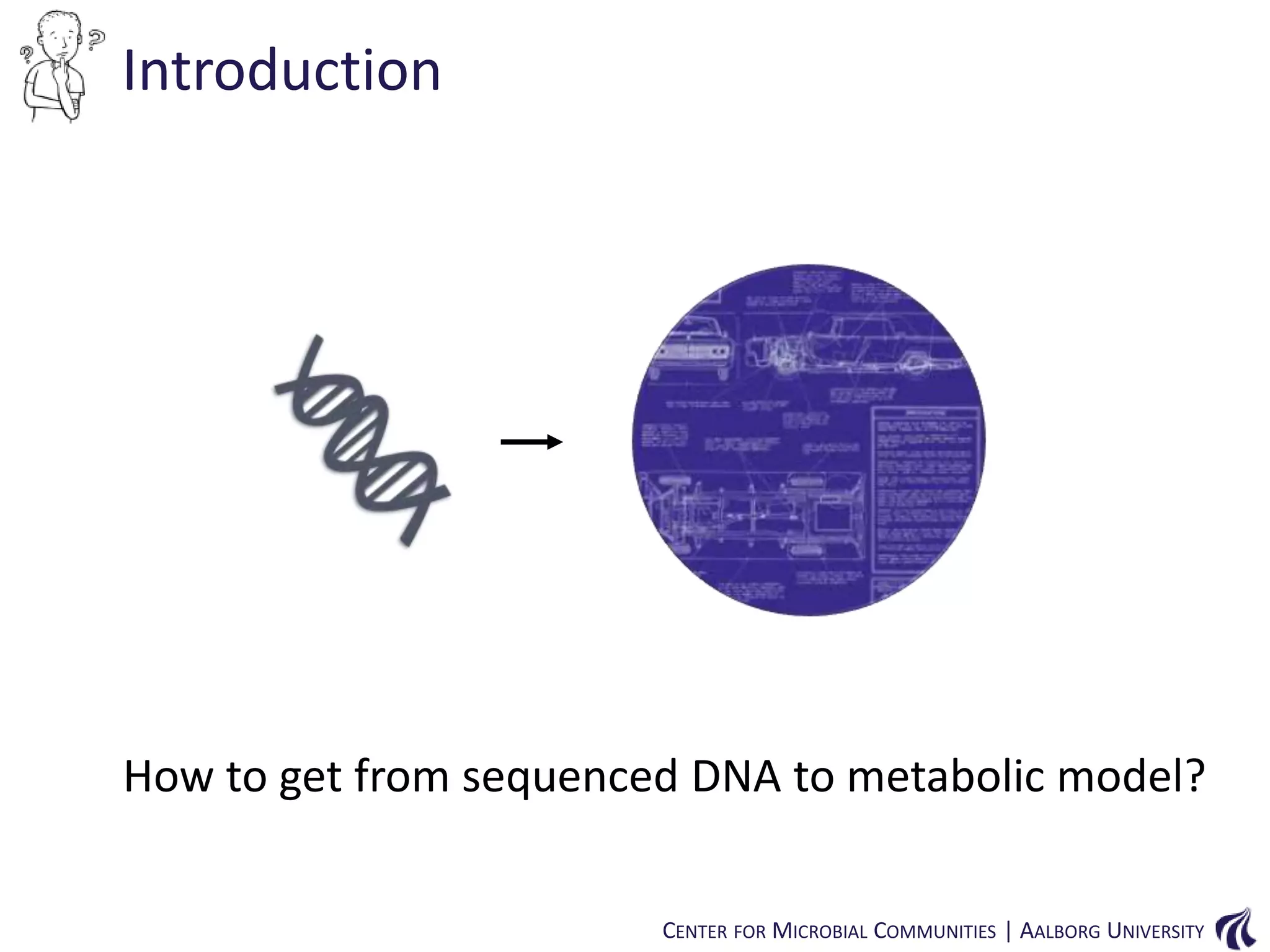
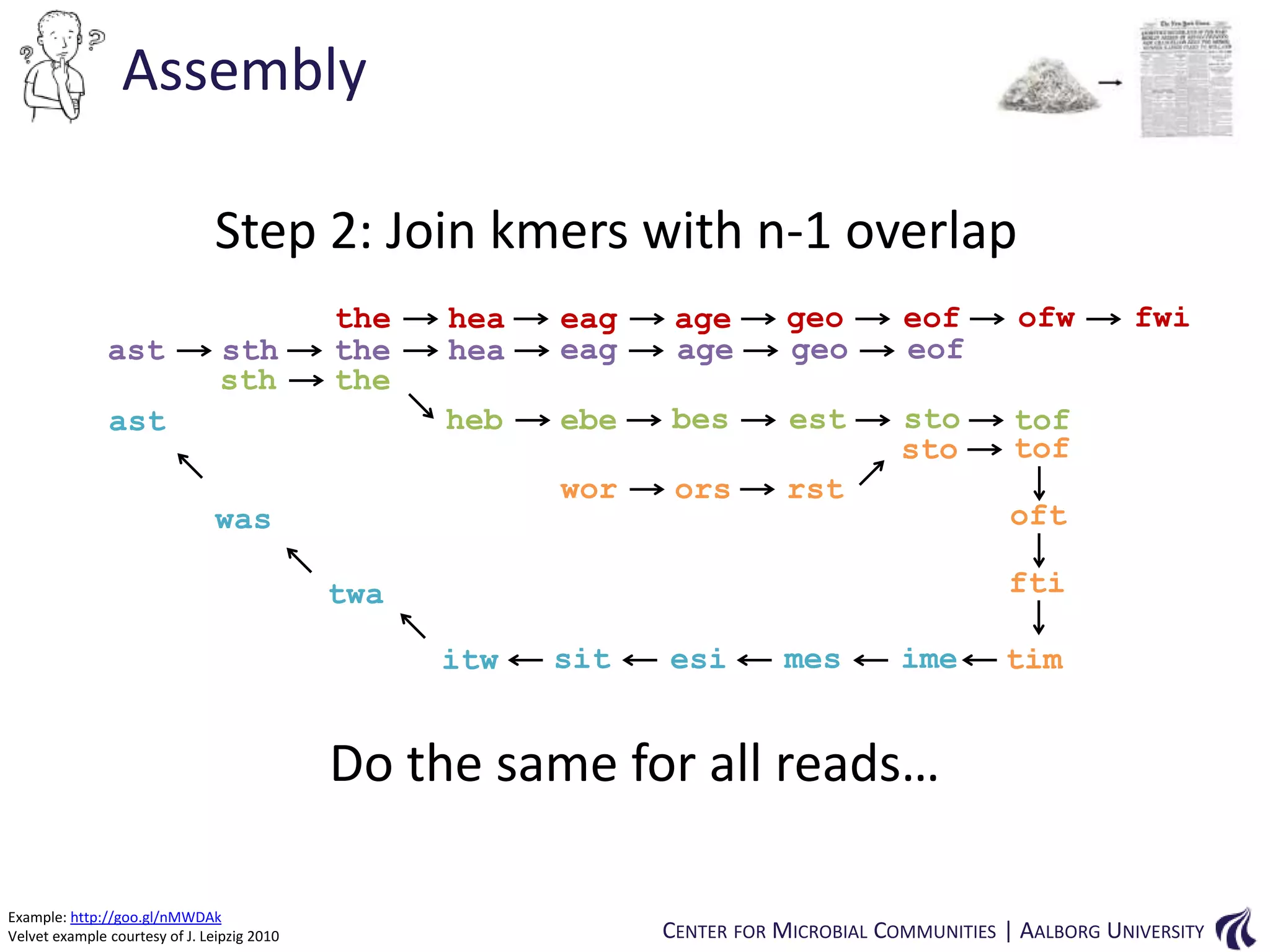
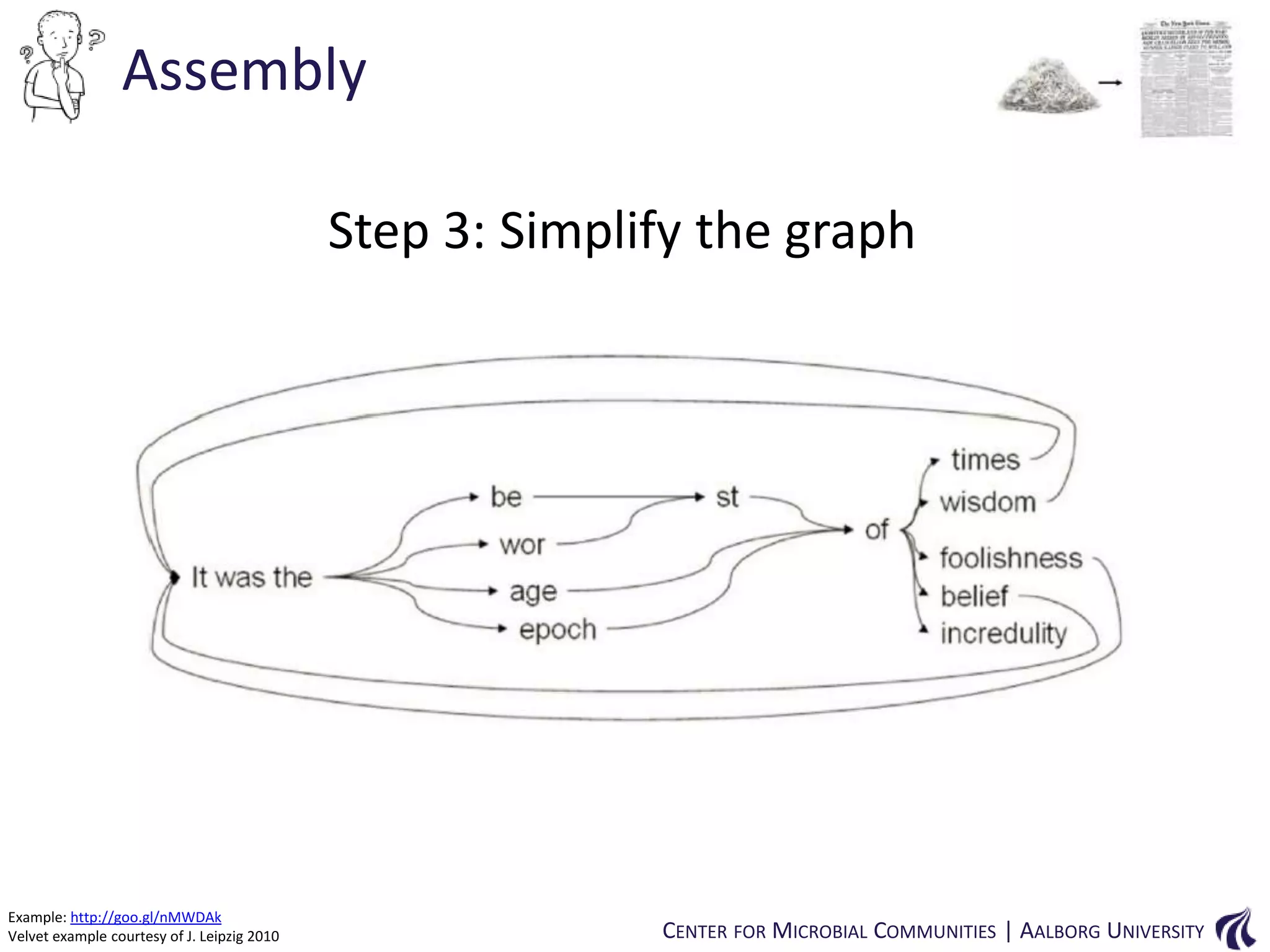
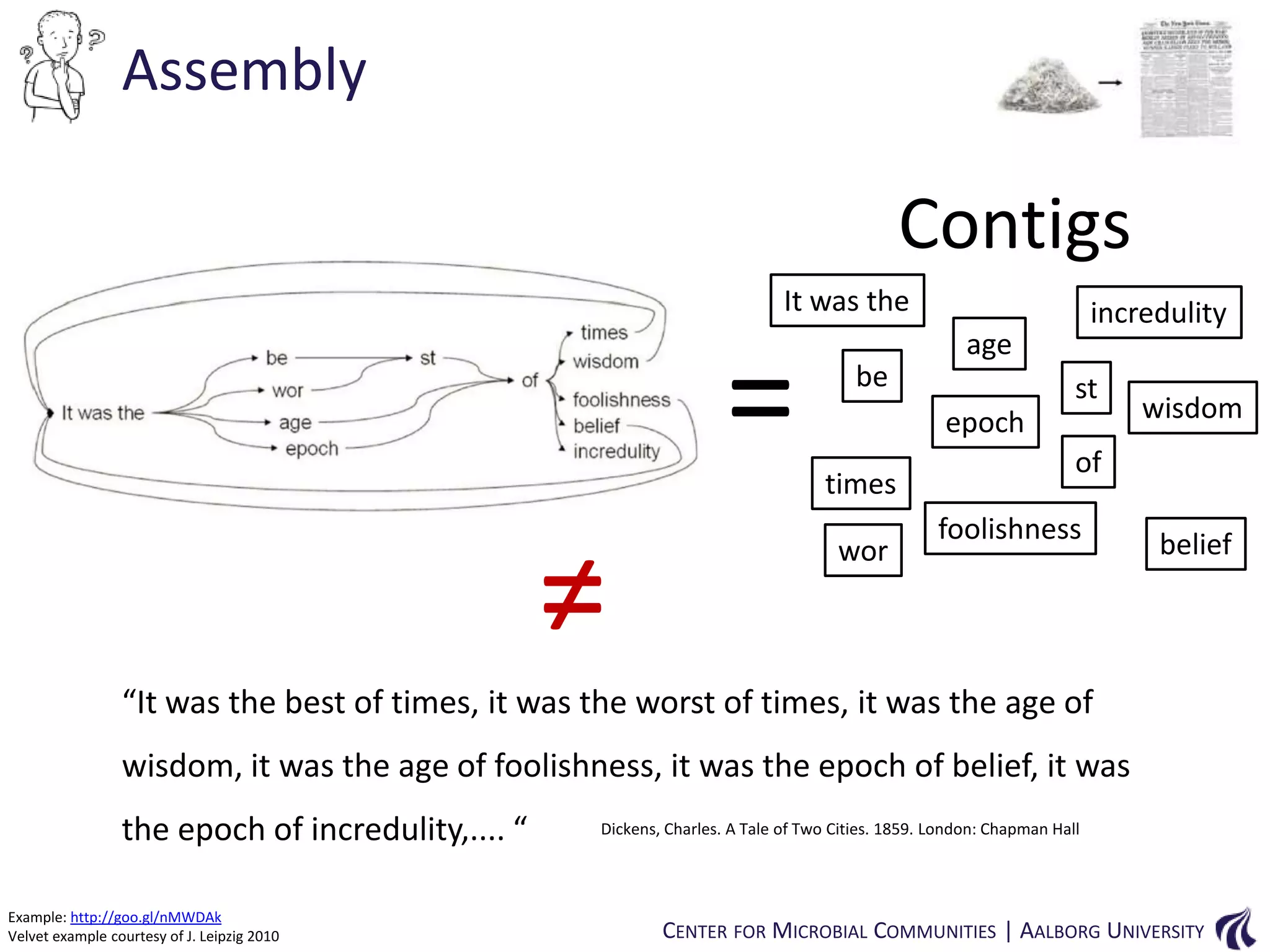
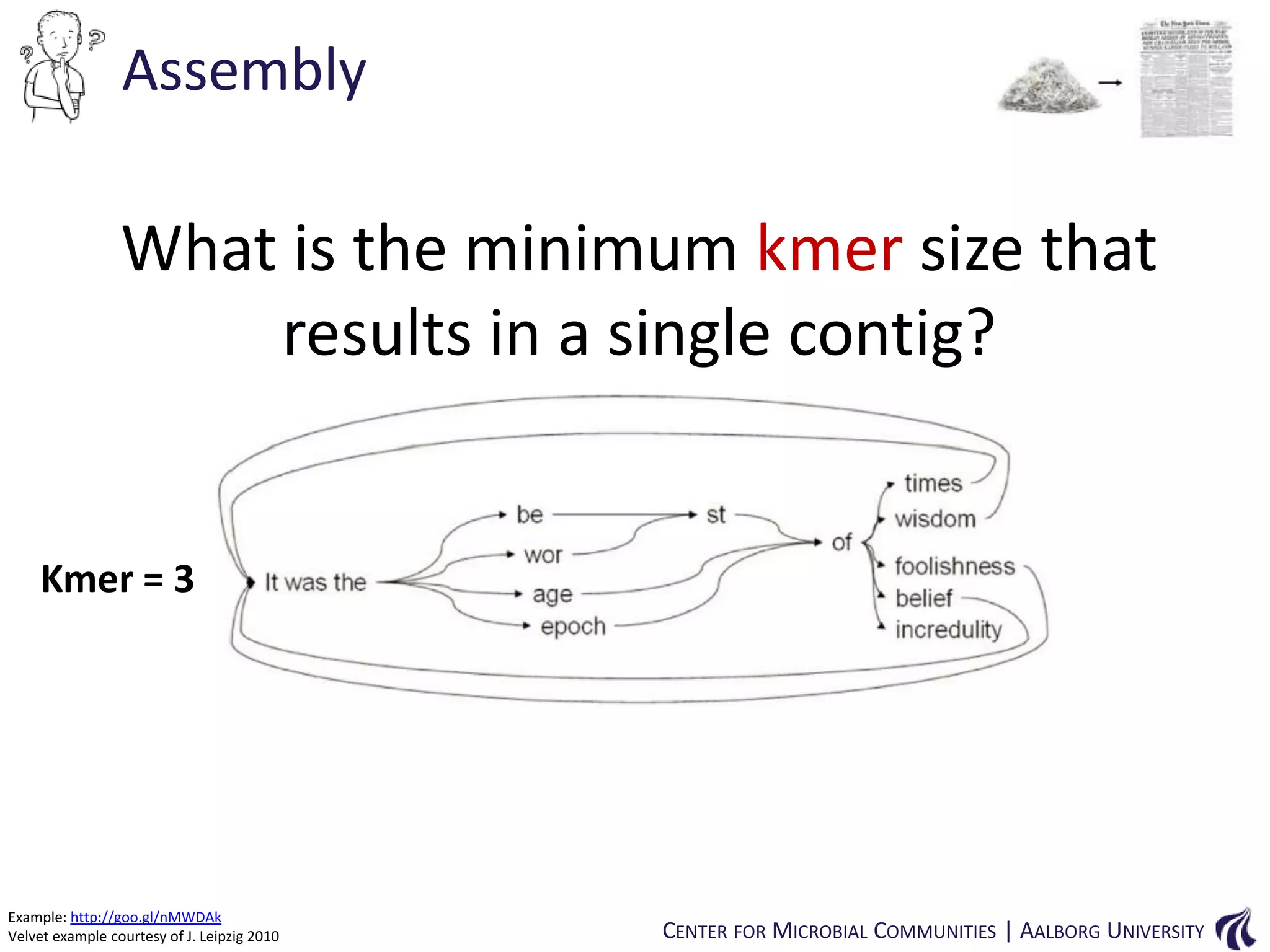
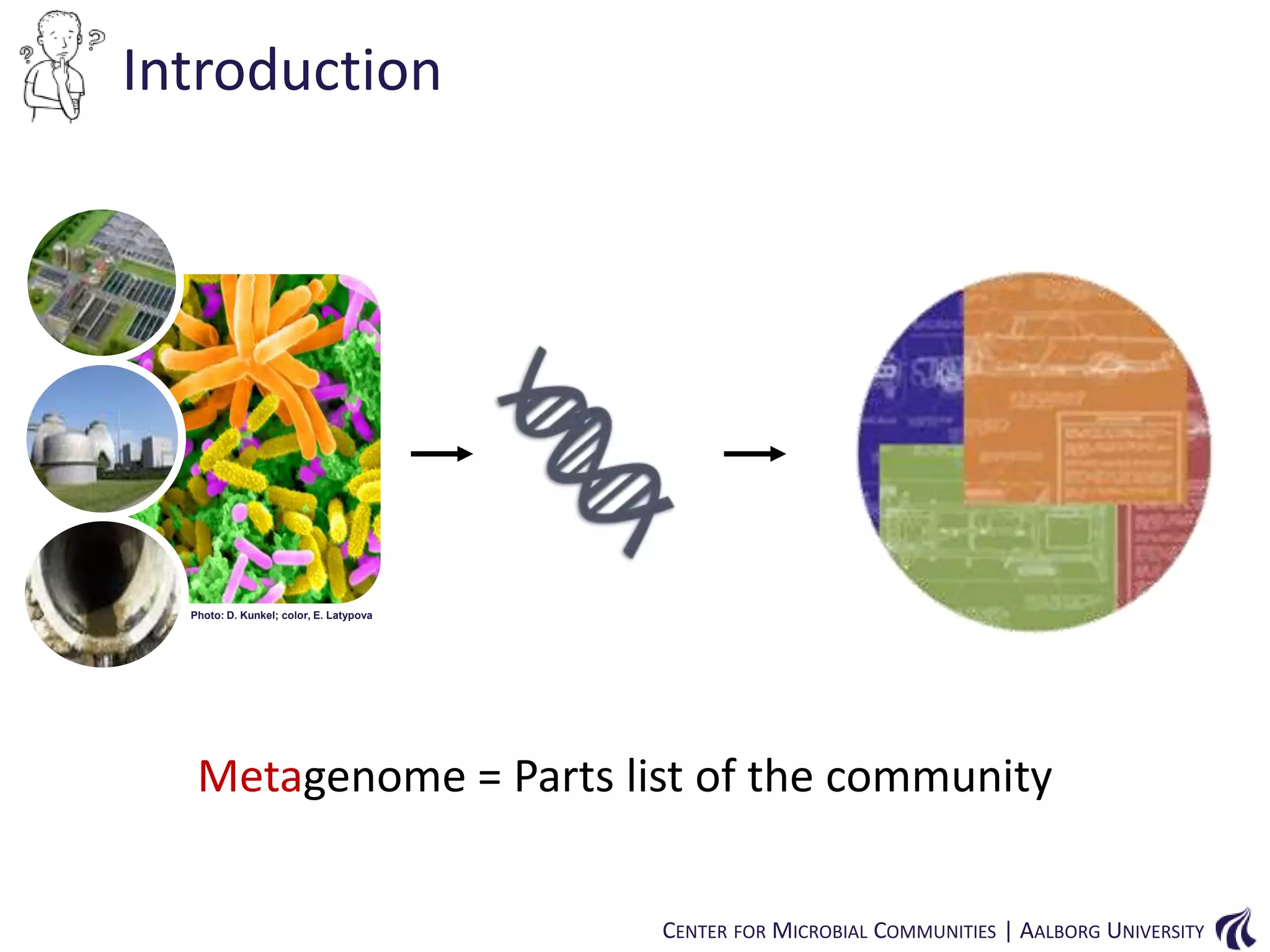
This document provides an overview of genomics and metagenomics. It begins with an introduction to genomics, describing genome assembly, validation, and metabolic reconstruction. It then covers metagenomics, discussing its history, pitfalls, and potentials. Key points include that genomics analyzes the parts list of a single genome, while metagenomics analyzes the collective genomes of an entire microbial community. Metagenomics has been used to explore novel sequences from various environments, perform comparative analyses between ecosystems, and extract genomes from low-abundance species.

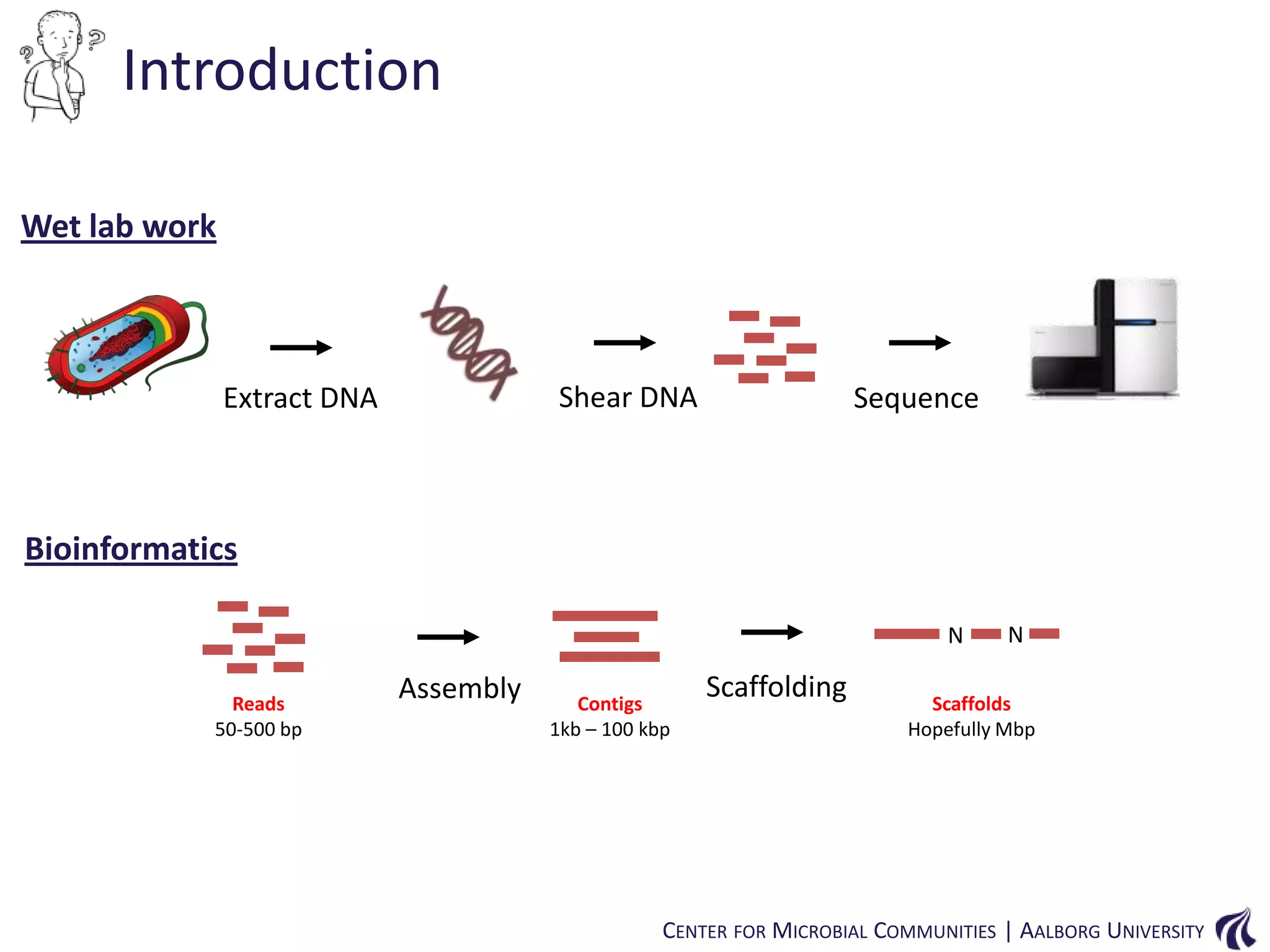
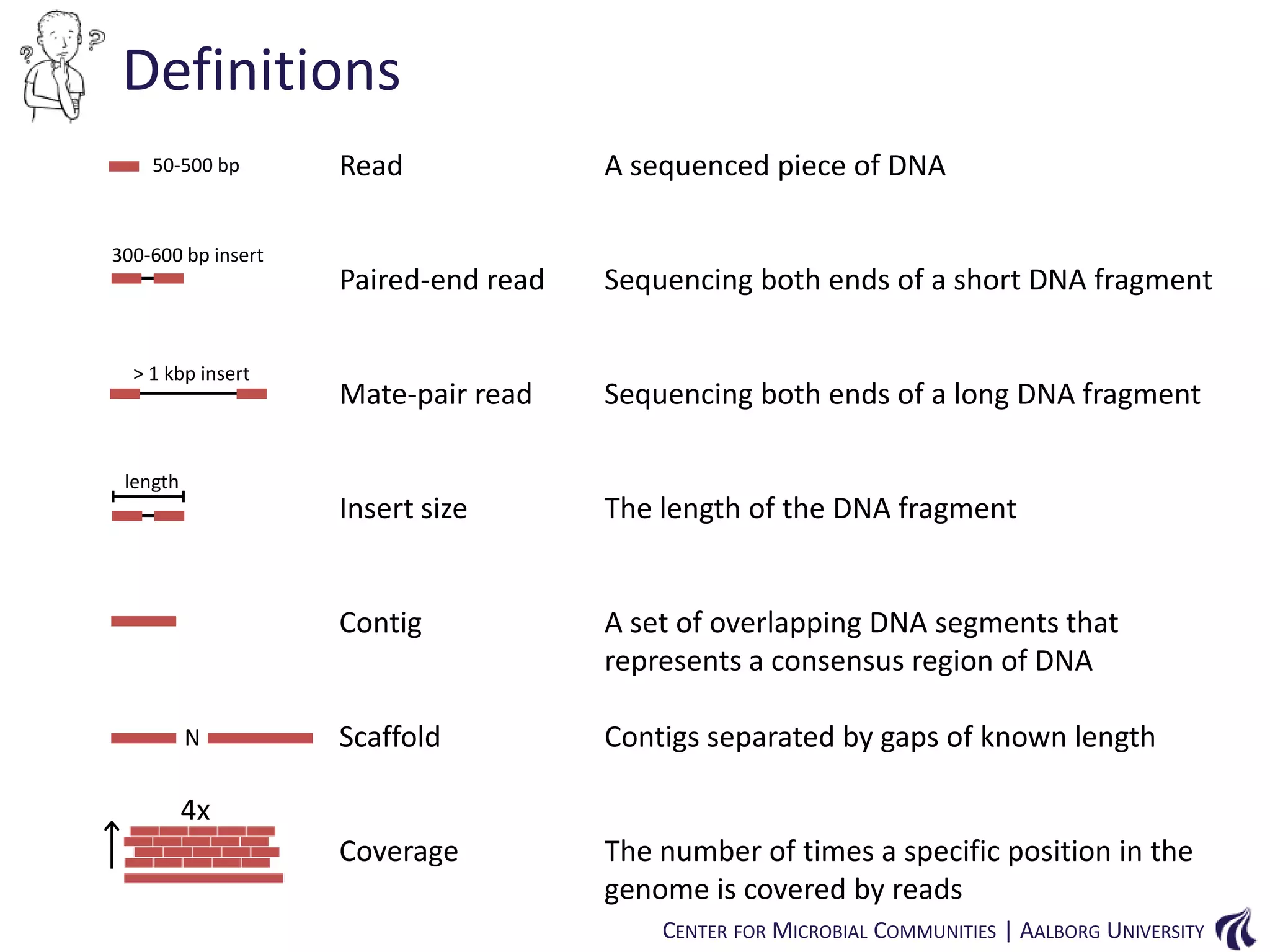
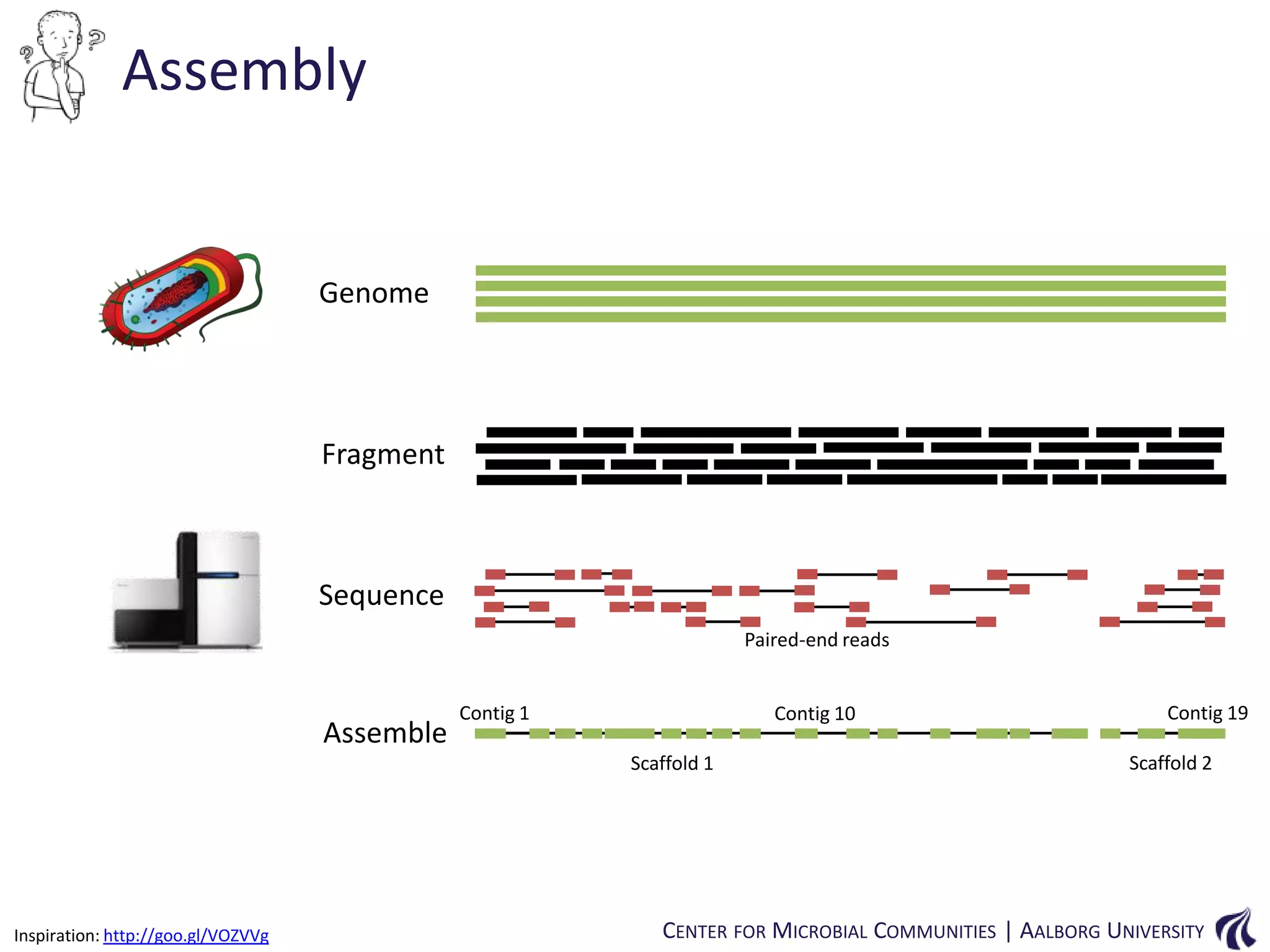



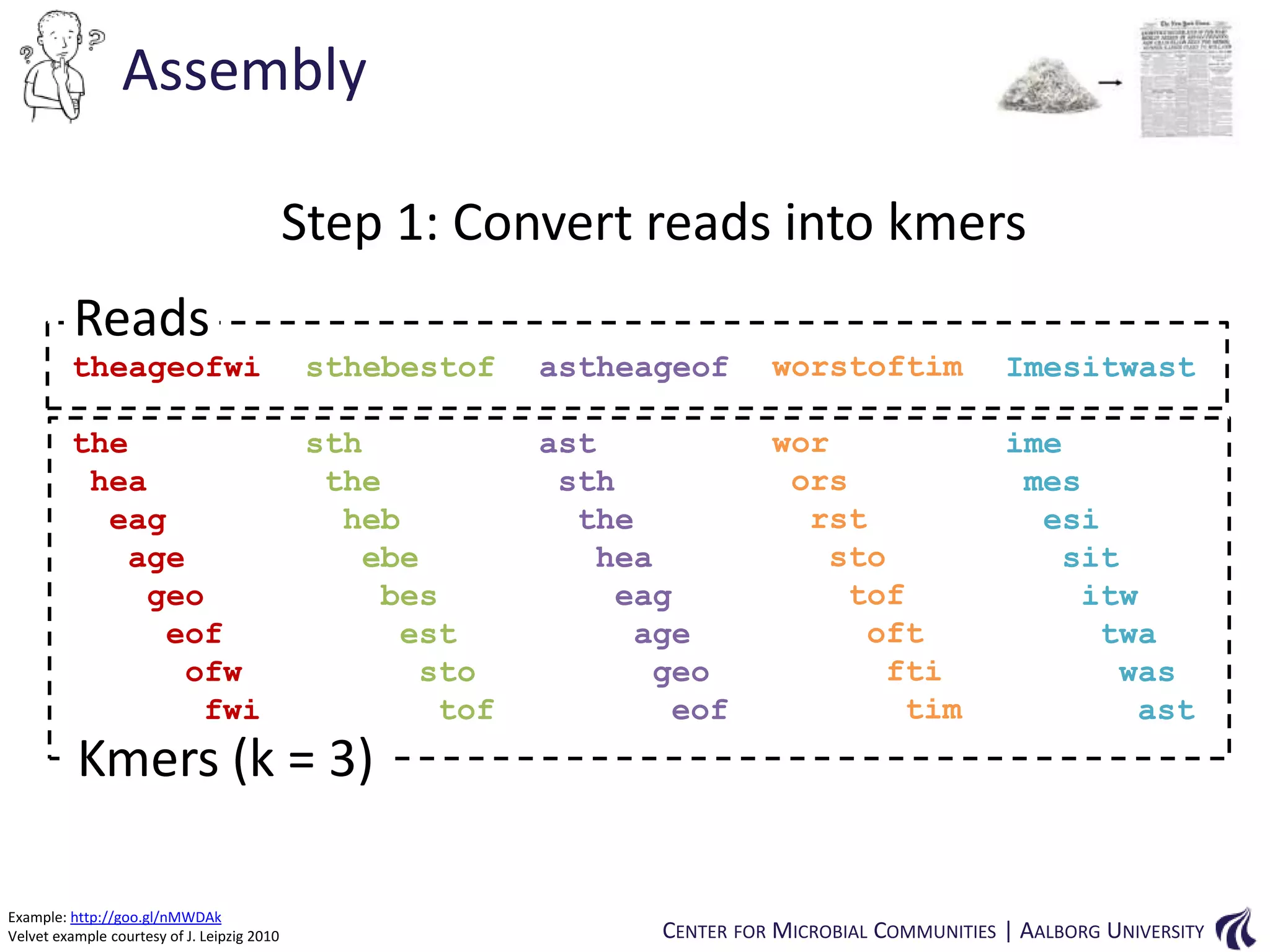






![Introduction
”...functional analysis of the collective genomes of soil
microflora, which we term the metagenome of the soil.”
- J. Handelsman et al., 1998
PubMed: metagenom*[Title/Abstract]
CENTER FOR MICROBIAL COMMUNITIES | AALBORG UNIVERSITY](https://image.slidesharecdn.com/2013-131029070115-phpapp01/75/2013-10-29-albertsen-genomics-metagenomics-35-2048.jpg)
![Introduction
”...functional analysis of the collective genomes of soil
microflora, which we term the metagenome of the soil.”
- J. Handelsman et al., 1998
Sequencing costs
PubMed: metagenom*[Title/Abstract]
http://www.genome.gov/sequencingcosts/
CENTER FOR MICROBIAL COMMUNITIES | AALBORG UNIVERSITY](https://image.slidesharecdn.com/2013-131029070115-phpapp01/75/2013-10-29-albertsen-genomics-metagenomics-36-2048.jpg)