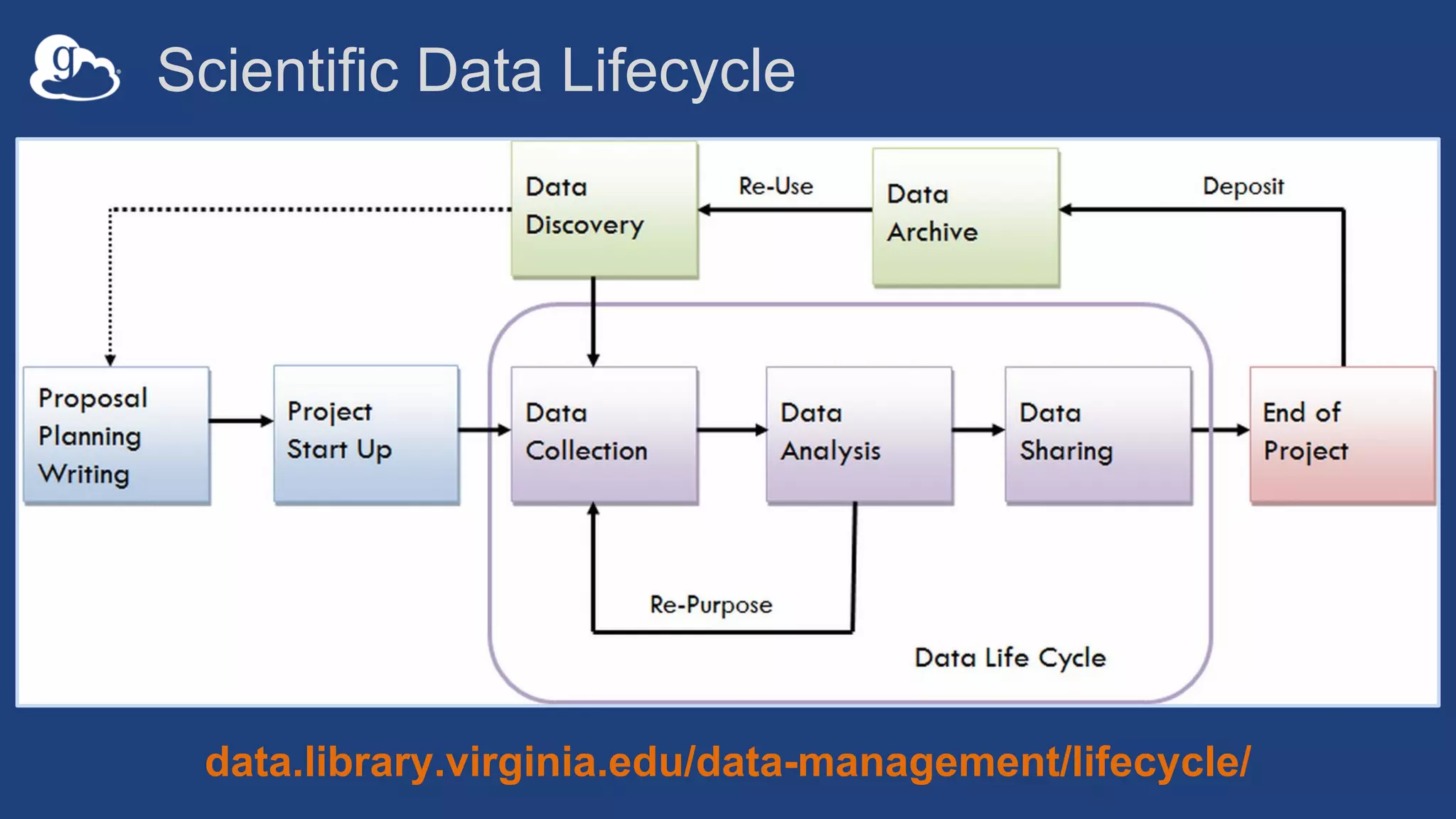
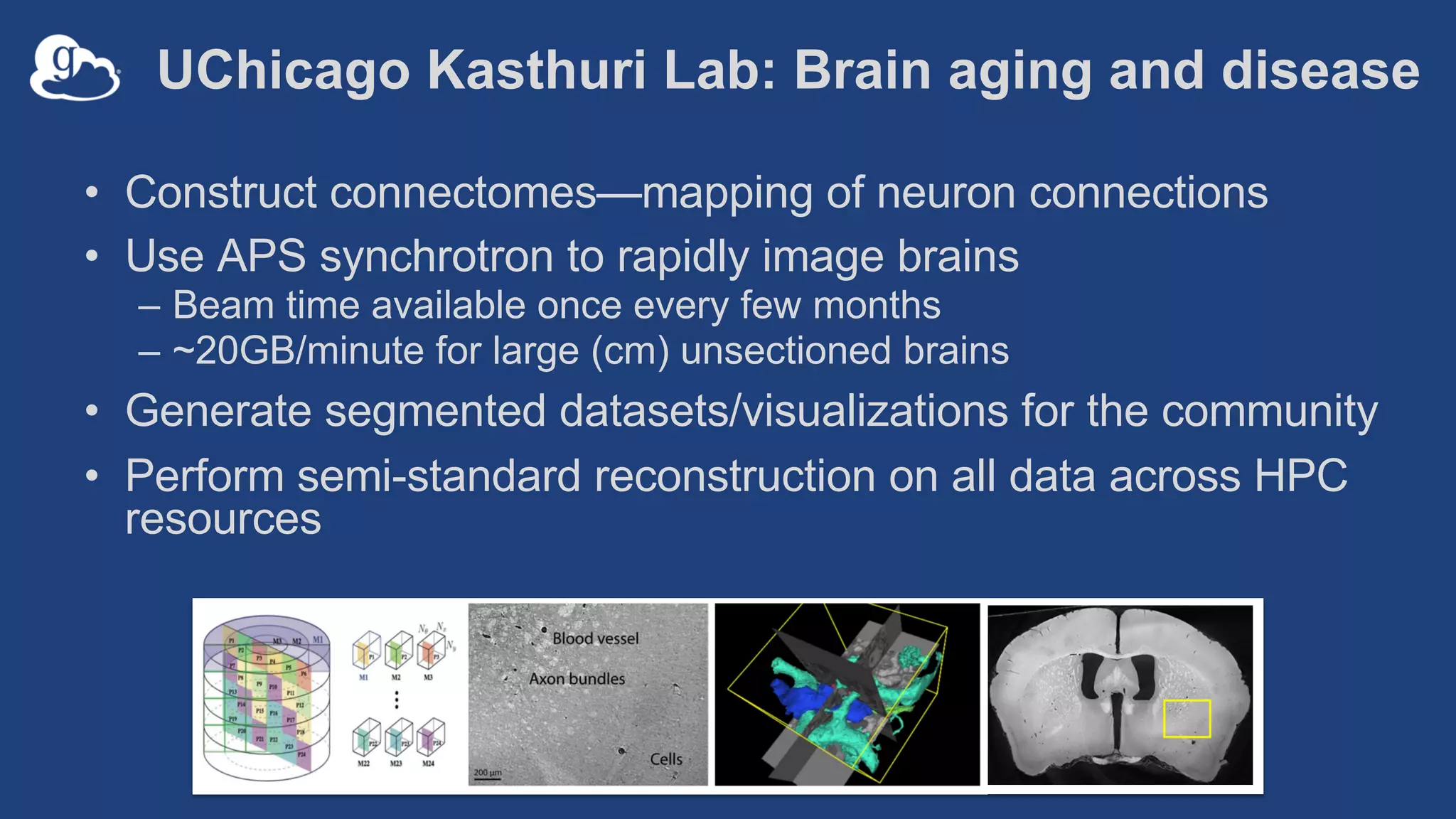
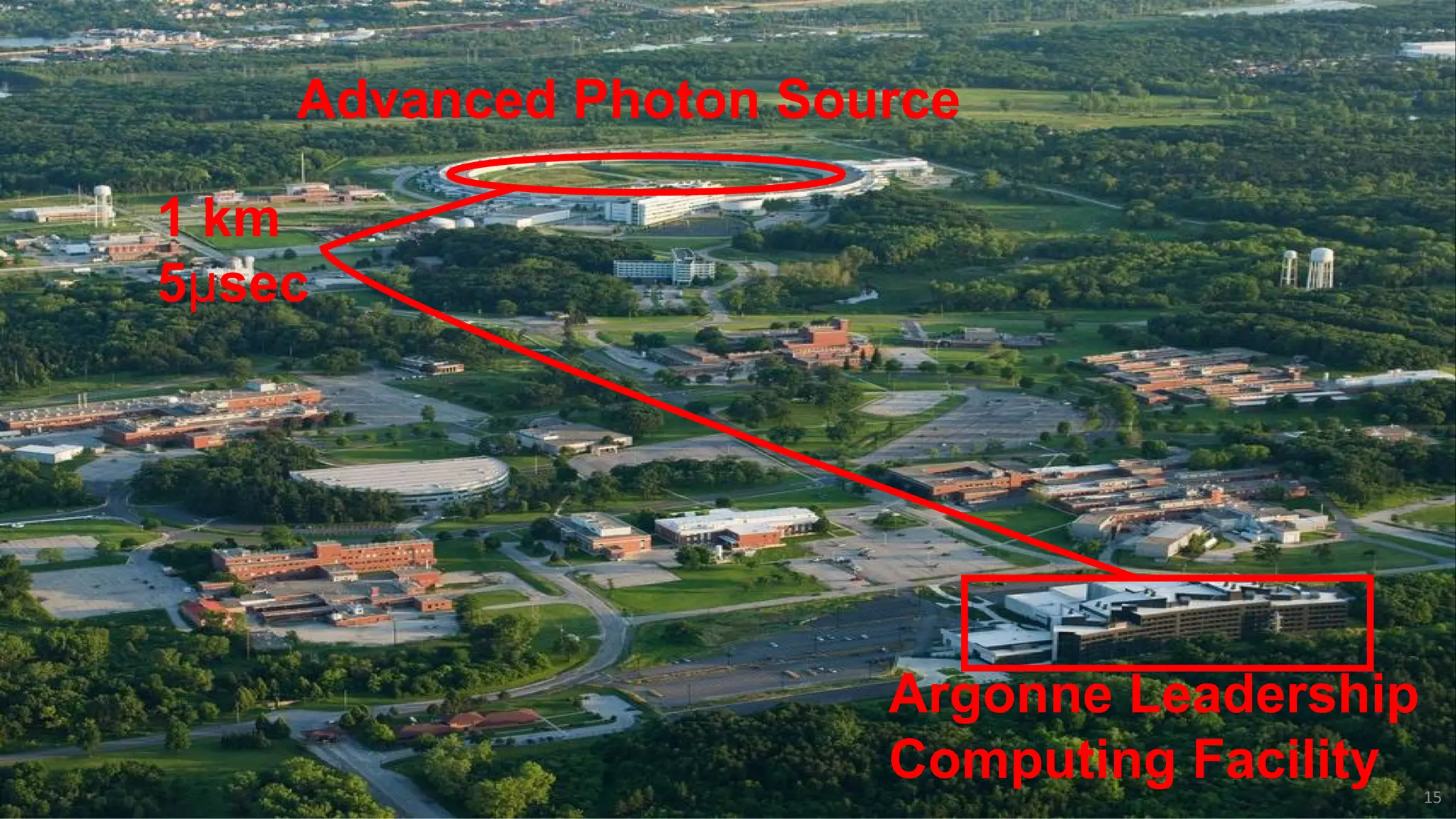
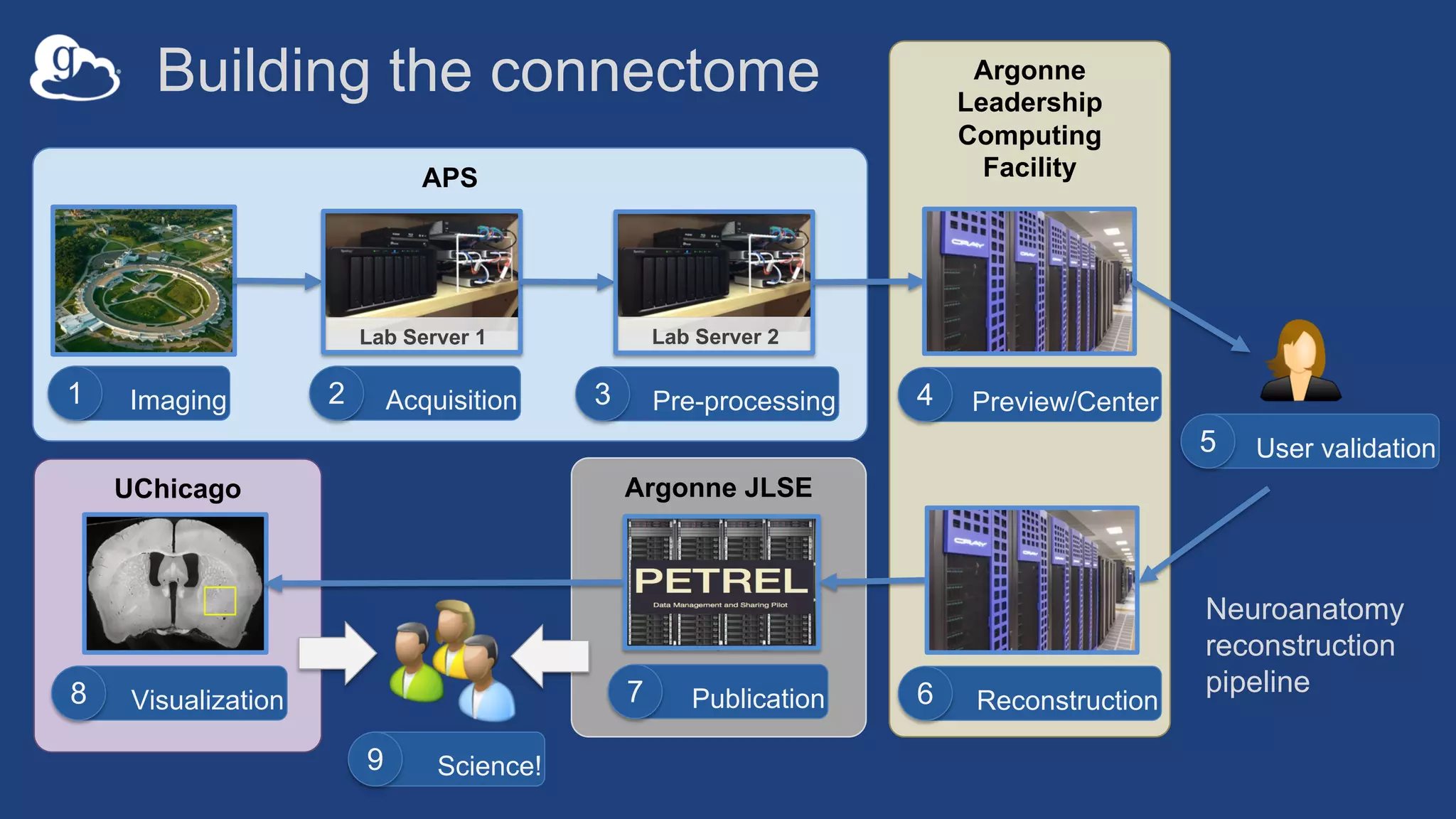
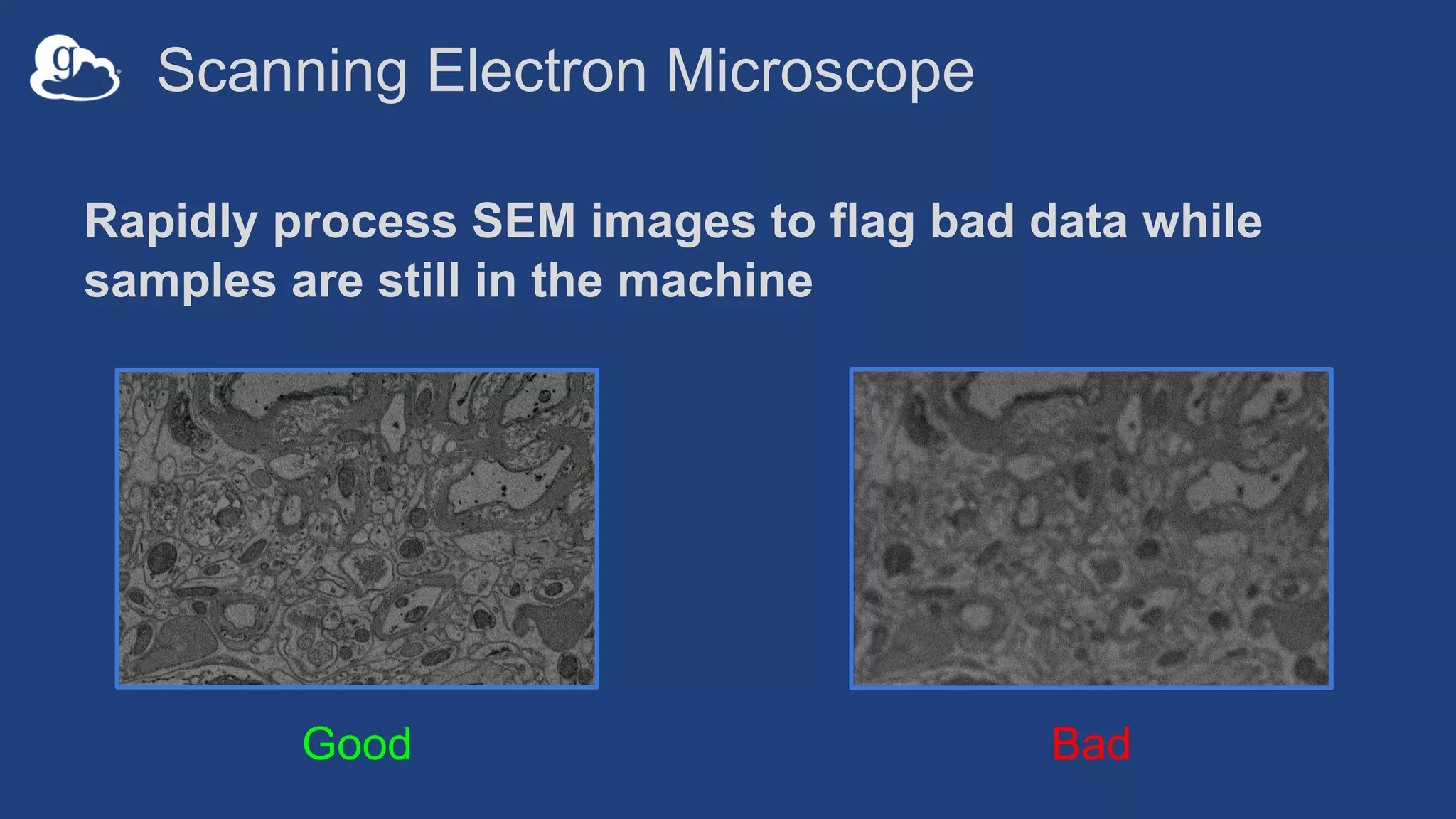
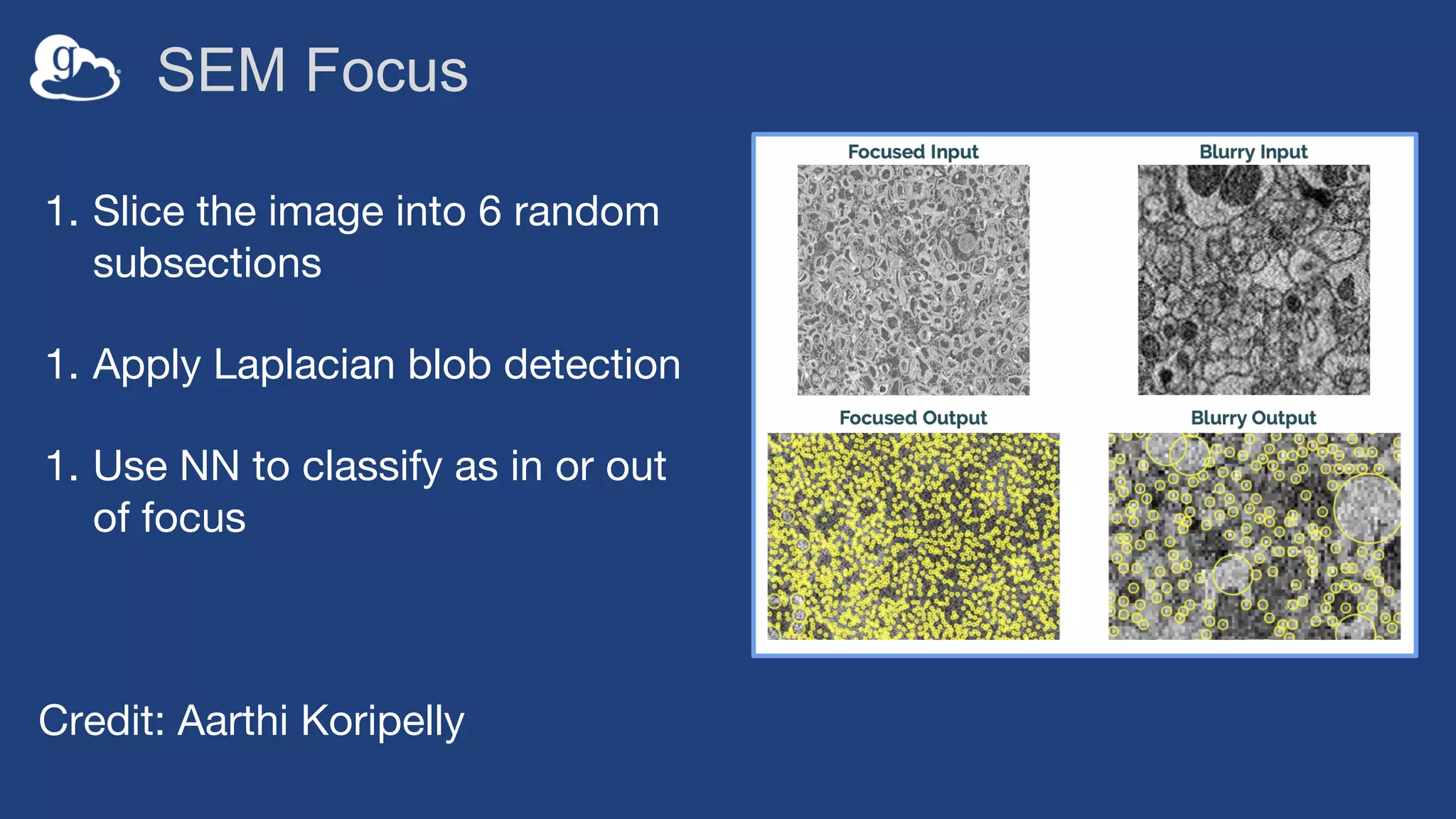
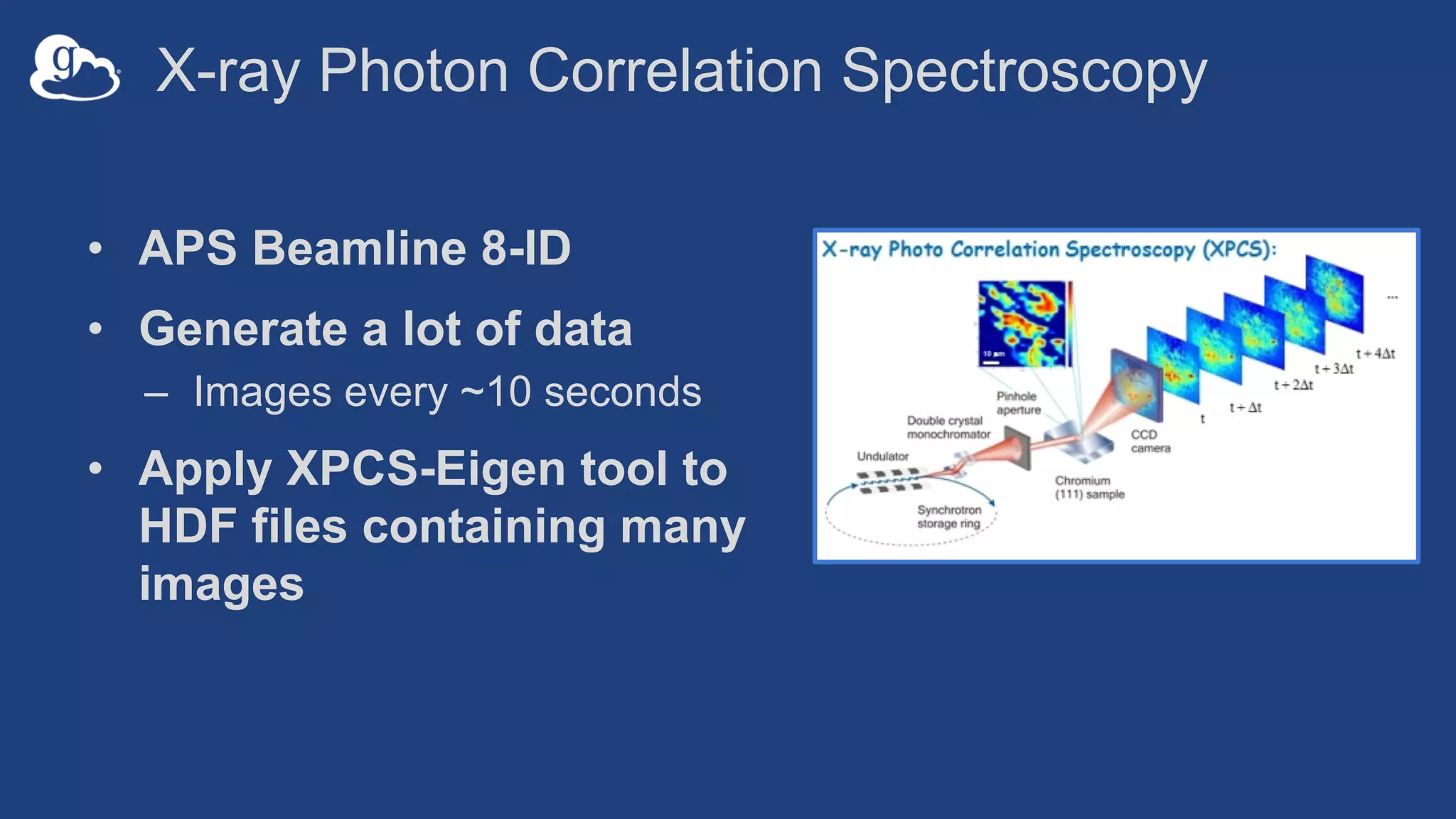
This document discusses using Globus to automate the management and analysis of large scientific instrument data. It provides examples of challenges with managing large datasets from the Event Horizon Telescope and applying Globus services and automation to help address these challenges. Specific use cases discussed include building connectomes from microscopy data and applying deep learning to flag bad scanning electron microscope images. The document emphasizes that automation needs transparency, results need to be easily findable, and leveraging specialized services can help.