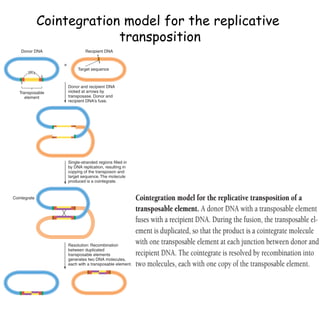
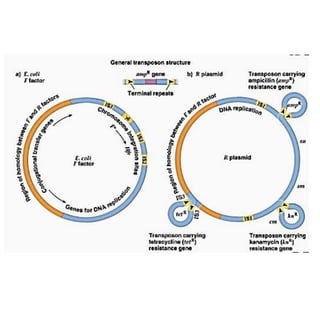
The document discusses transposons, including their structures and mechanisms of transposition. It provides details about two specific transposons:
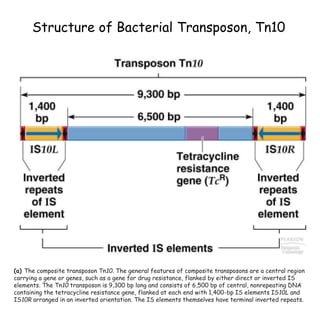
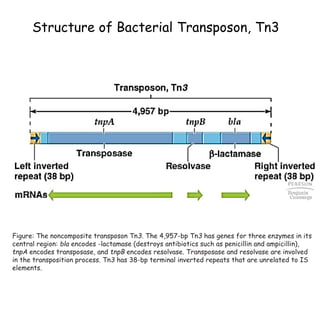
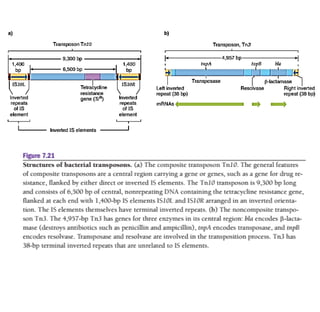
1) Tn10 is a bacterial transposon composed of a central resistance gene flanked by inverted IS elements. Tn3 is a noncomposite transposon with genes for antibiotic resistance and transposition enzymes.
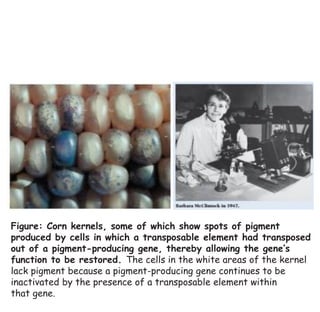
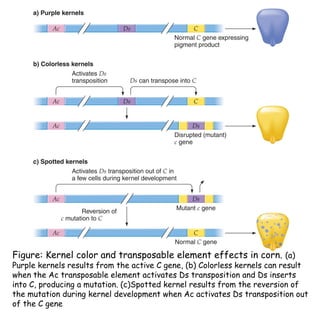
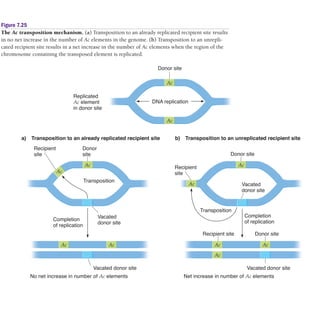
2) Ac-Ds elements in corn include the autonomous Ac element and non-autonomous Ds elements. Ac activates Ds transposition, which can insert into pigment genes and cause color patterns in kernels.
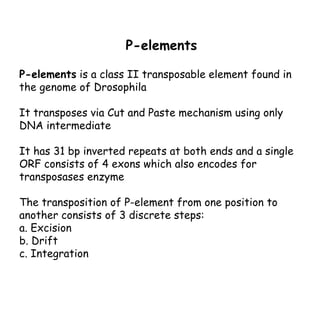
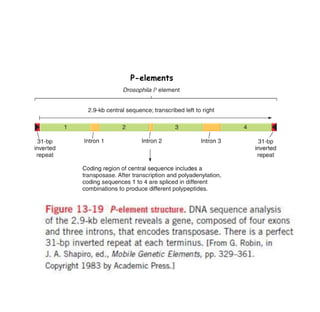
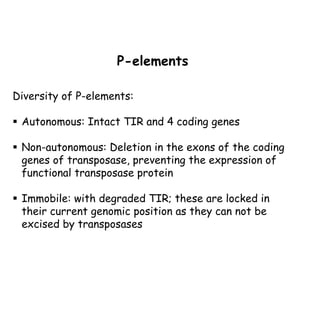
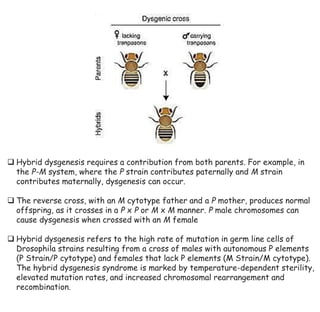
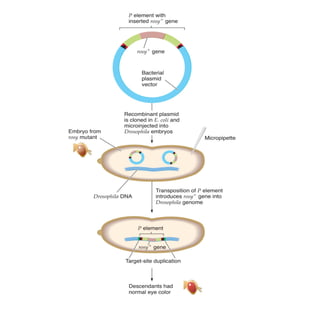
3) P elements in Drosophila vary in length and autonomy. Autonomous elements encode transposase while non-autonomous elements rely