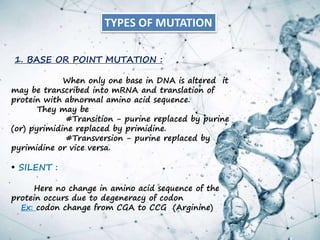
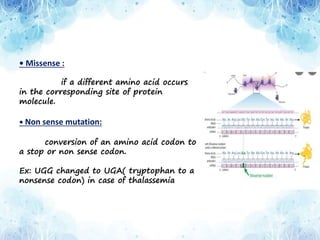
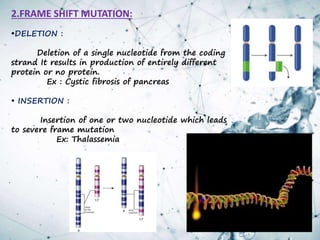
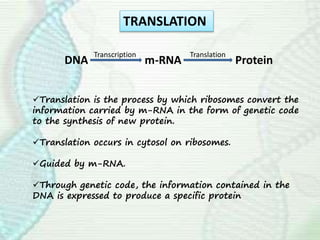
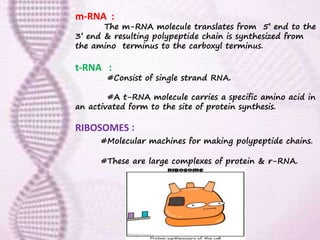
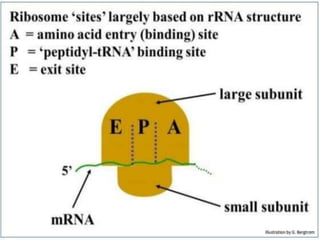
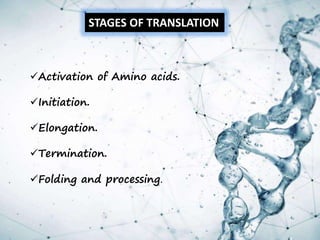
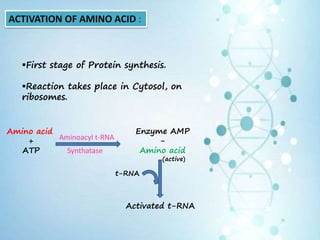
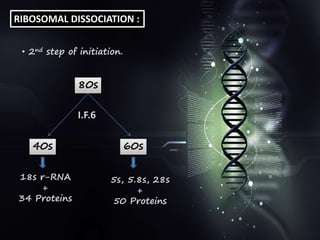
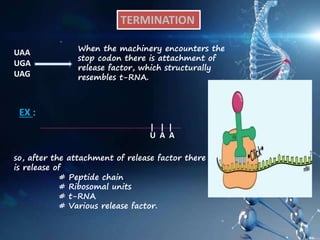
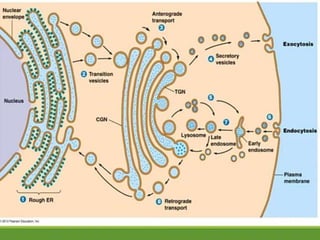
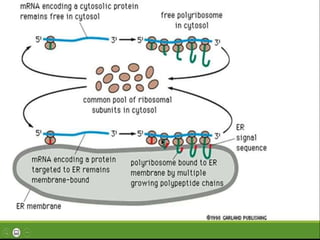
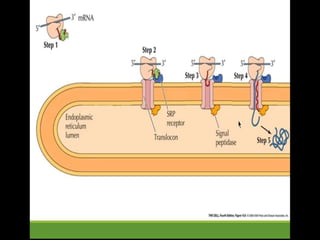
This document provides an overview of translation. It defines key terms like genetic code, codon, mutation and wobble hypothesis. It describes the basic requirements for translation like mRNA, tRNA and ribosomes. The stages of translation including activation, initiation, elongation, termination and post-translation modification are summarized. Protein targeting and sorting mechanisms to organelles are also outlined. Finally, some inhibitors of protein synthesis and disorders associated with defects in protein targeting are mentioned.

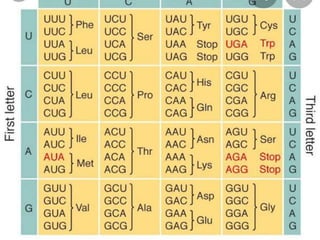
![SALIENT FEATURES OF GENETIC CODE
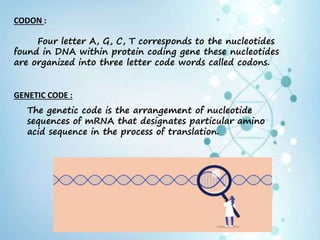
1) Number of Codons : 64 Codon sequences.
2) Directionality : 5’ – 3’
3) Stop/Termination/
Nonsense codons : # UAA
# UAG
# UGA
*Terminate polypeptide chain.
*Stops protein synthesis
4) Initiation Codon : [AUG] Initiates protein synthesis.
Dual function
of AUG : Initiator of codon
Synthesis of amino acid
methionine](https://image.slidesharecdn.com/translation-210801163823/85/Translation-in-eukaryotes-5-320.jpg)