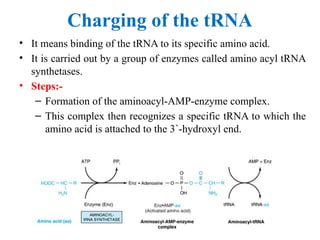
The document provides an overview of the genetic code, detailing the codons, their functions, and characteristics such as degeneracy, unambiguity, and universality. It explains mutations, including point and frameshift mutations, and their effects on protein synthesis, along with the translation process consisting of initiation, elongation, and termination. Additionally, the document discusses post-translational modifications required for protein activation and function.



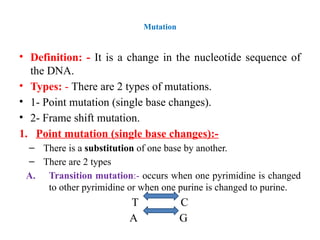
![B. Transversion mutation: - Occurs when a purine is
changed to either of the 2 pyrimidines or when a
pyrimidine is changed to either of the 2 purines.
• Effects of point mutation:-
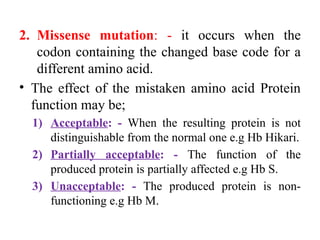
1. Silent mutation: - It means that the codon containing the
changed base still code for the same amino acid.
– It is due to degeneracy of the genetic code:- i.e the changed
base is the 3rd
base of the codon.
– e.g In Hb Bristol [it has aspartic acid at position 67 that is coded
by 2 codons GAU and GAC].](https://image.slidesharecdn.com/geneticcodemutation-241121043709-628cb809/85/Genetic-code-mutation-types-and-effects-pptx-8-320.jpg)

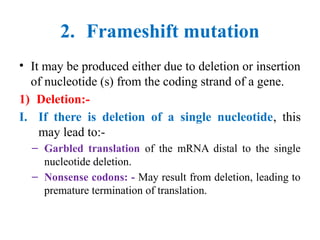

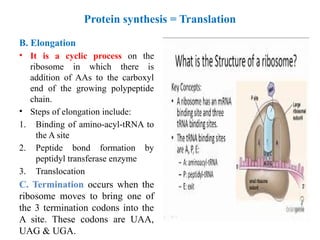
![Protein synthesis = Translation
• Protein synthesis: - Is the translation of
mRNA nucleotides sequence into sequence
of amino acids of specific protein.
• The message is read from 5 to 3 .
ˋ ˋ
• Steps of protein synthesis include initiation,
elongation and termination.
A. Initiation:-
– Requirements of initiation: - [tRNA,
ribosome, mRNA, amino acids, GTP and
ATP, and at least 10 eukaryotic initiation
factors (eIFc)].
– Initiation includes 4 stages:-
1. Ribosomal dissociation
2. Formation of the 43s pre-initiation
complex
3. Formation of the 48s initiation complex
4. Formation of the 80s initiation complex](https://image.slidesharecdn.com/geneticcodemutation-241121043709-628cb809/85/Genetic-code-mutation-types-and-effects-pptx-14-320.jpg)