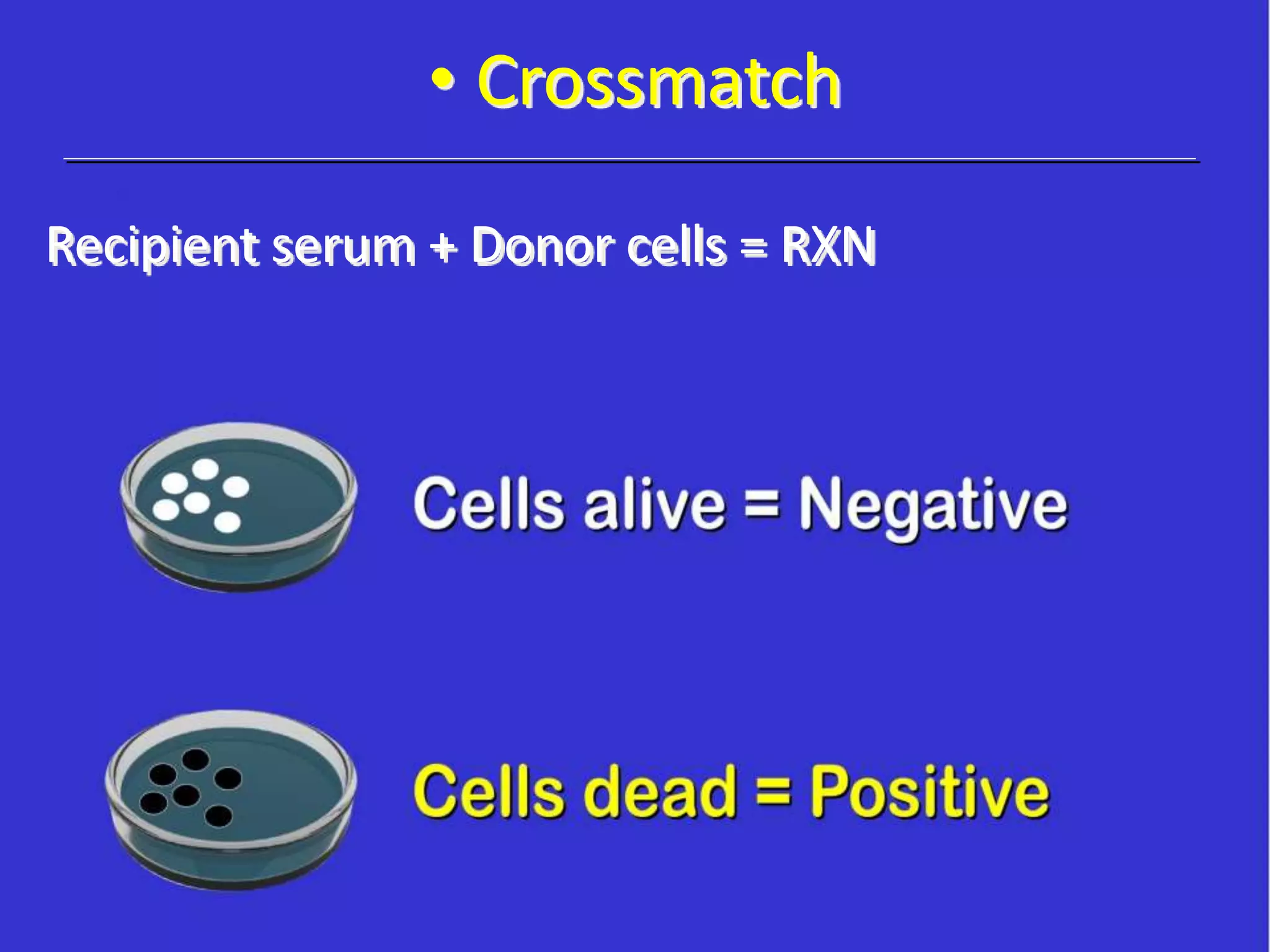
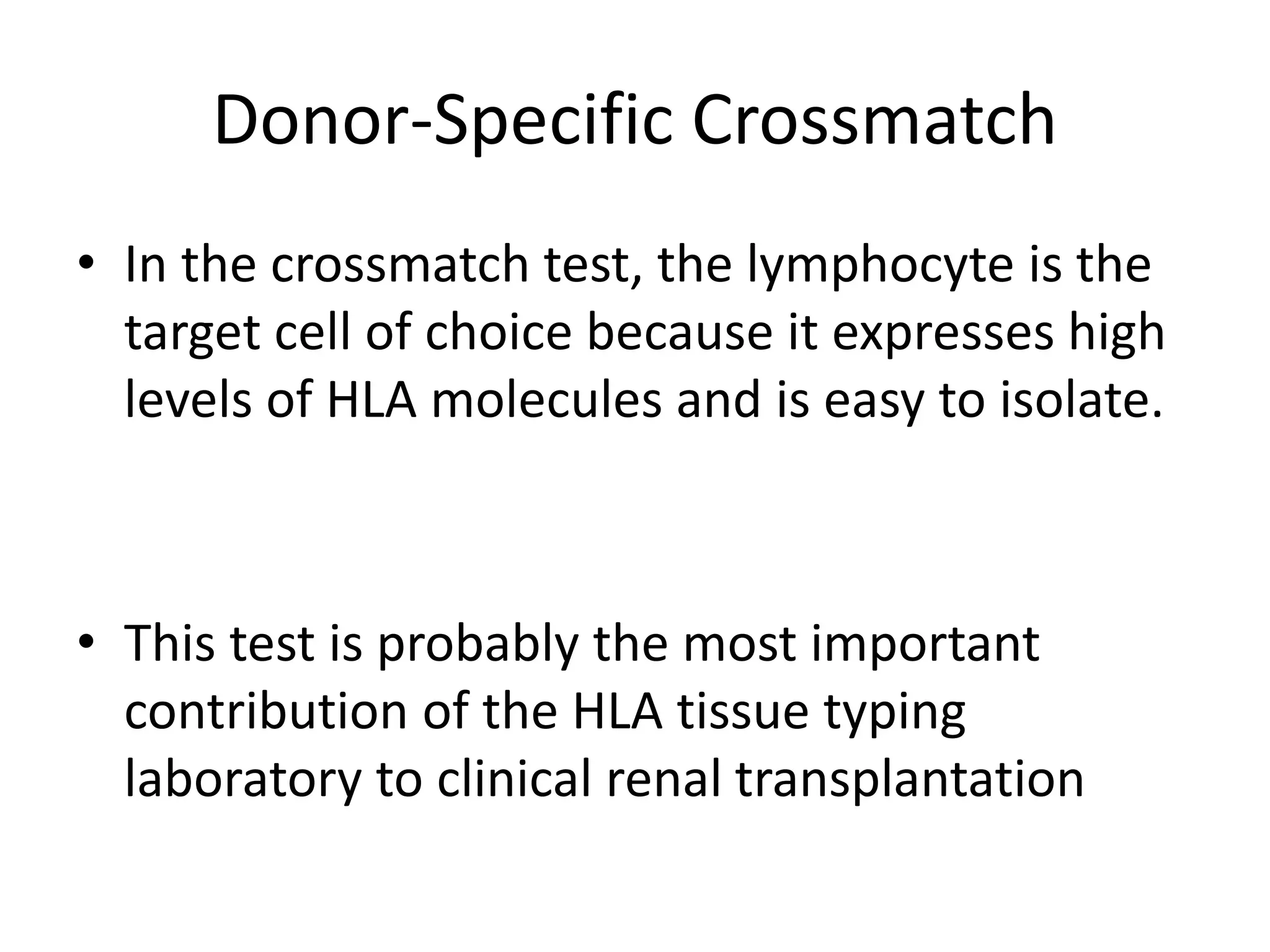
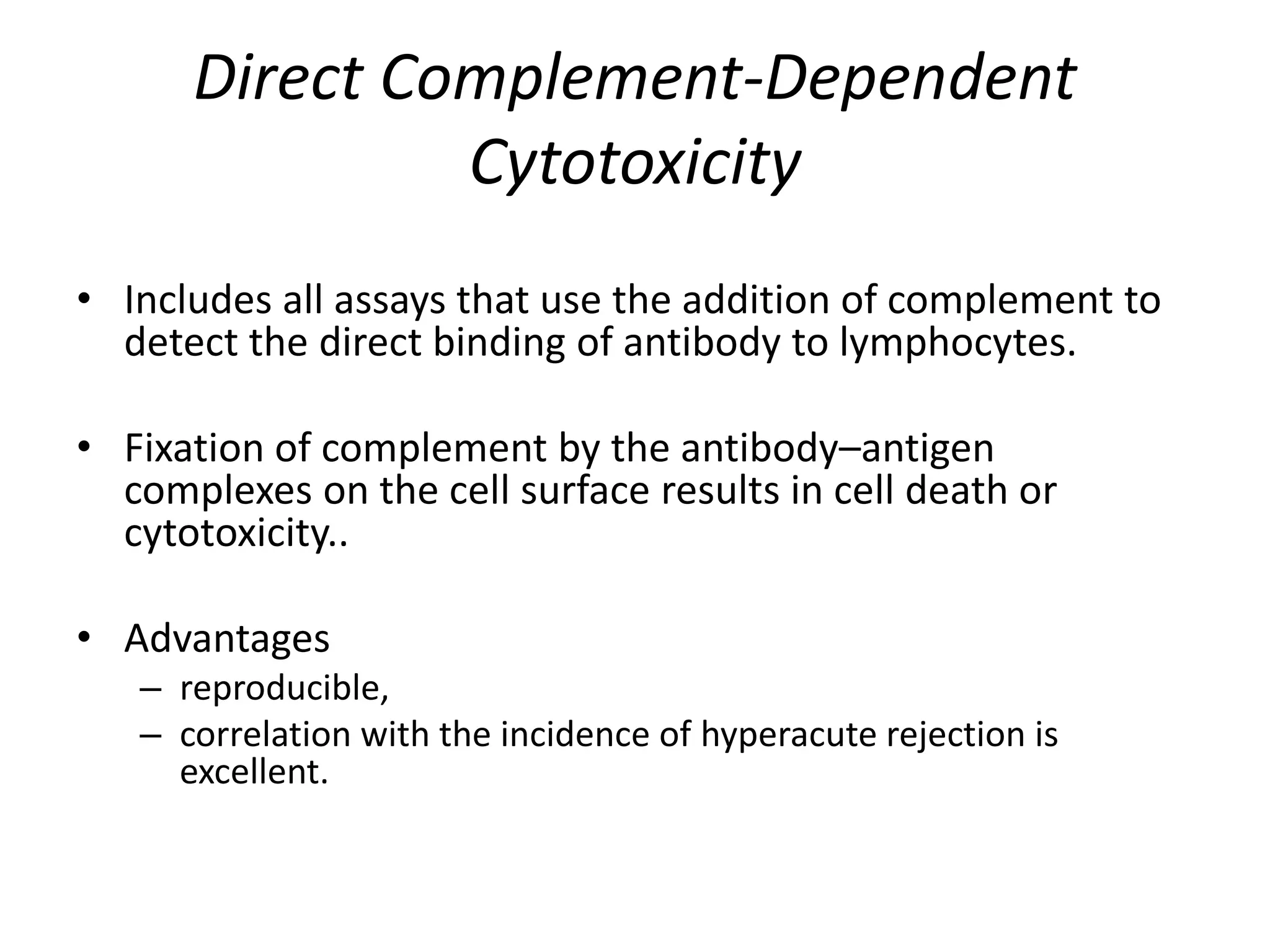
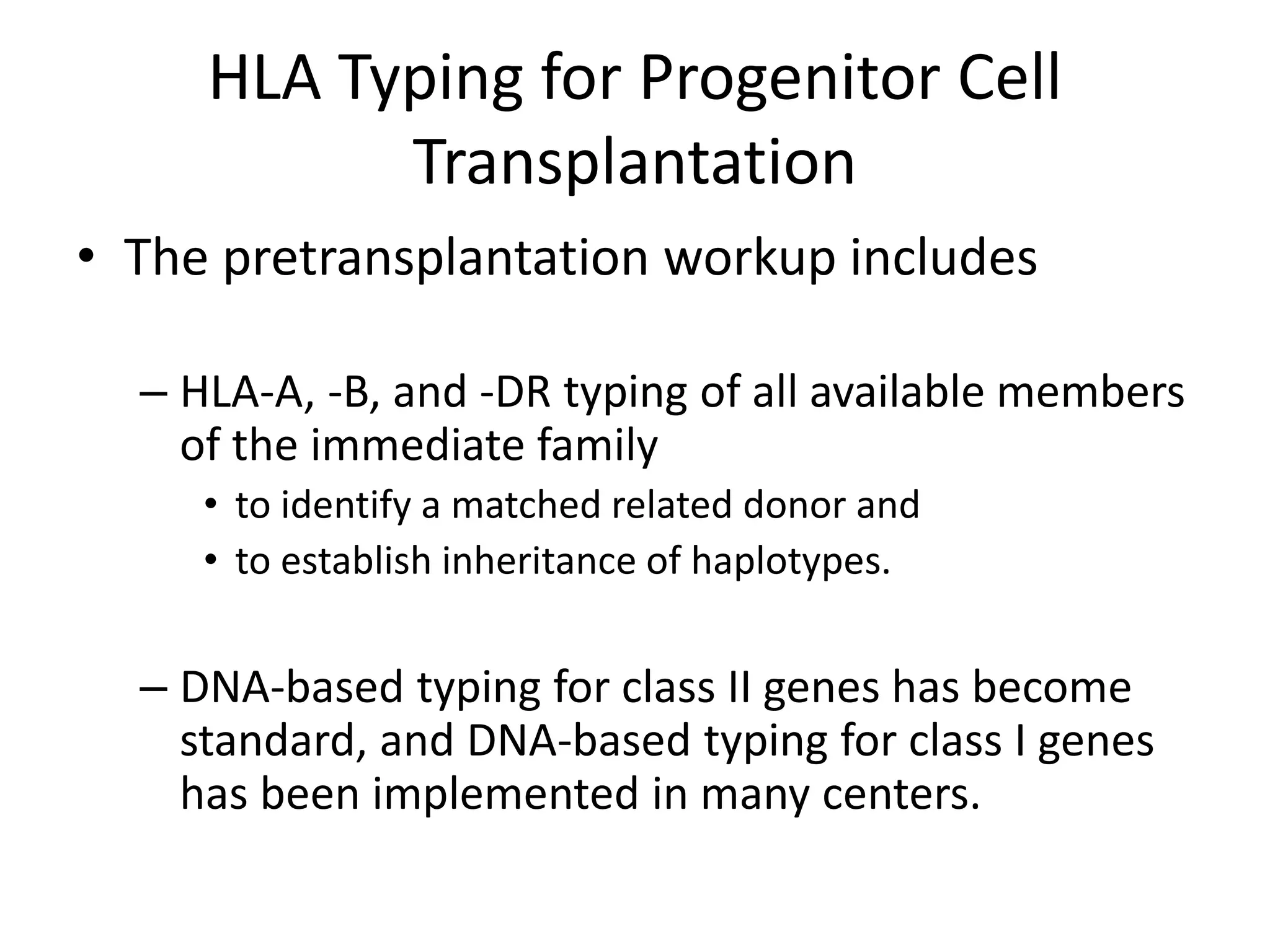
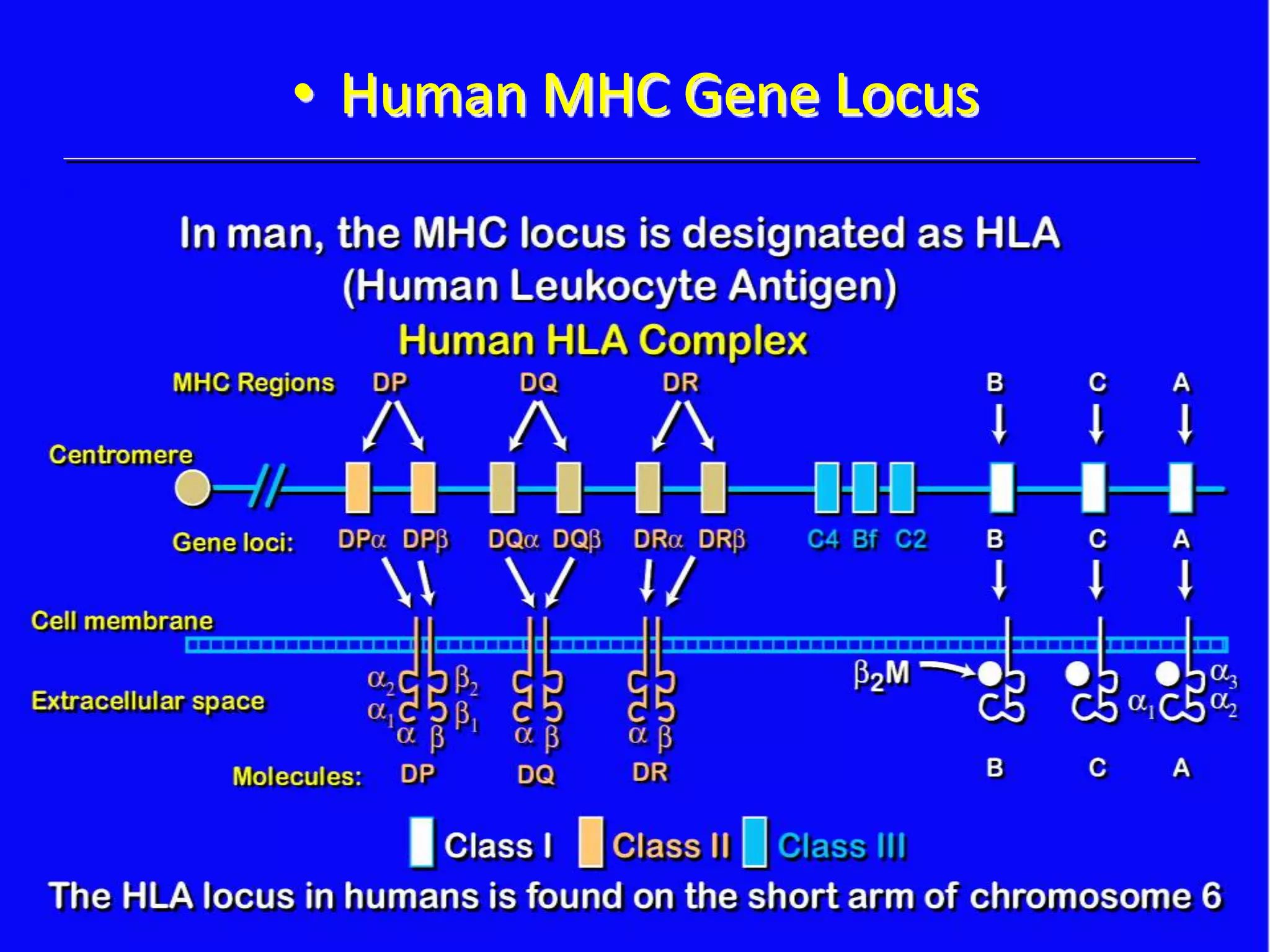
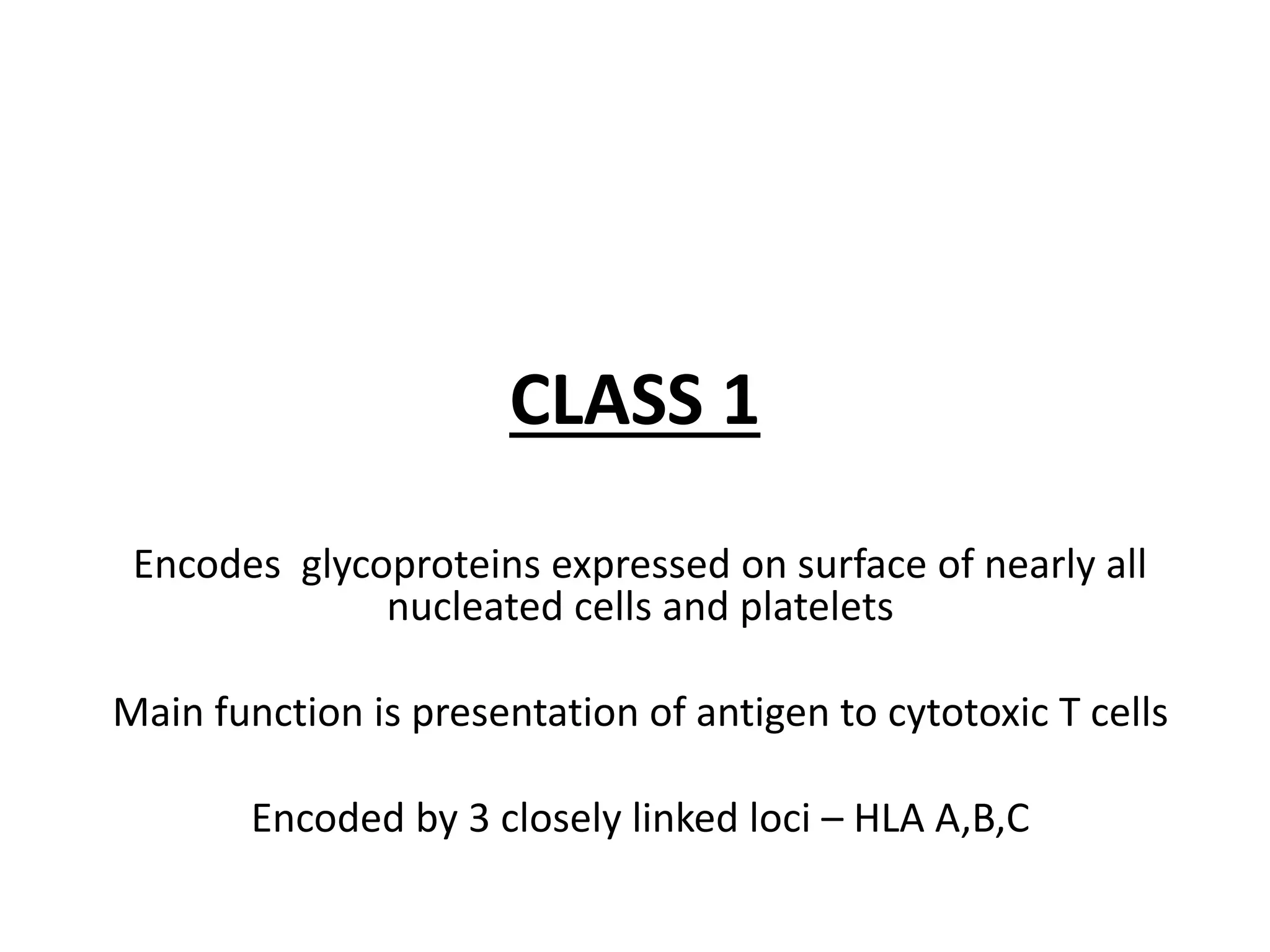
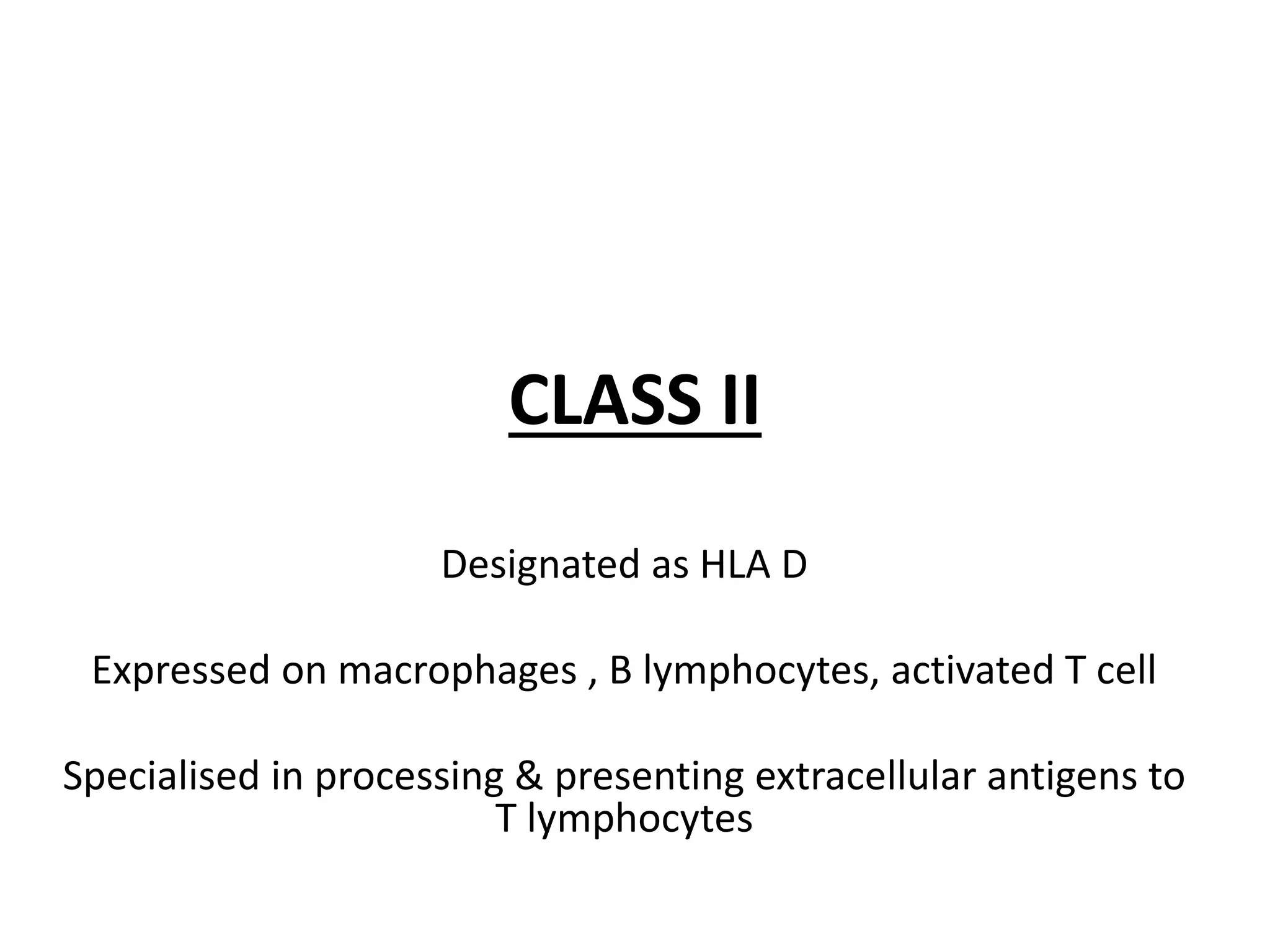
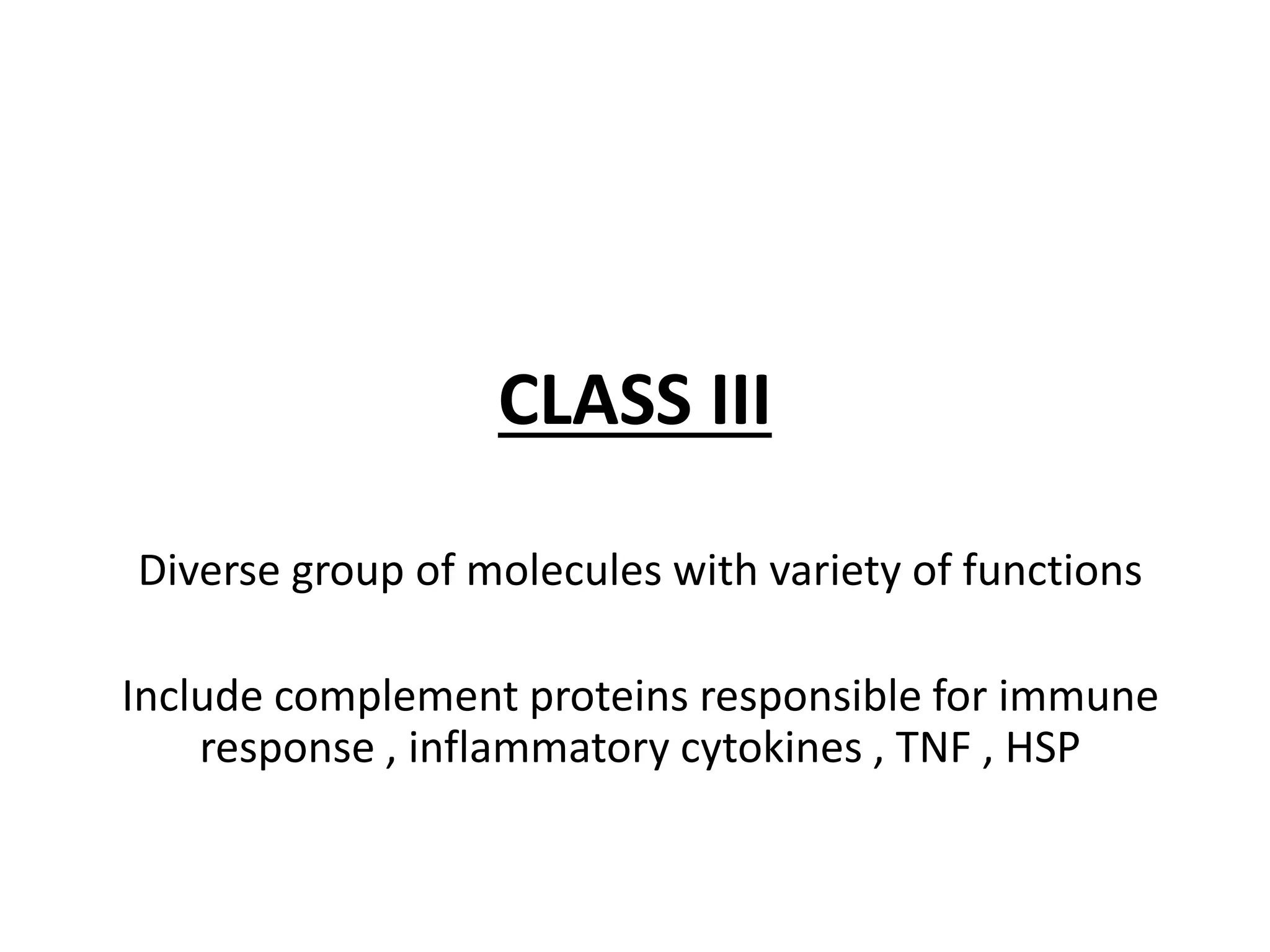
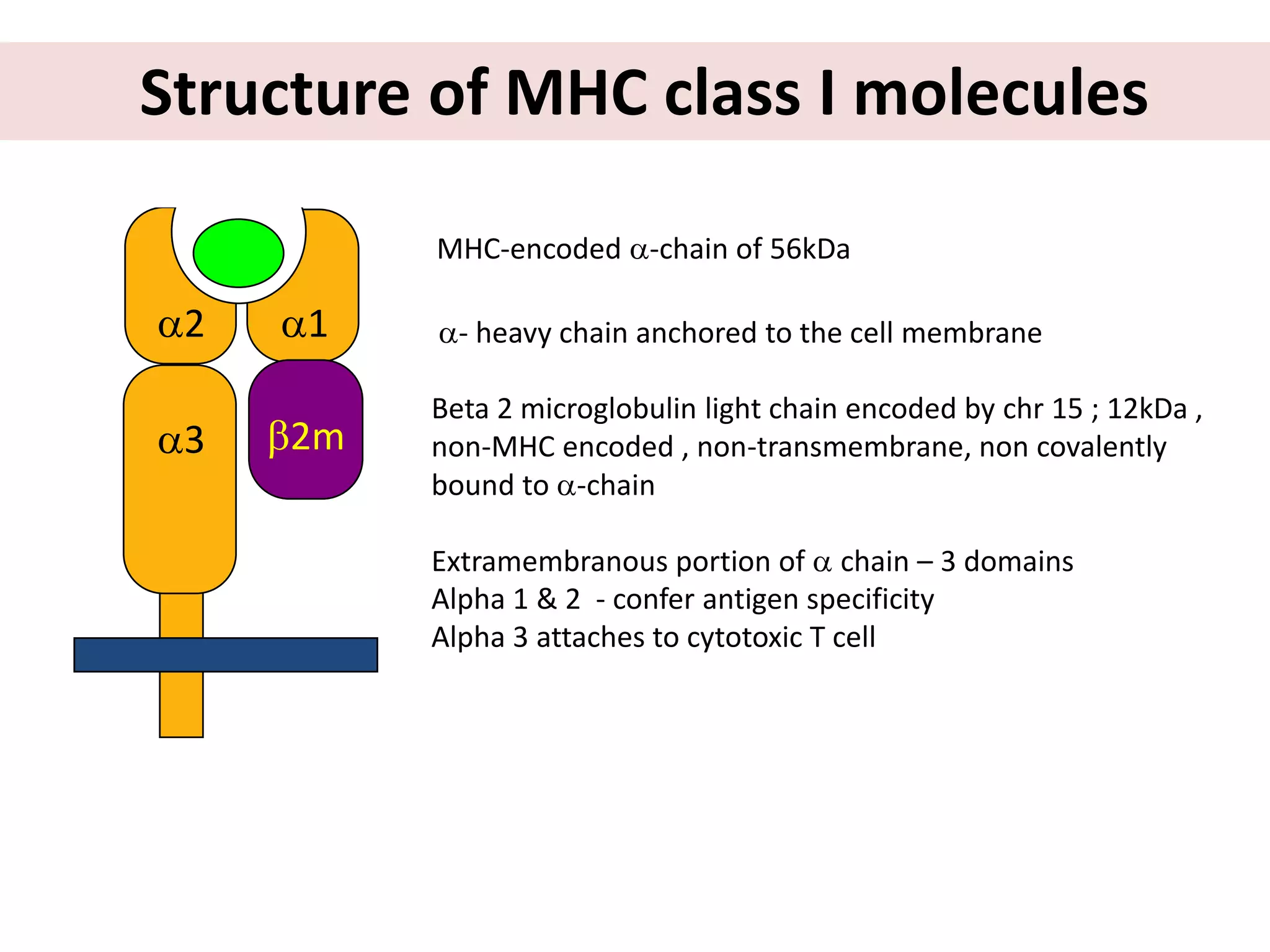
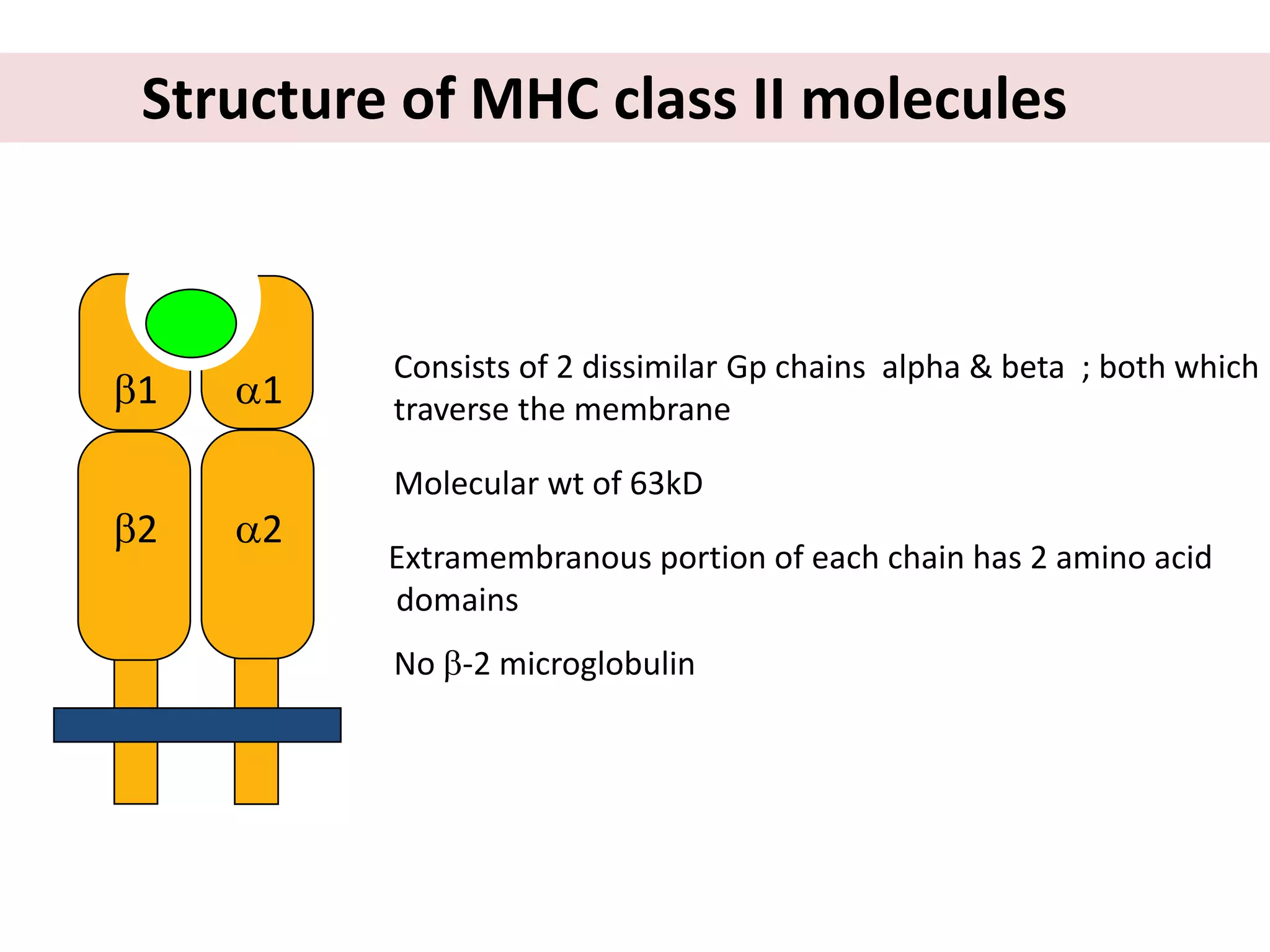
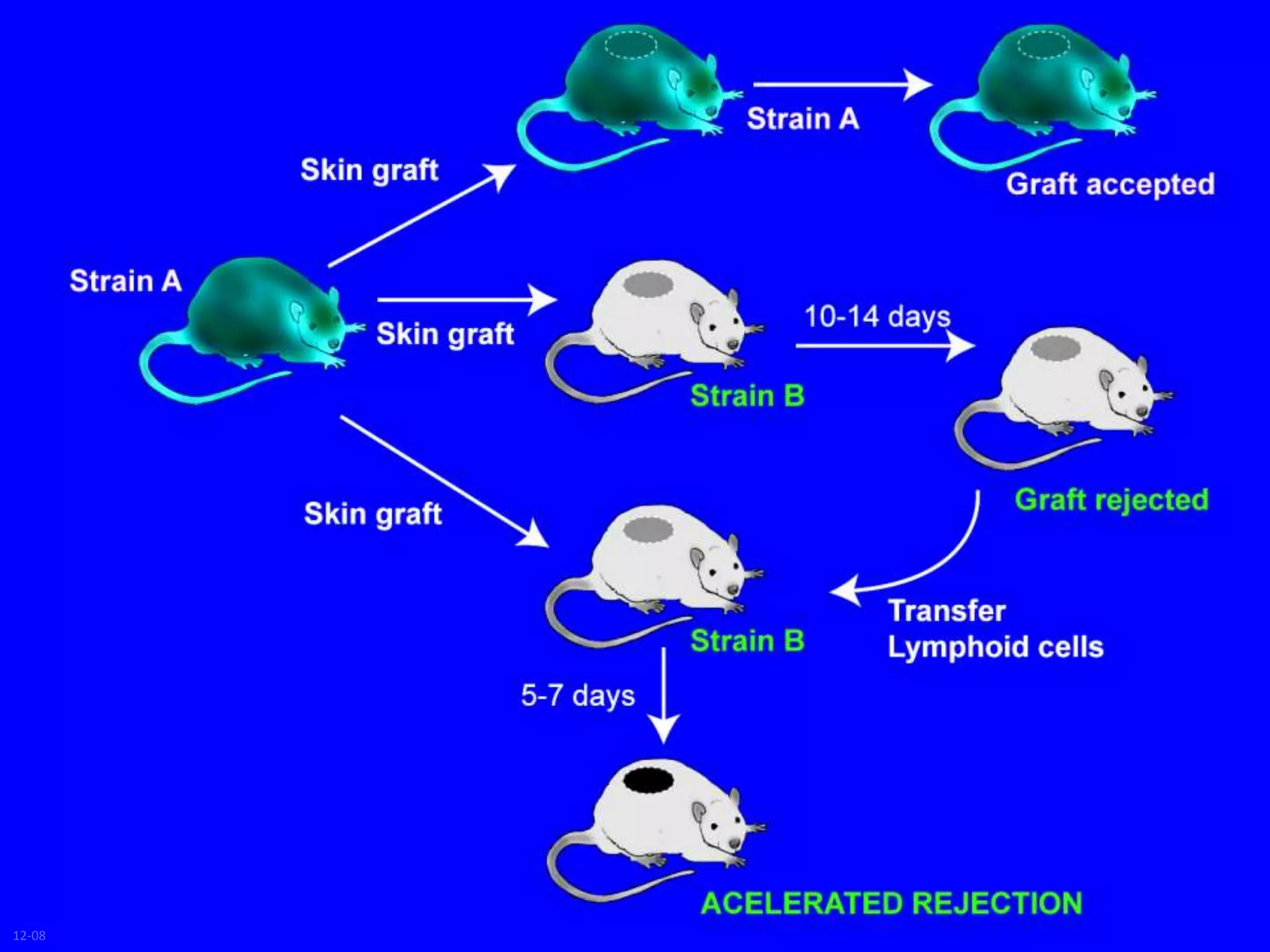
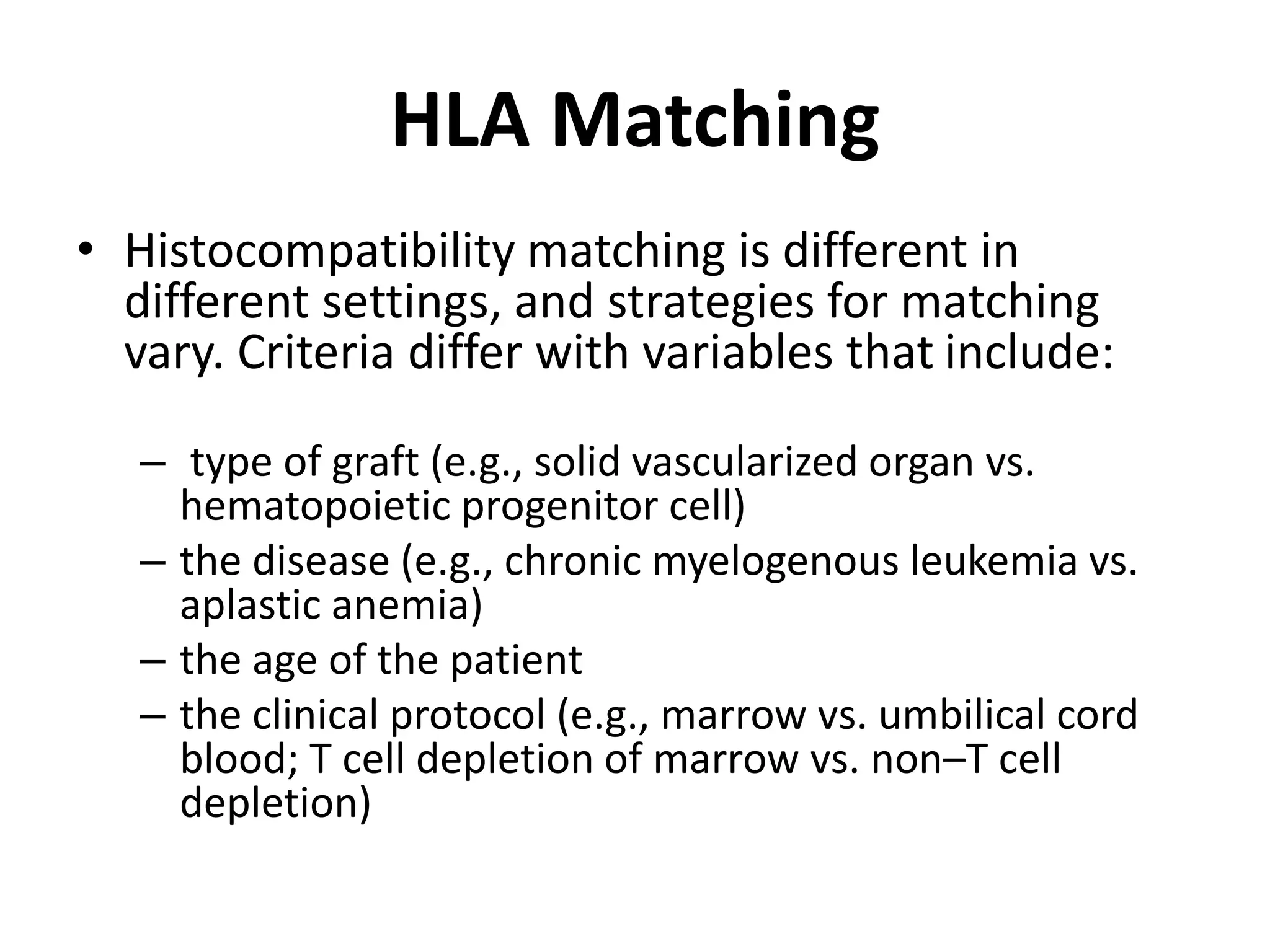
This document discusses human leukocyte antigen (HLA) gene locus and HLA matching in organ transplantation. It contains the following key points:
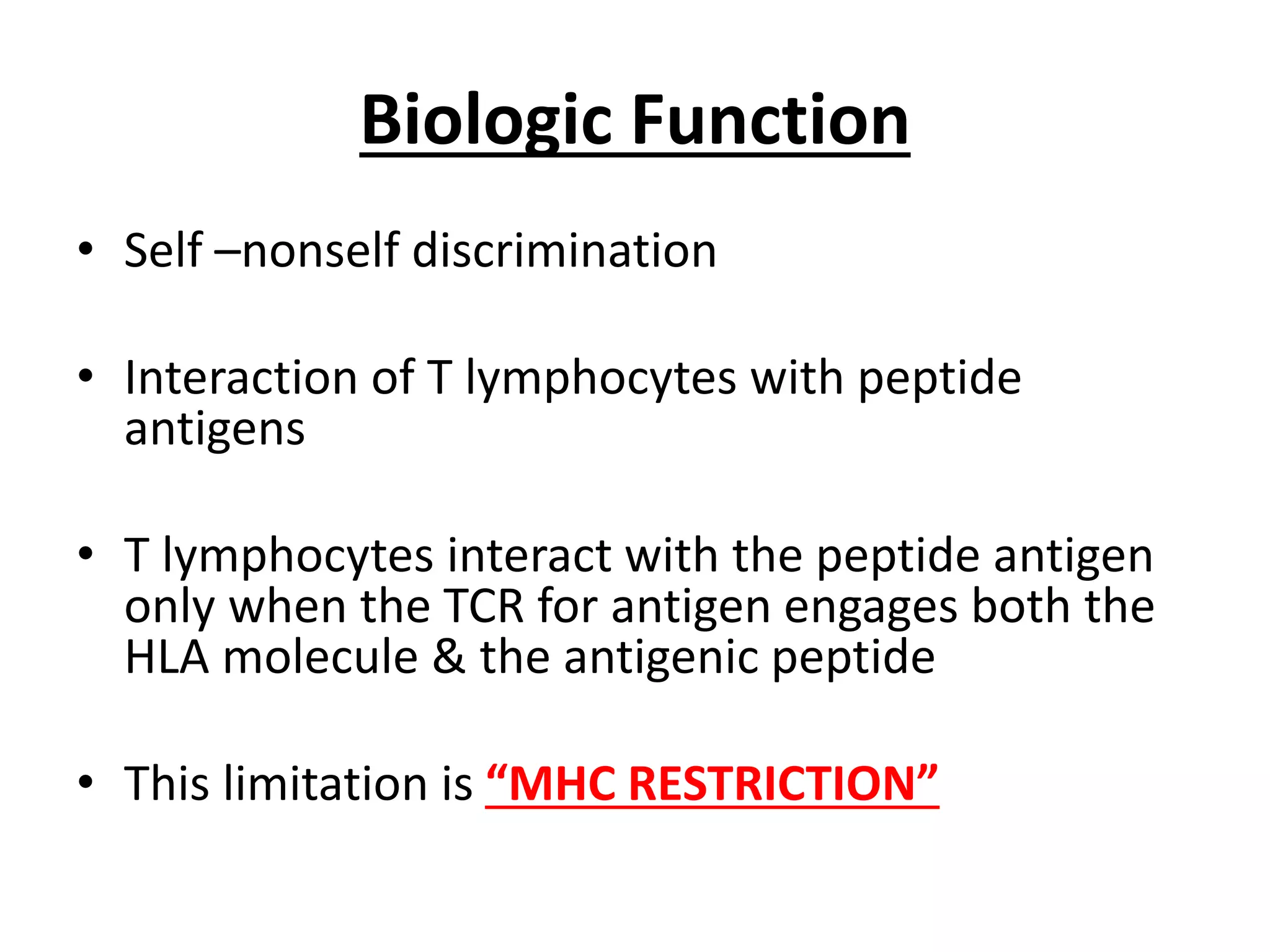
1. HLA genes encode antigen-presenting molecules that are crucial for the immune system's recognition of self vs. non-self. Close HLA matching between donor and recipient is important for transplant success.
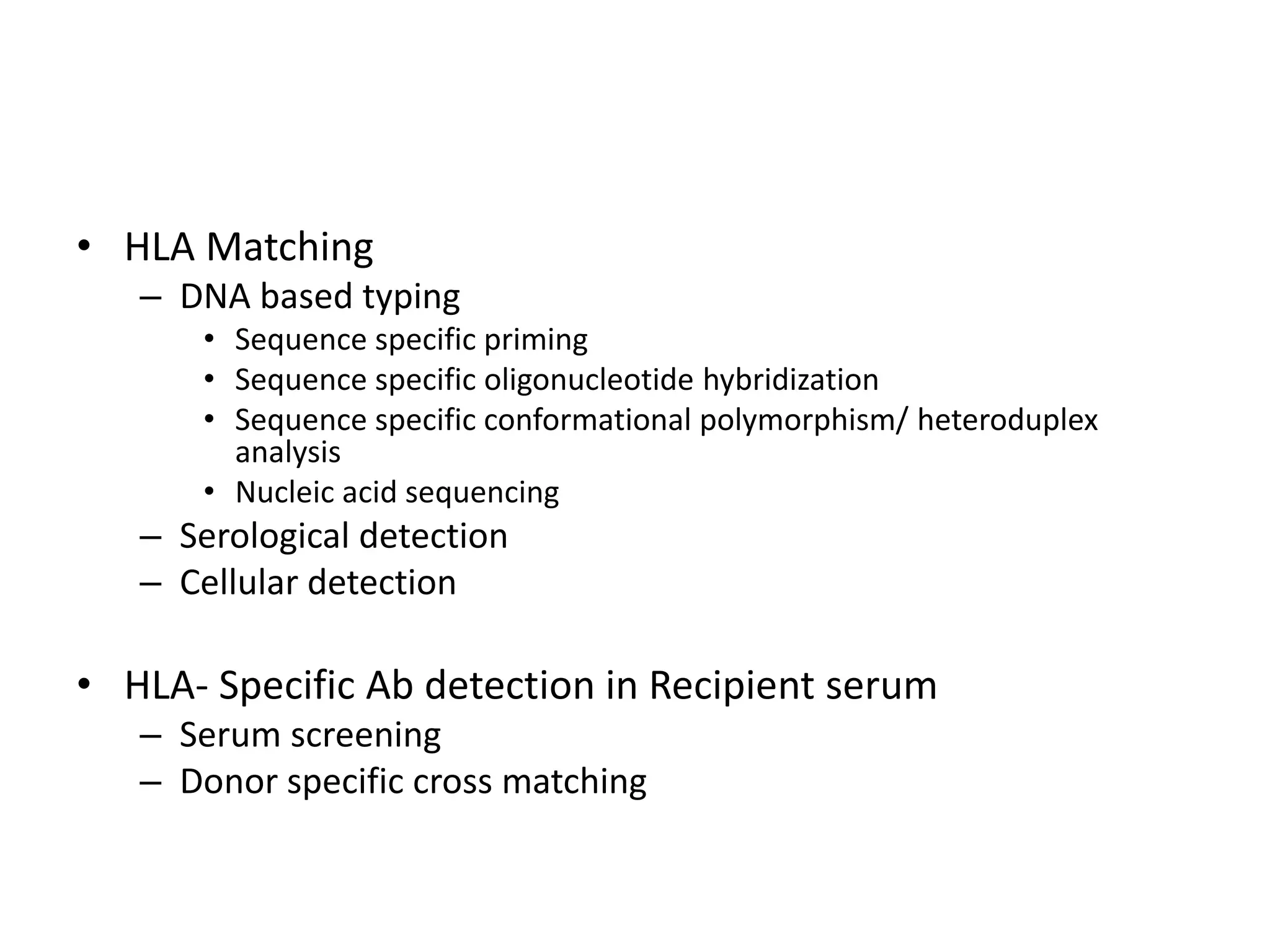
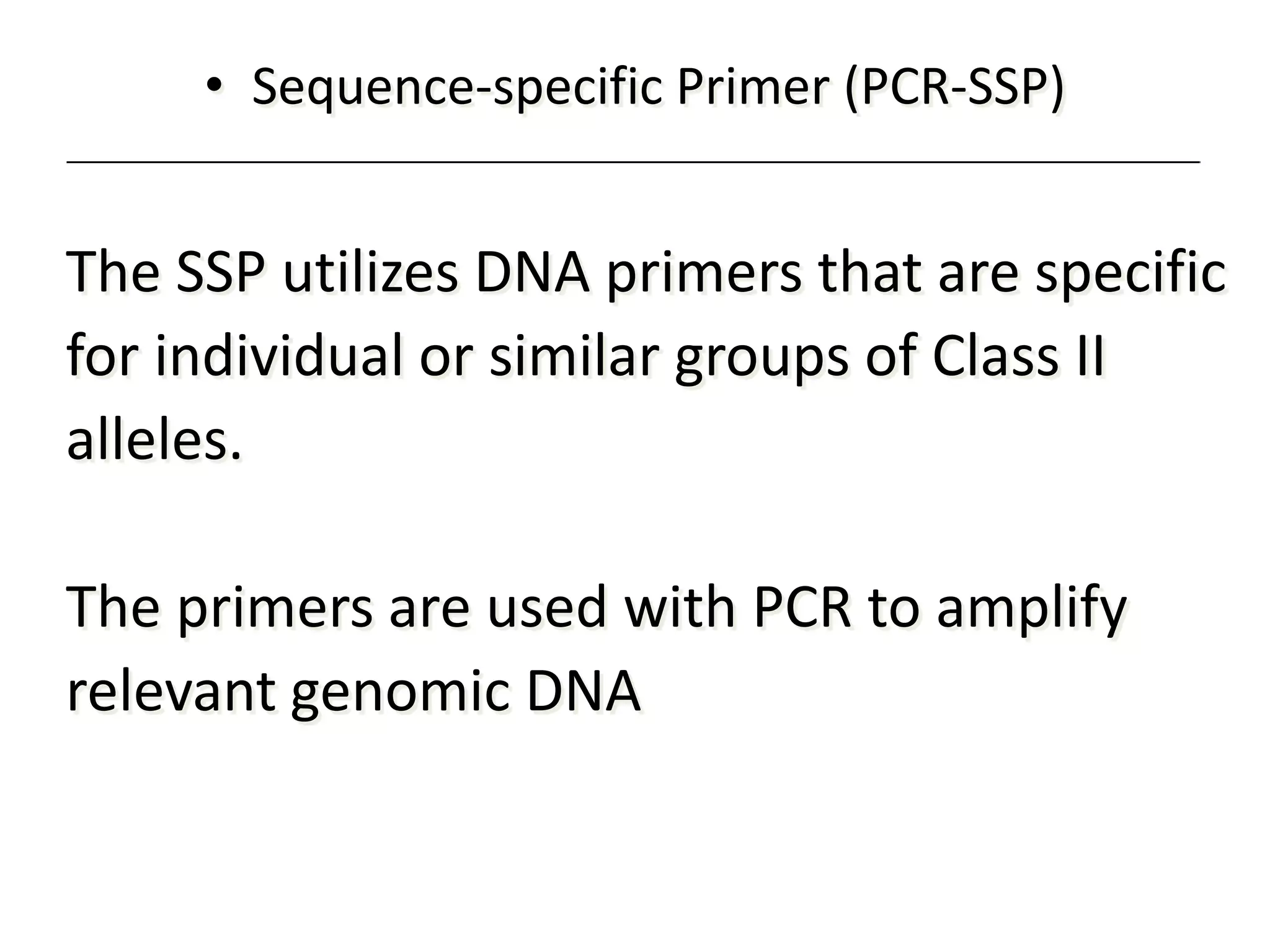
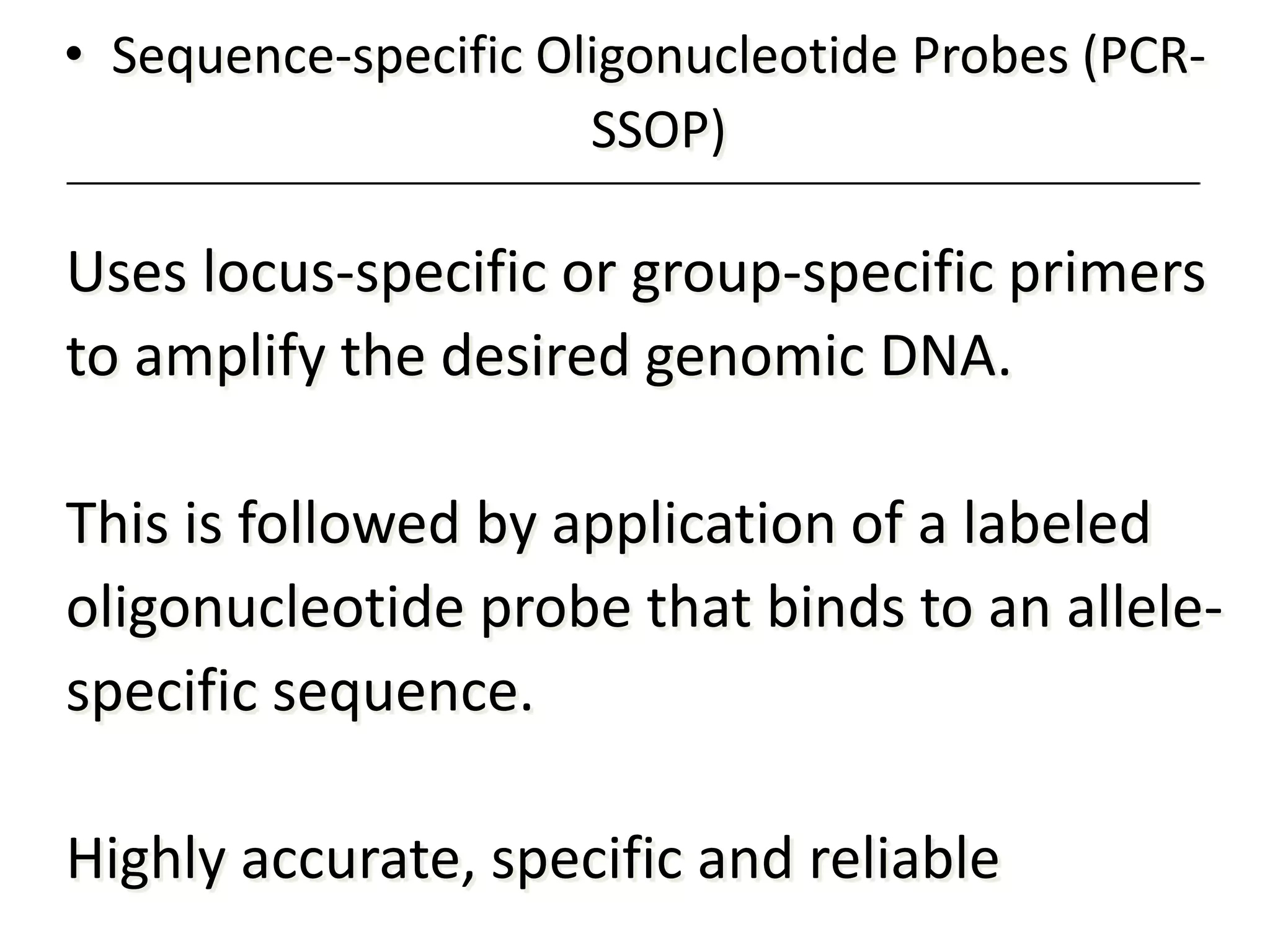
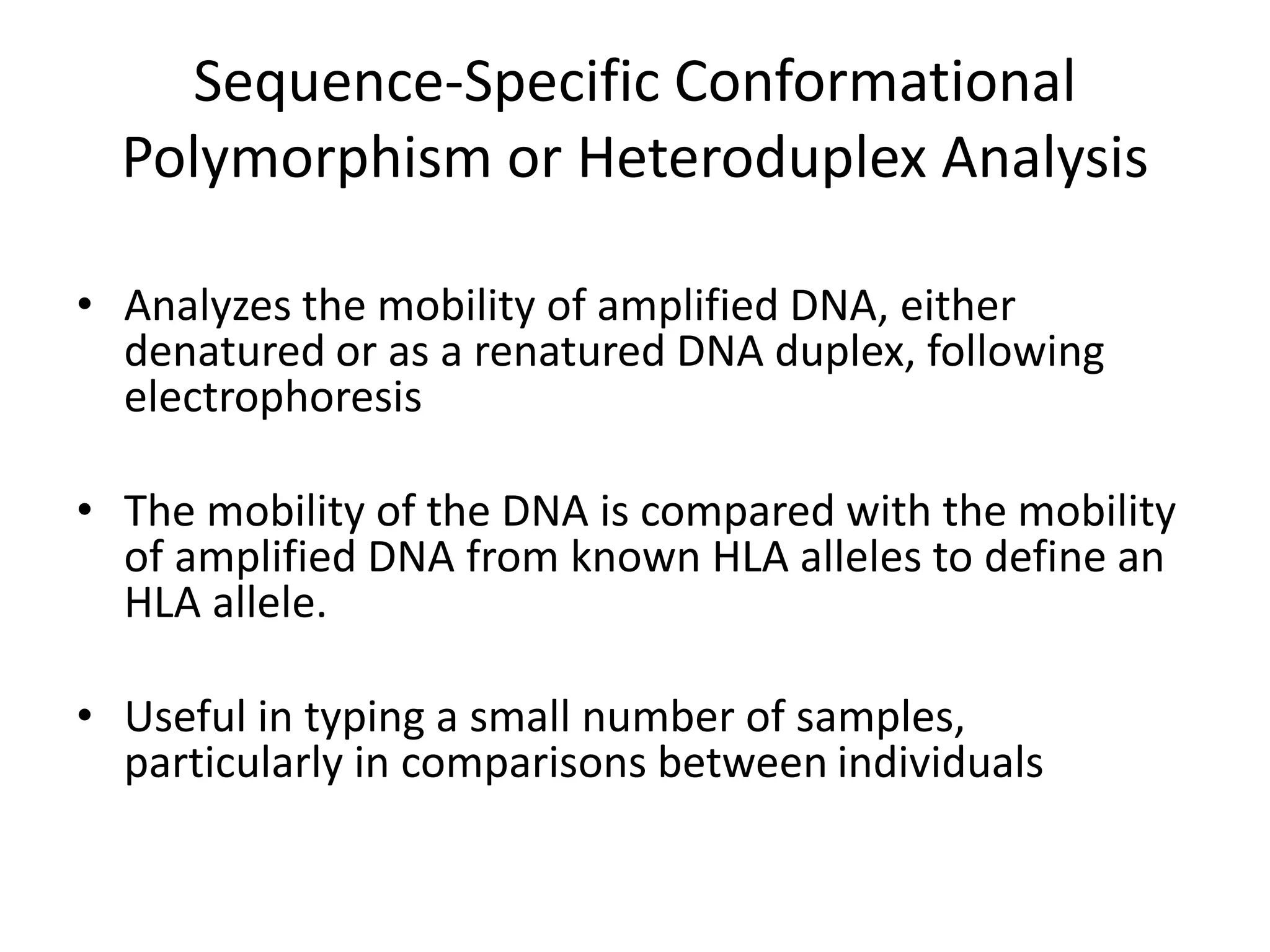
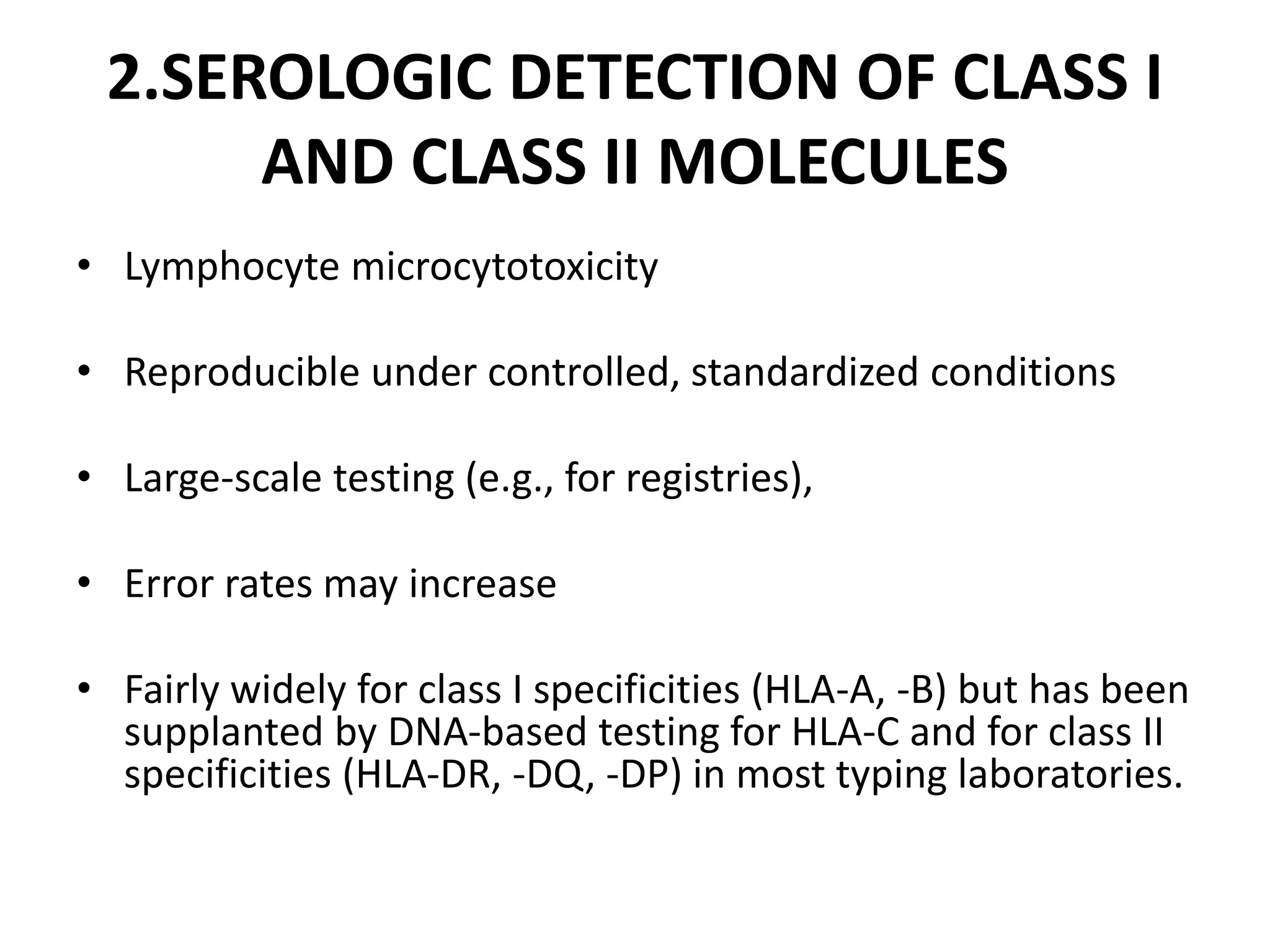
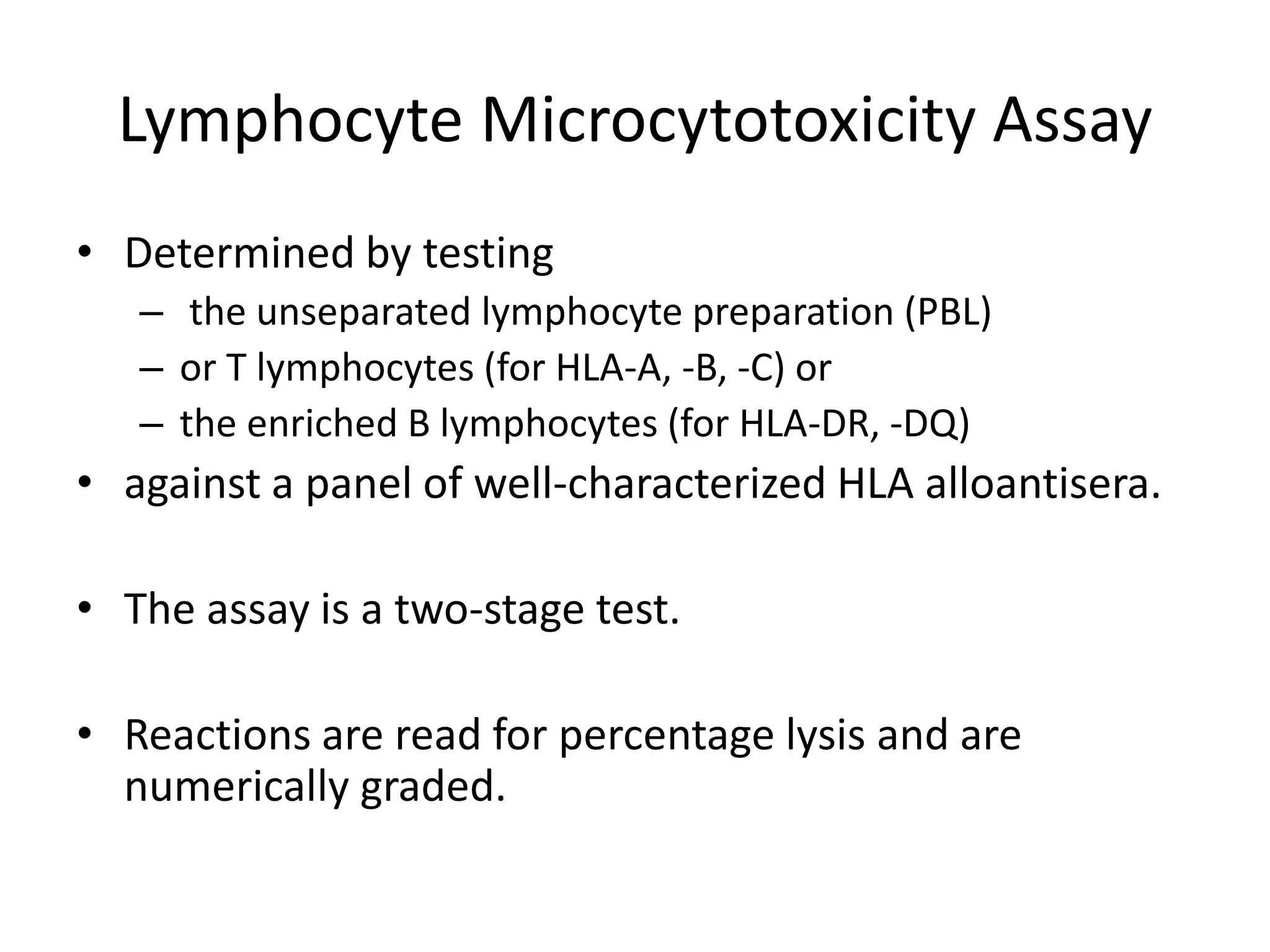
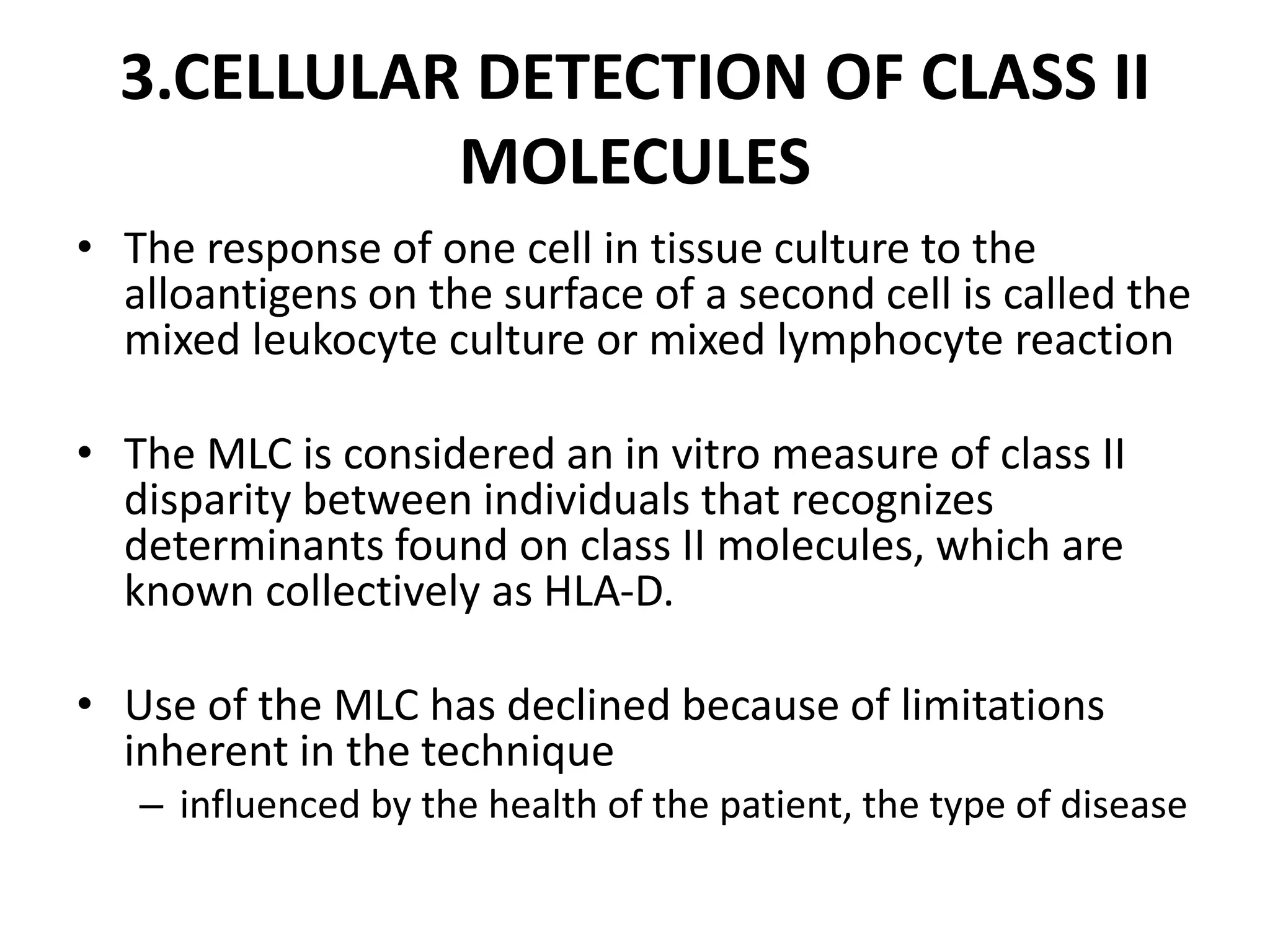
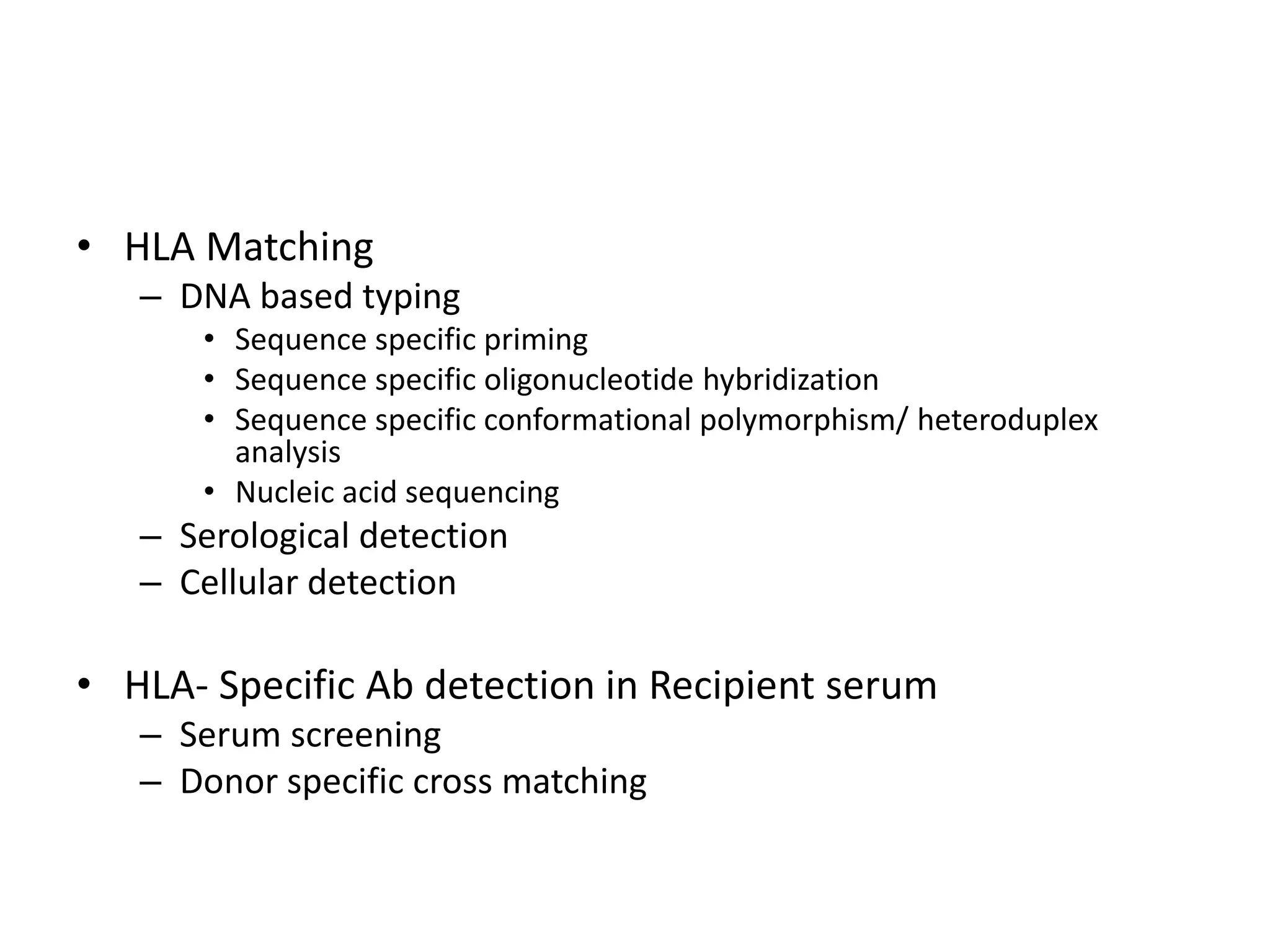
2. Modern HLA typing techniques include DNA-based methods like sequence-specific priming and probing, as well as serological detection methods using lymphocyte microcytotoxicity assays.
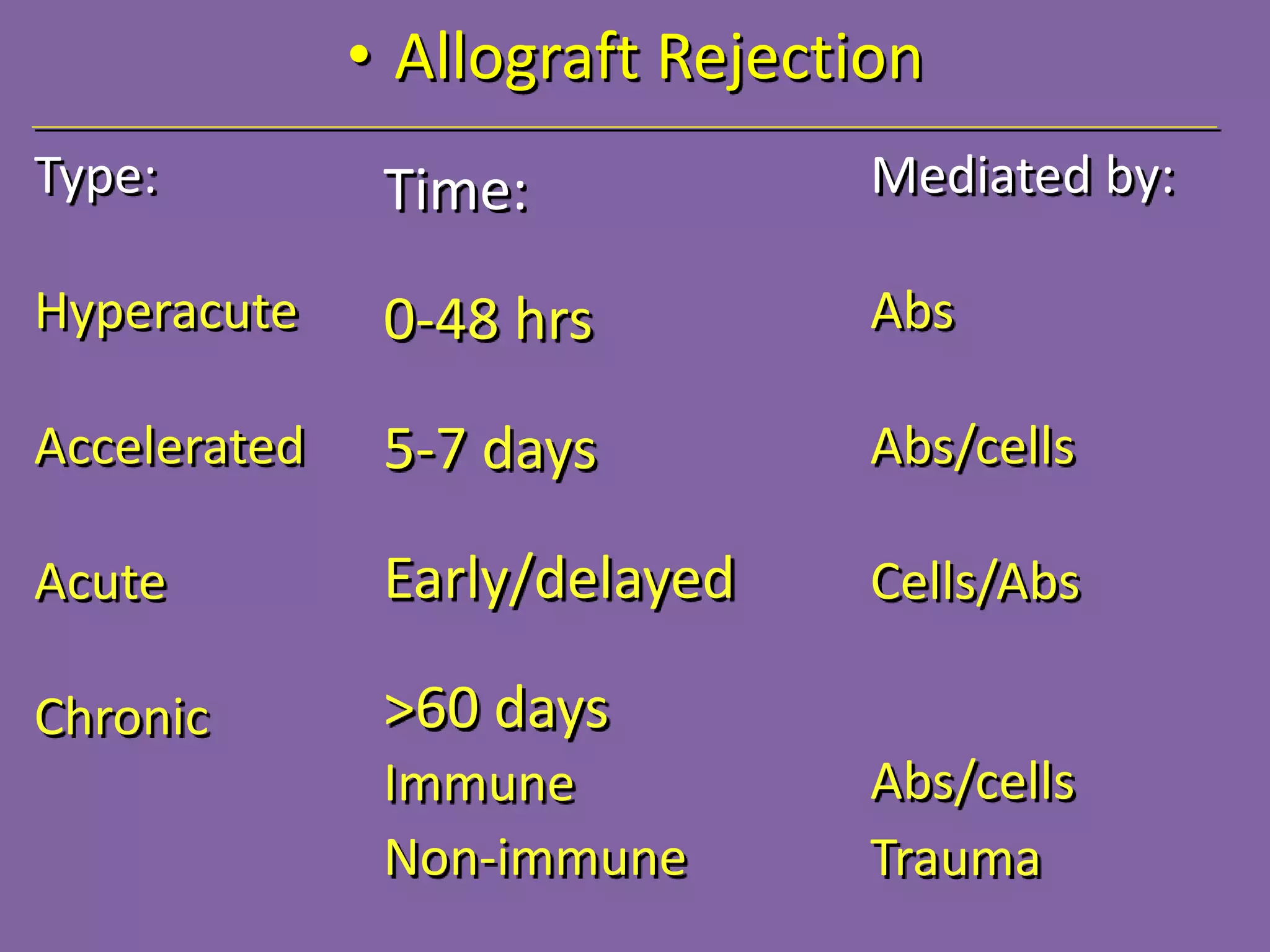
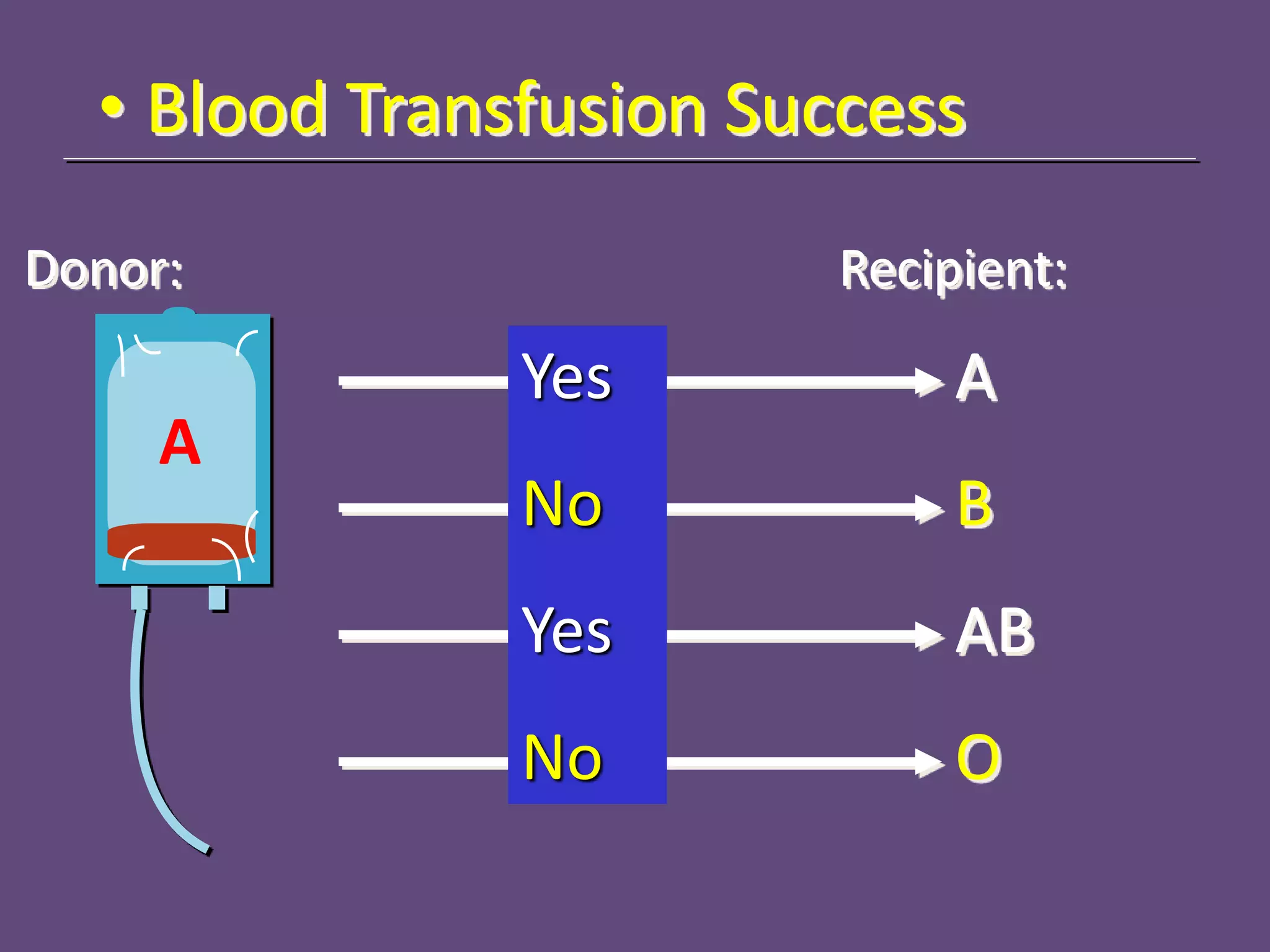
3. Achieving an accurate HLA match and minimizing recipient immune response requires histocompatibility testing of HLA genes as well as immunosuppressive therapy after transplantation.










![Nucleic Acid Sequencing
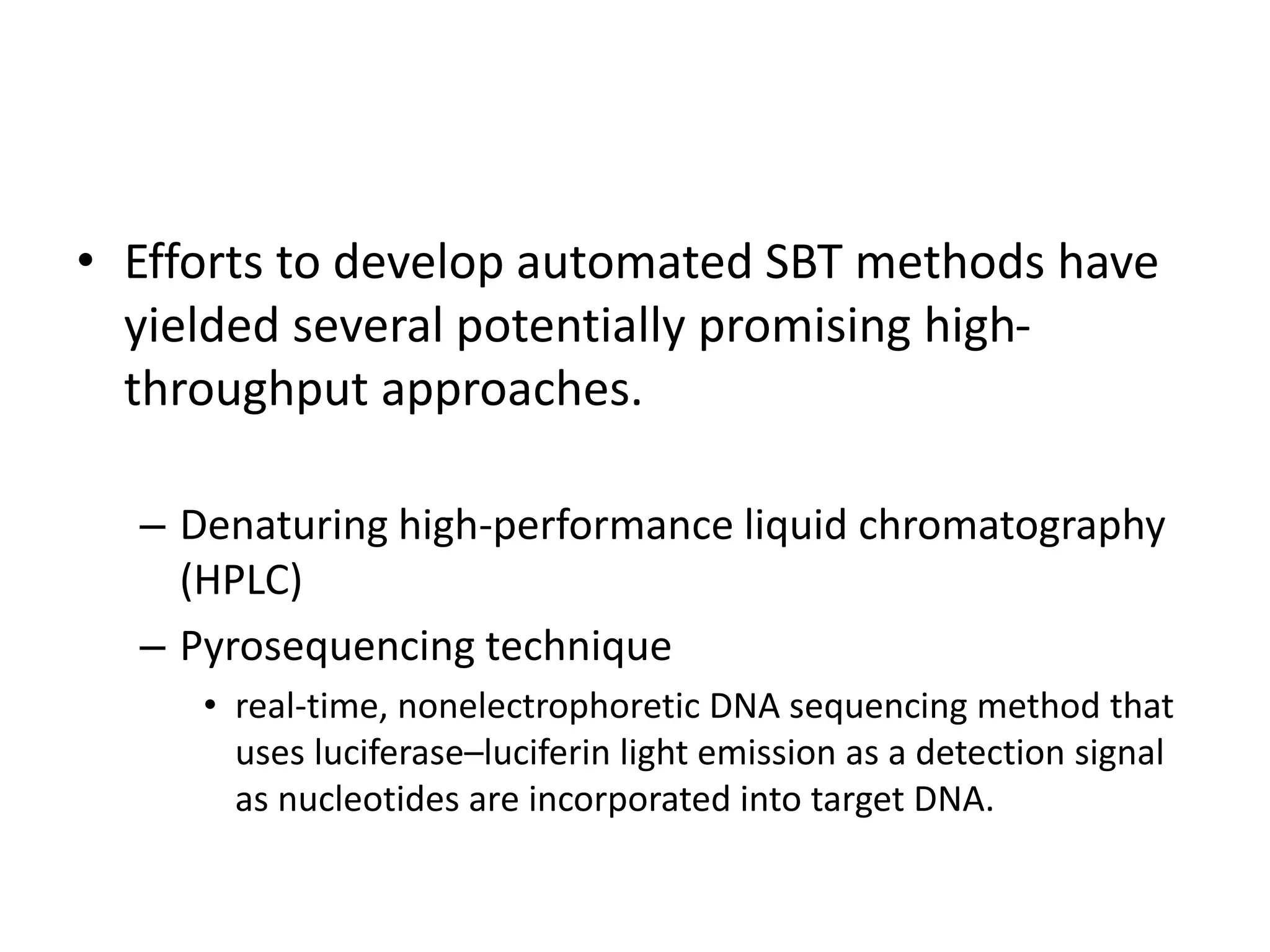
• Involves direct determination of the DNA sequences of the
HLA alleles carried by an individual (sequence-based typing
[SBT]).
• Identified following PCR amplification to separate the
alleles based on SSPs, or as a mixture of two alleles.
• Sequencing is labor-intensive and highly complex
• Determine the HLA match at an allele level between
hematopoietic progenitor cell transplant patients and their
prospective](https://image.slidesharecdn.com/organtransplantation-140501052049-phpapp02/75/Tests-In-Organ-Transplantation-32-2048.jpg)

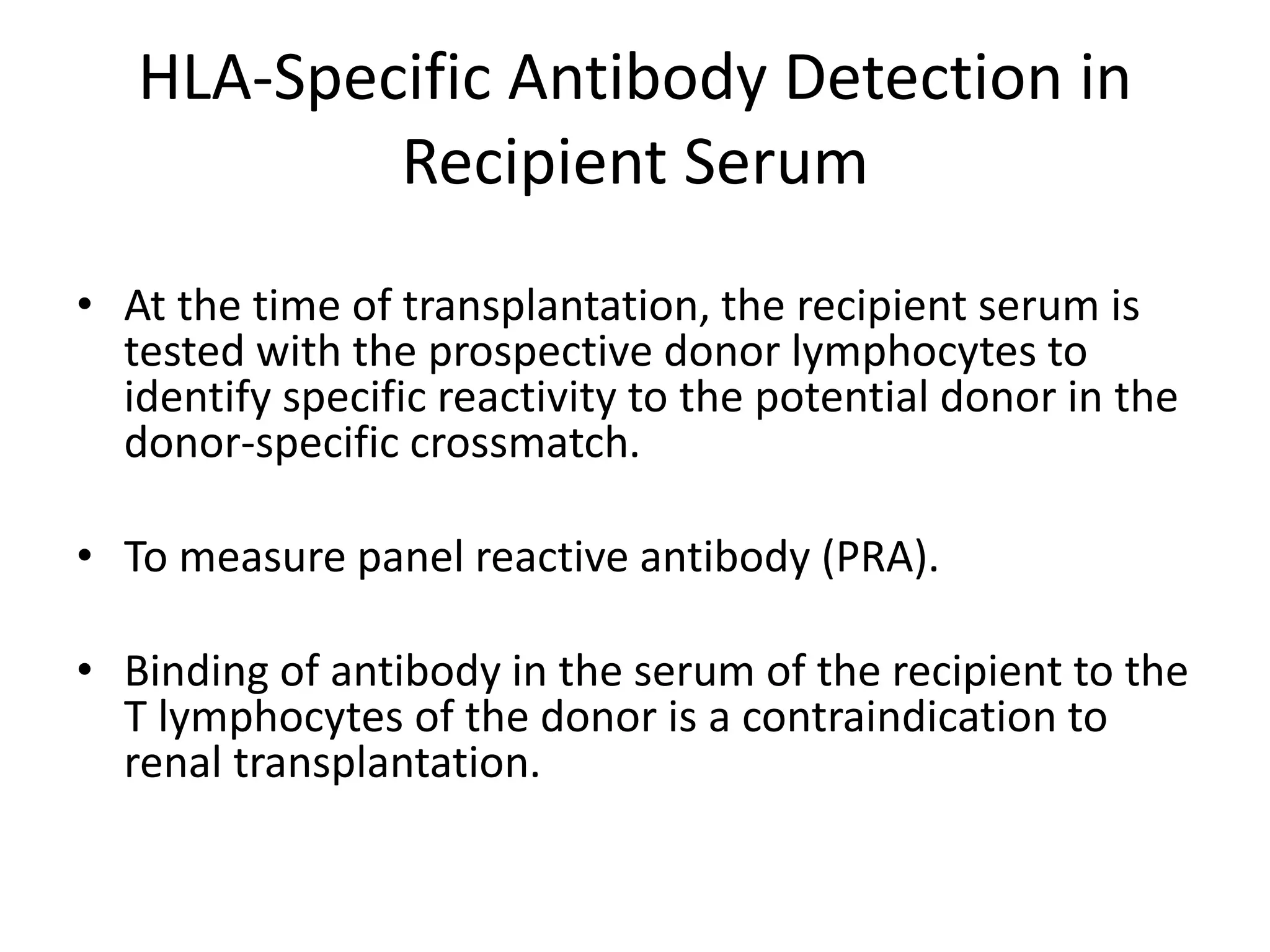
![Serum Screening (PRA)
• The goals of a PRA screen are as follows:
• 1. To identify the level of presensitization of the patient to
HLA antigens
• 2. To identify the HLA specificity of the antibodies to predict
the HLA antigen(s) to be avoided when donors are selected.
• 3. To identify patients with irrelevant antibodies (e.g.,
immunoglobulin [Ig]M autoantibodies)
– to avoid false-positive readings at the time of the donor-specific
crossmatch.](https://image.slidesharecdn.com/organtransplantation-140501052049-phpapp02/75/Tests-In-Organ-Transplantation-41-2048.jpg)