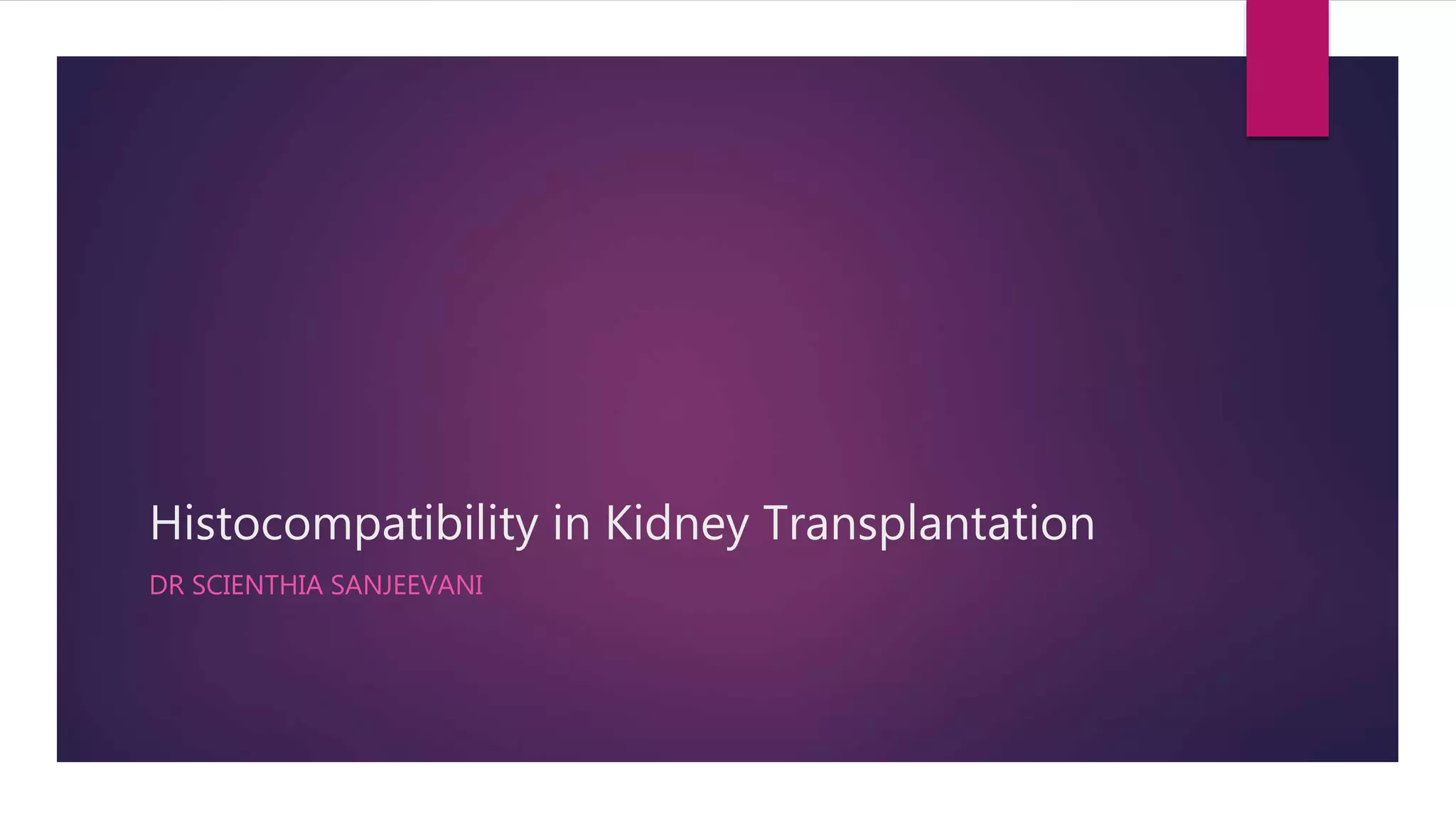
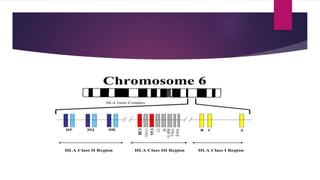
This document discusses histocompatibility in kidney transplantation. It describes the discovery of human leukocyte antigens (HLAs) in 1958 and how they are encoded on chromosome 6. It then summarizes HLA Class I and Class II loci, how they present antigens, and the techniques used for HLA typing, including comparing CDC, FCXM, SPI, SAB, and epitope matching methods. The document also discusses HLA-specific allosensitization, antibody detection and risk assessment, and the virtual crossmatch.