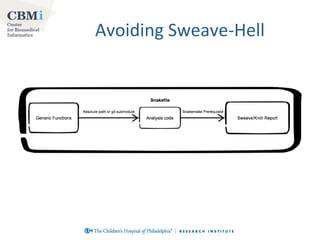
This document discusses the advantages of using Make and Snakemake for bioinformatics workflows compared to other options like shell scripts and Perl. It outlines key features of Make like its focus on files, ability to restart safely, and support for parallelization. The document then introduces Snakemake as building upon Make's strengths while addressing weaknesses like lack of control flow and cluster support. It provides examples of using Snakemake with cluster resources and utilities like logs, dry-runs and workflow diagrams. Finally, it discusses best practices like versioning, caching, integration with R, leaving paper trails and ensuring reproducibility.