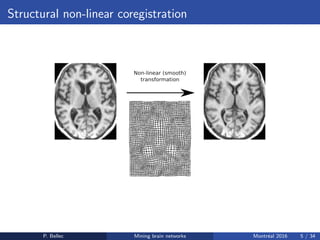
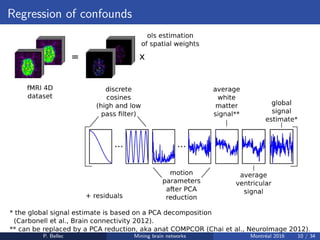
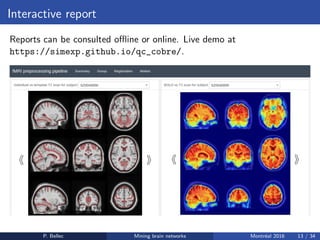
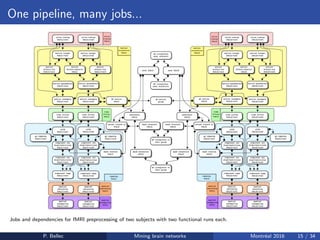
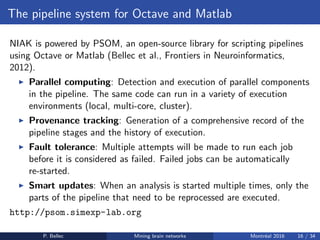
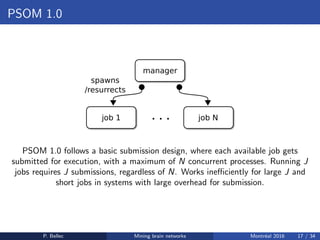
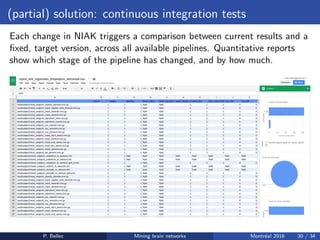
The Neuroimaging Analysis Kit (NIAK) is a software package designed for data mining of brain networks in large fMRI datasets, featuring a catalog of complete workflows and integration with systems like CBRAIN. It supports both MINC and NIfTI file formats and emphasizes quality control and pipeline efficiency through advanced features like a Docker container for deployment and continuous integration testing. NIAK aims for robust methodologies, scalability for large datasets, and continuous updates to improve processing capabilities.