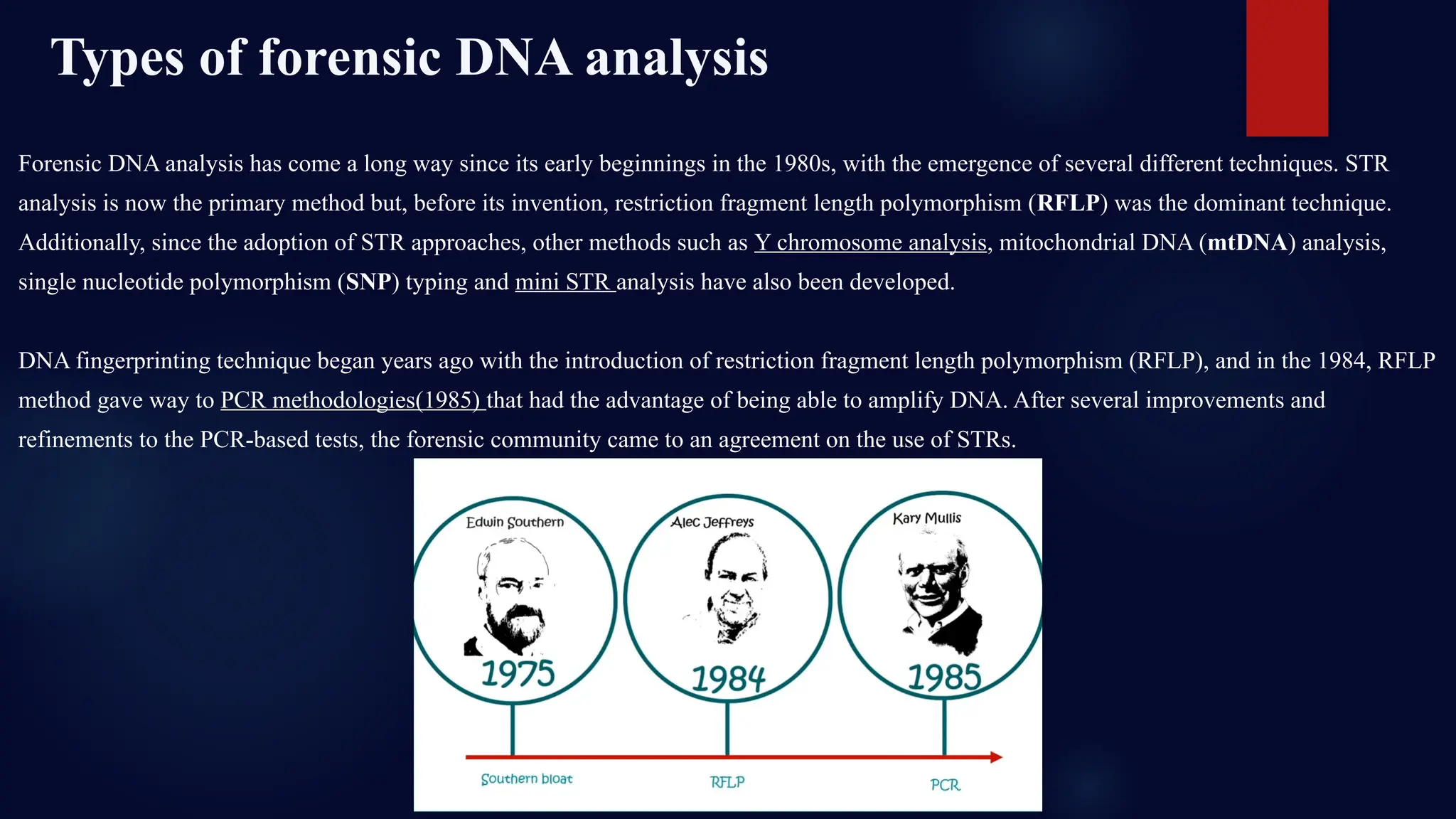
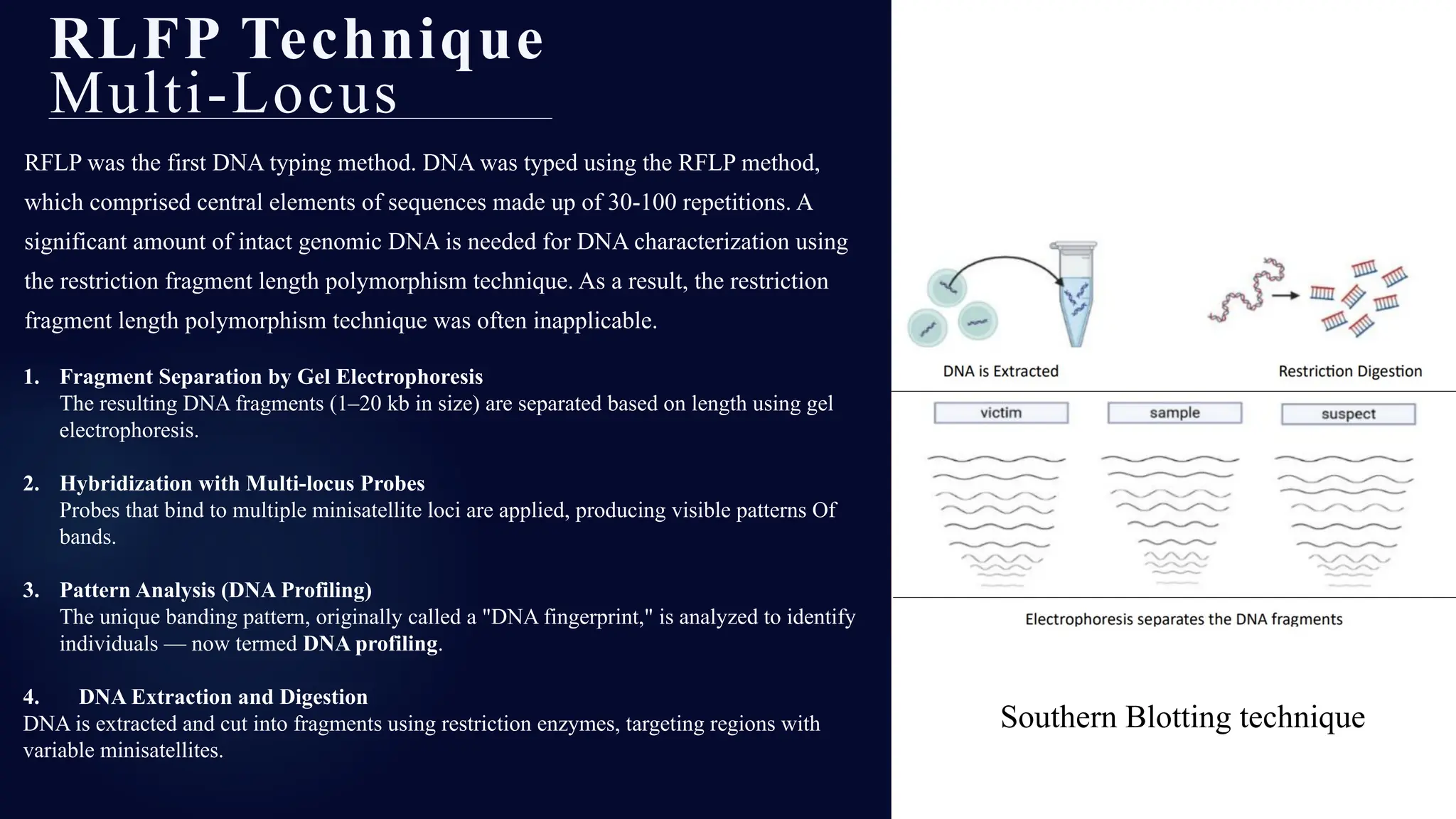
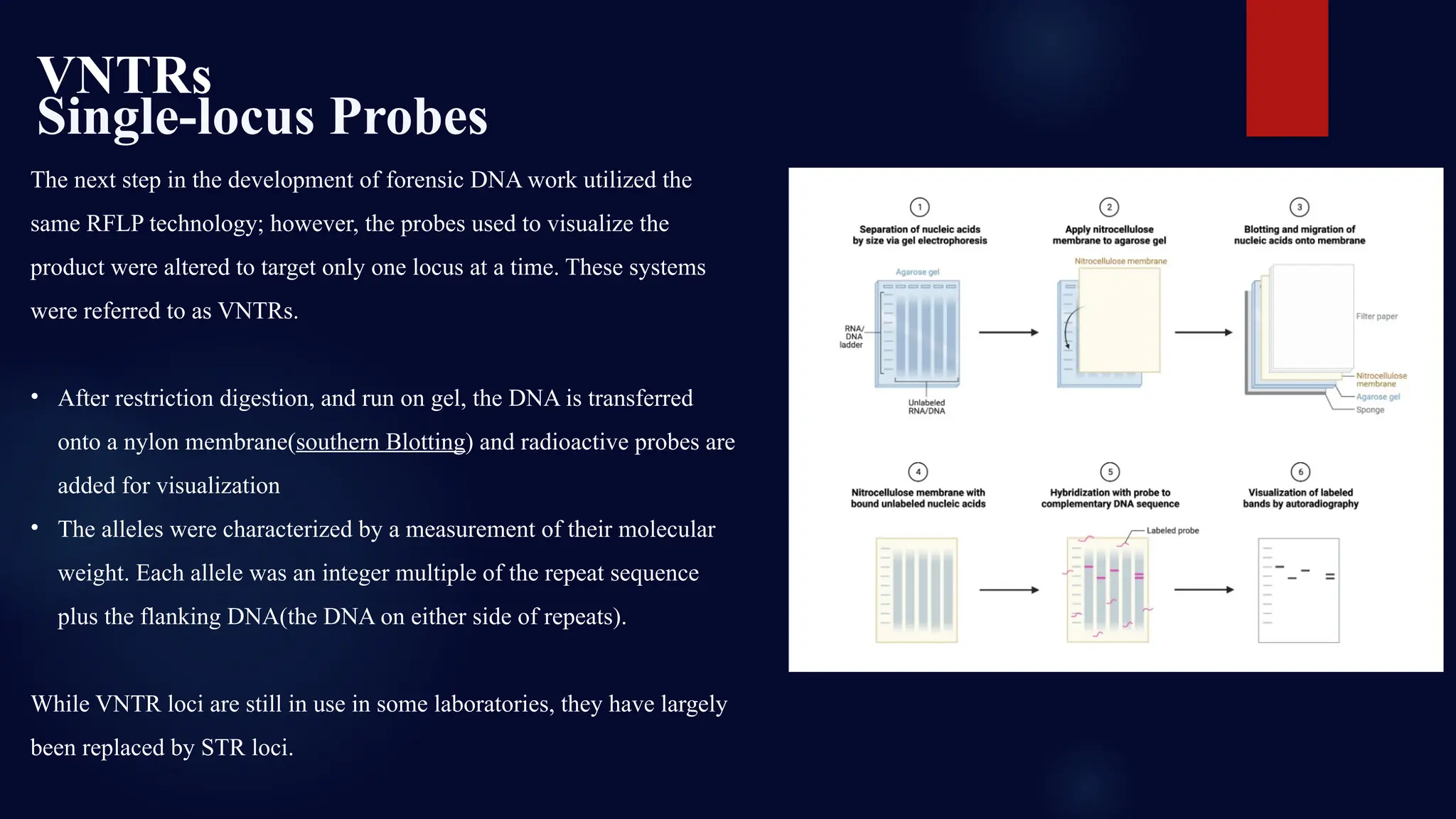
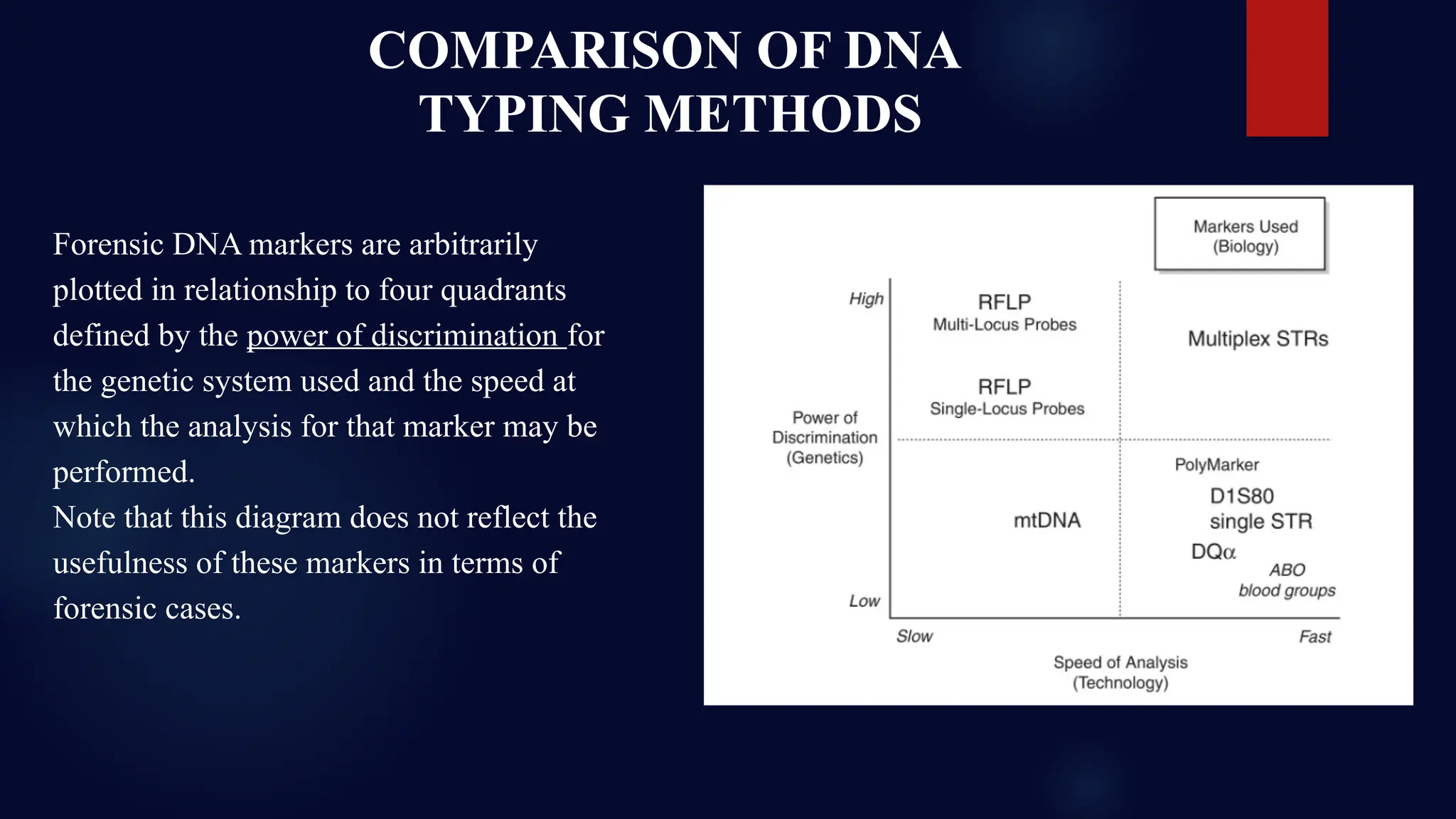
This presentation dives deep into the powerful world of DNA profiling and its essential role in modern forensic science. Beginning with the history of DNA fingerprinting, pioneered by Sir Alec Jeffreys in 1985, the presentation traces the evolution of forensic DNA analysis from the early days of RFLP (Restriction Fragment Length Polymorphism) to today's highly efficient STR (Short Tandem Repeat) typing methods.
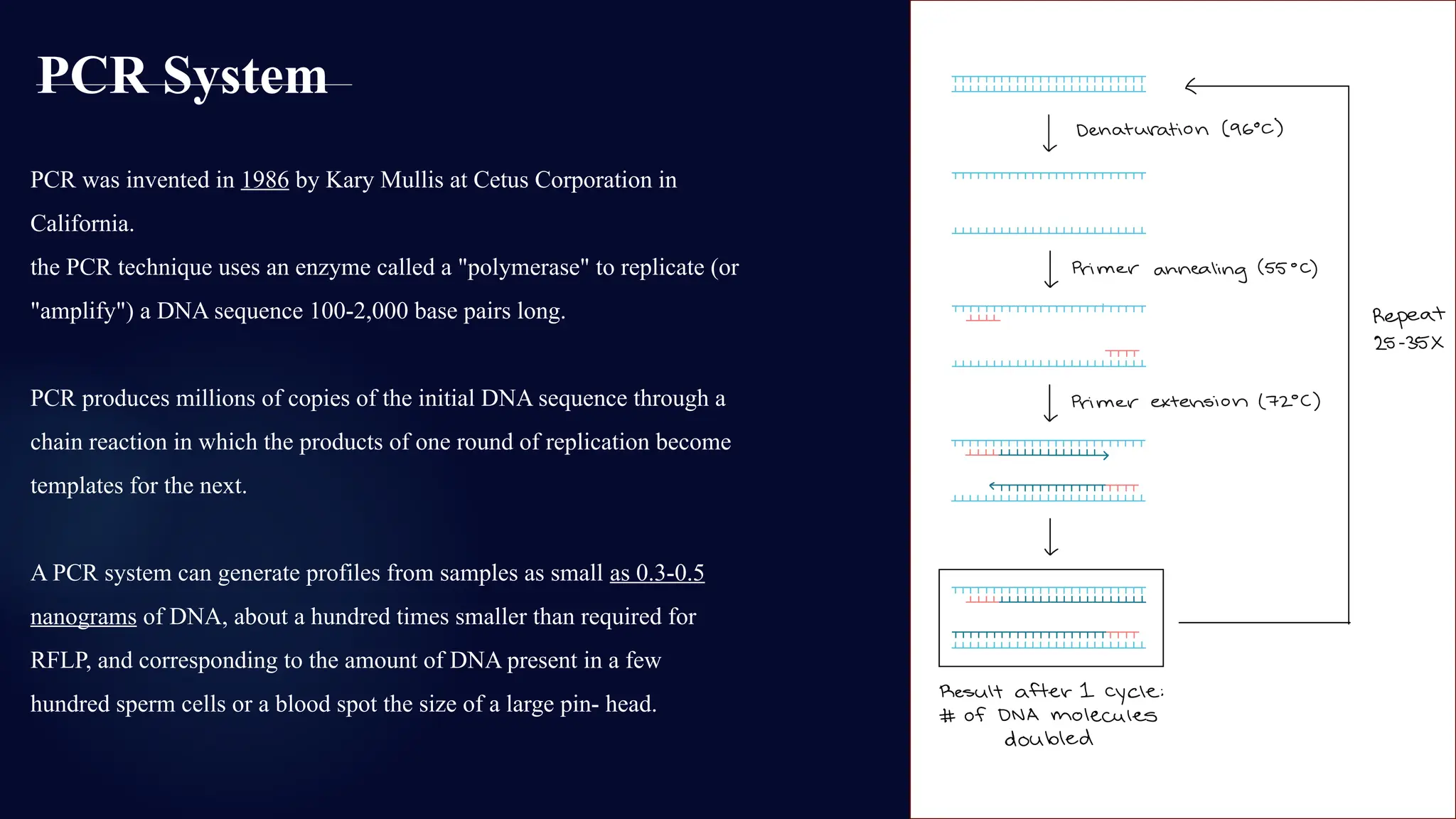
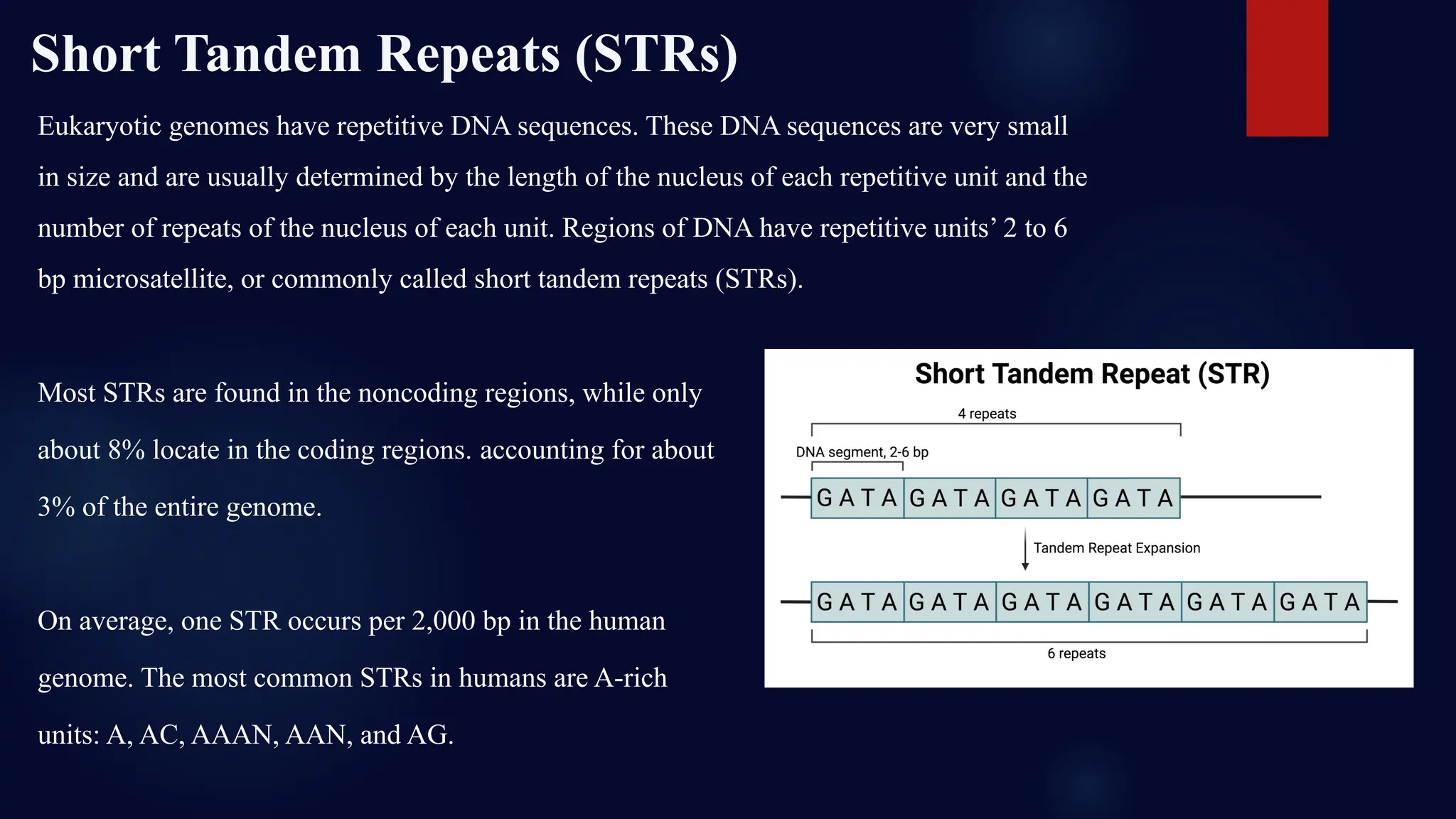
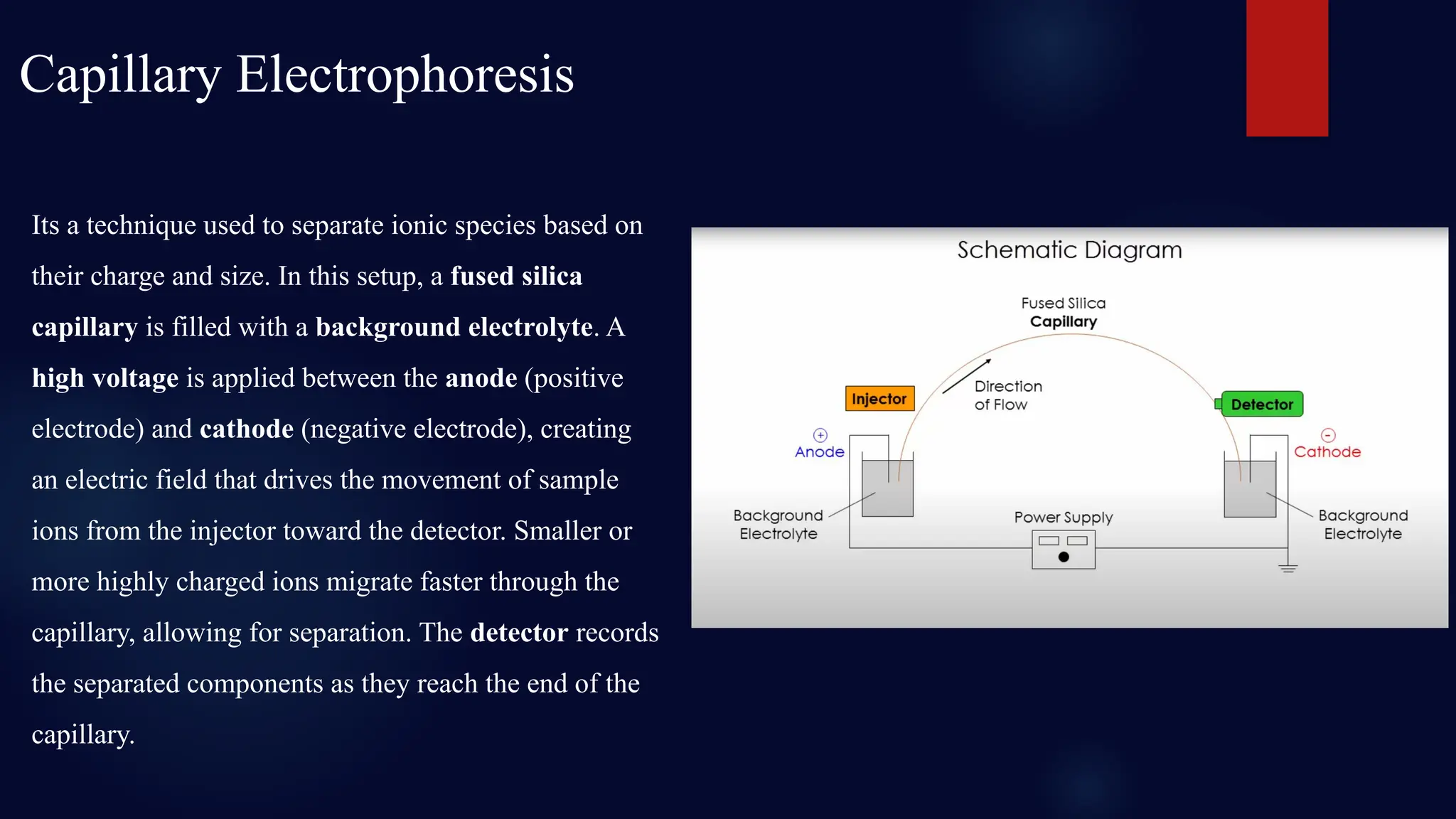
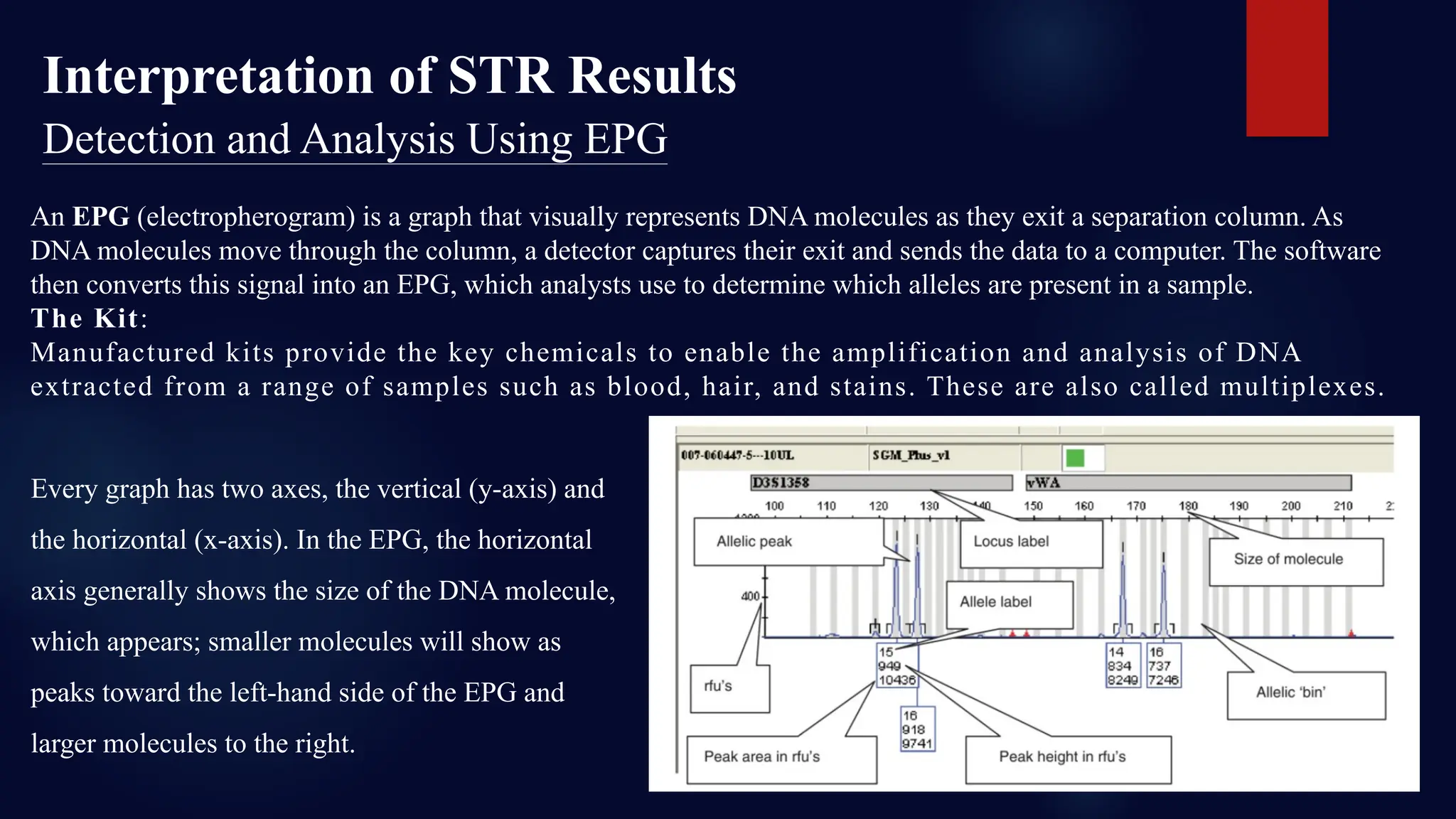
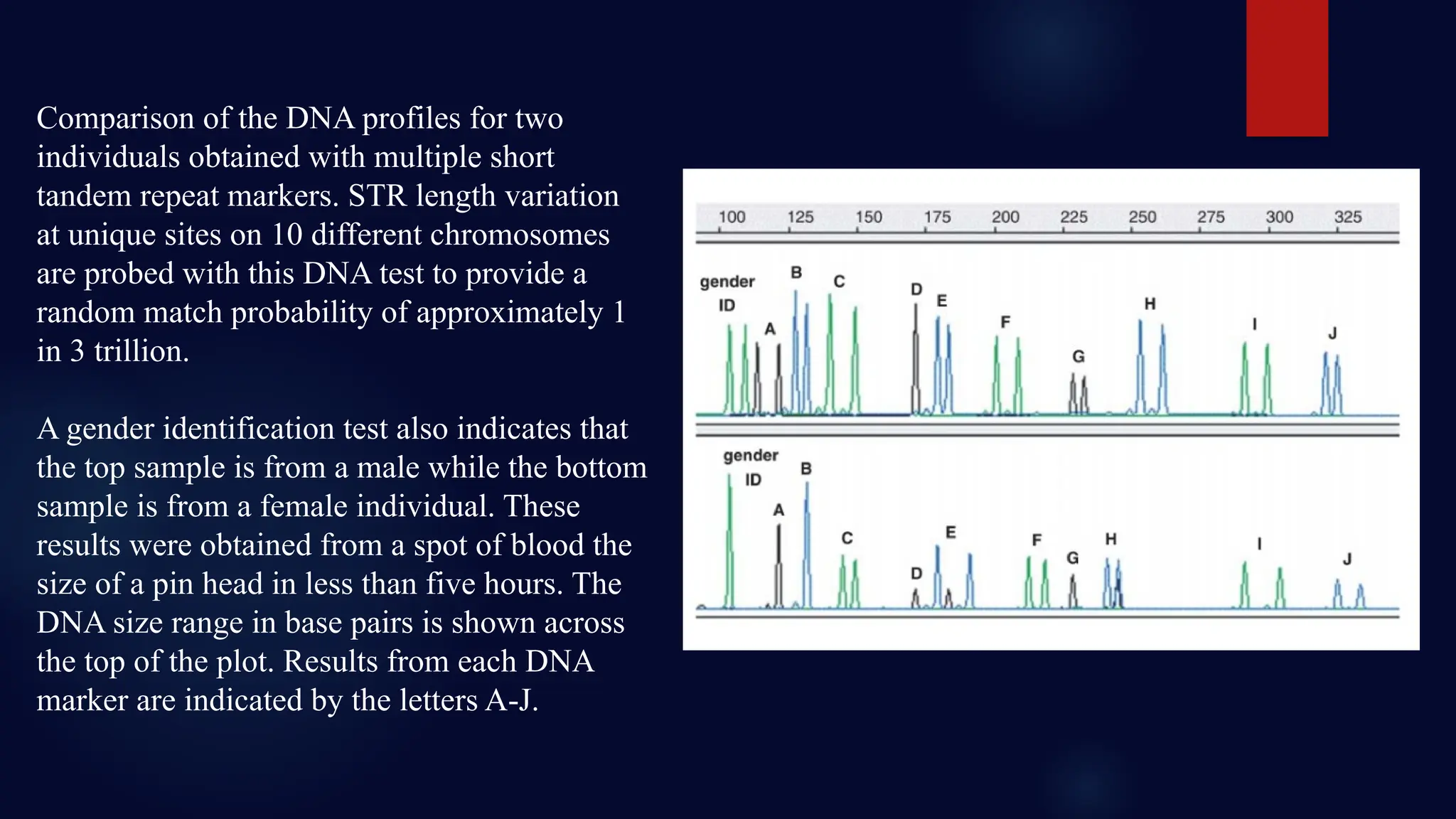
You will learn about the key steps involved in STR analysis, including DNA extraction, amplification using PCR, capillary electrophoresis, and allele interpretation using electropherograms (EPGs). Detailed slides explain how STR markers, classified by repeat unit length and structure, are analyzed for human identification with remarkable precision—even from minute or degraded biological samples.
The presentation also introduces other DNA typing techniques such as Y-chromosome STR analysis, mitochondrial DNA (mtDNA) profiling, and SNP typing, alongside a comparative view of their strengths and limitations.
Real-world forensic applications are explored, from crime scene investigations, missing persons identification, and disaster victim recovery, to paternity testing and cold case resolution. Ethical considerations are addressed, emphasizing the need for informed consent, privacy protections, and responsible DNA database management.
Whether you're a forensic science student, a researcher, or simply curious about genetic identification methods, this presentation offers a comprehensive and clear overview of how STR typing works, its scientific basis, and its vital role in modern-day justice.