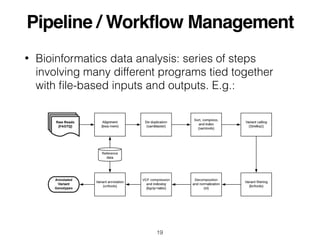
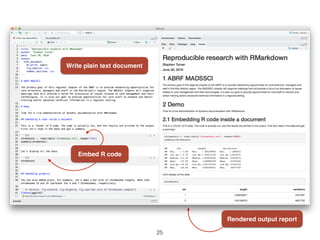
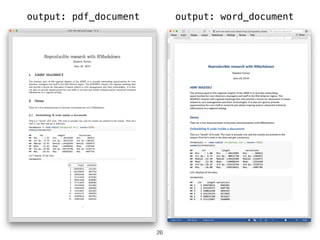
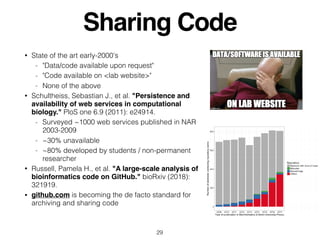
The document outlines challenges and tools for improving rigor and reproducibility in bioinformatics, highlighting the rapid advancements in genomics and the complexities of bioinformatics software and workflows. It emphasizes the importance of robust documentation, code sharing, and the use of technologies like conda and Docker for managing bioinformatics analyses. Various resources and practices are suggested to enhance reproducibility in research, including dynamic documentation methods and workflow management systems.
![We Are in the Middle of a
New Movement in Genomics
• Genomics/bioinformatics advancing at grueling pace
- New questions
- New study designs
- New technologies, new [fill-in-the-blank]-seq
• New movements have:
- Leaders / method developers / early adopters
- First followers
- Everybody else
• New technology leads to more reproducibility issues
2
CORES!](https://image.slidesharecdn.com/2018-06-20-abrf-rigor-reproducibility-190508205625/85/2018-ABRF-Tools-for-improving-rigor-and-reproducibility-in-bioinformatics-2-320.jpg)