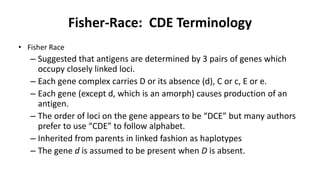
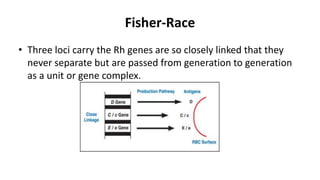
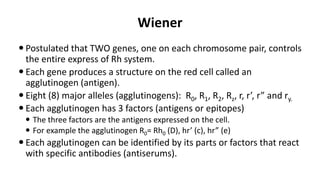
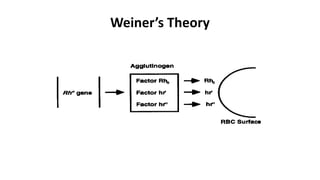
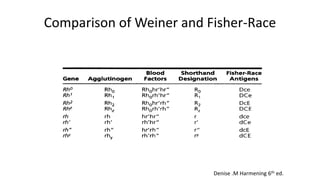
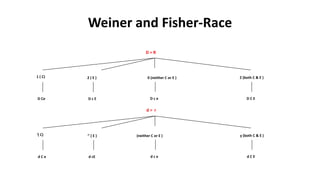
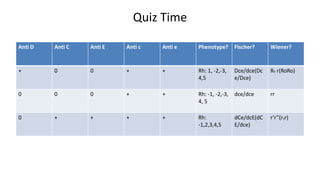
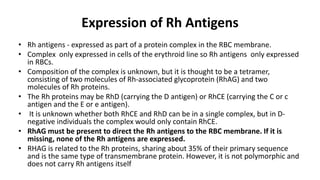
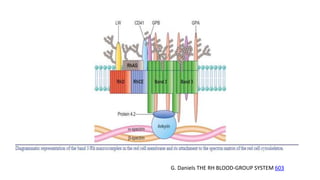
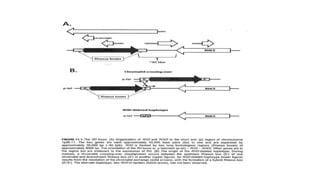
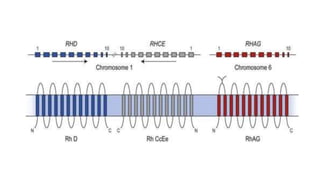
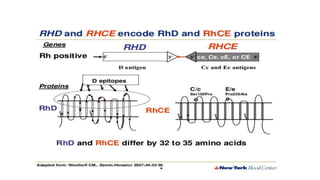
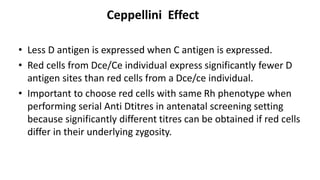
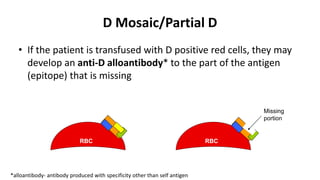
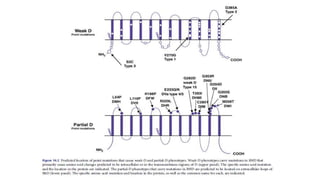
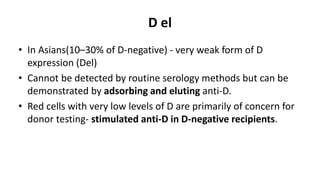
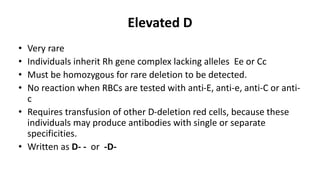
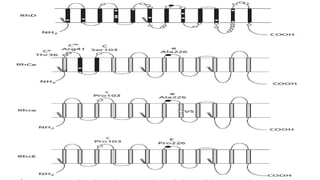
The Rh blood group system is one of the most complex blood group systems. It was discovered in 1939-1940 when it was found that a mother's serum contained antibodies that agglutinated her husband's red blood cells after she experienced a hemolytic transfusion reaction. This led to the discovery of the Rh antigen, especially the highly immunogenic D antigen. The Rh system has over 60 antigens and is determined by genes on chromosome 1. There are different nomenclature systems to describe Rh antigens and their inheritance including Fisher-Race, Wiener, Rosenfield, and ISBT systems. The most common Rh antigens are D, C, c, E, and e which are expressed on the red blood cell membrane as part of a