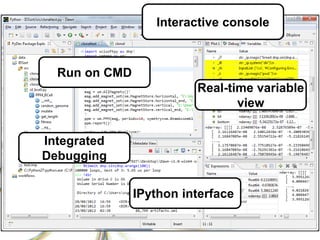
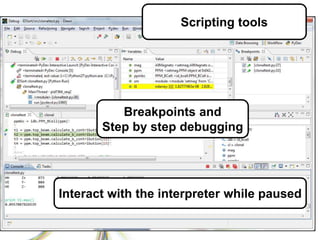
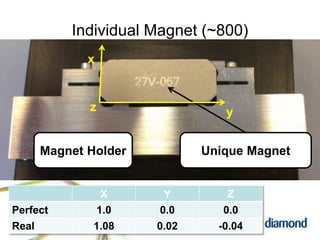
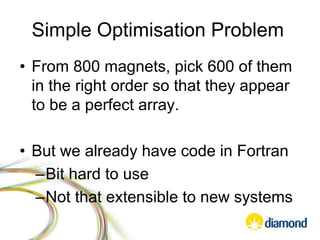
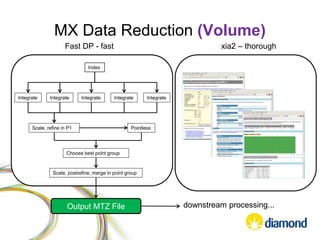
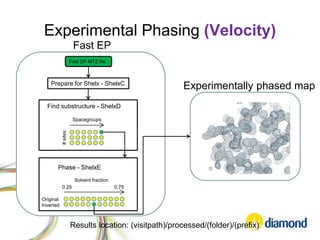
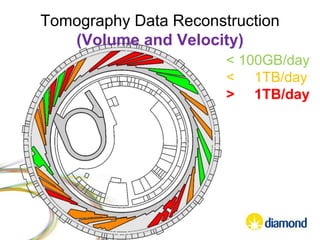
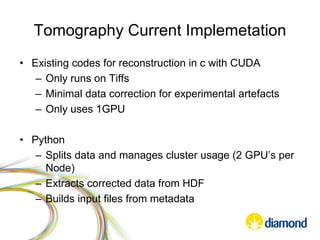
This document summarizes how Python is used for high throughput science at Diamond Light Source. It describes how Python has been implemented in their data acquisition tools, analysis workflows, data processing pipelines, and tomography reconstruction to handle the large data volumes and rates from their detectors and beamlines. Python modules, libraries like NumPy and SciPy, and frameworks like IPython and Dawn have been developed to make Python accessible for scientists and enable processing of big data on their clusters.
















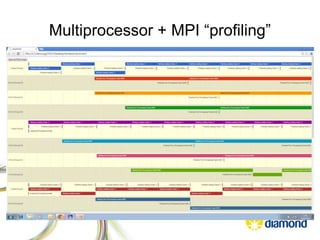
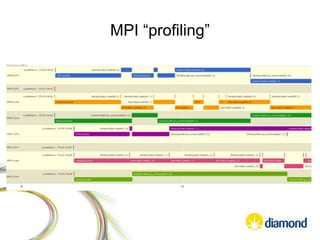
![Multiprocessor/MPI “profiling”
• Javascript
var dataTable = new google.visualization.DataTable()
• Python
import logging
logging.basicConfig(level=0,format='L
%(asctime)s.%(msecs)03d M' + machine_number_string +
' ' + rank_names[machine_rank] + ' %(levelname)-6s
%(message)s', datefmt='%H:%M:%S')
• Jinja2 templating to tie the 2 together](https://image.slidesharecdn.com/pythonforhighthroughputsciencebymarkbasham-140421104754-phpapp02/85/Python-for-High-Throughput-Science-by-Mark-Basham-43-320.jpg)


