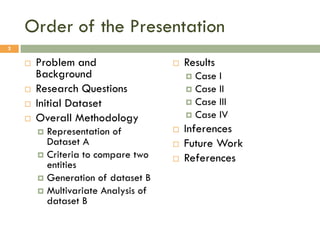
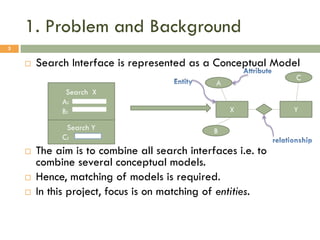
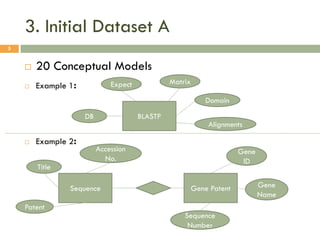
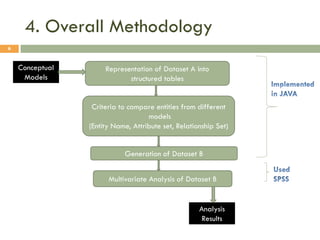
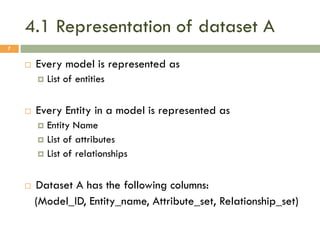
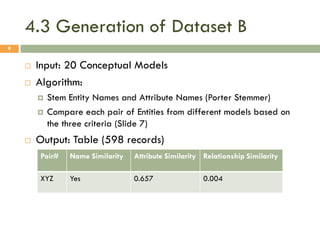
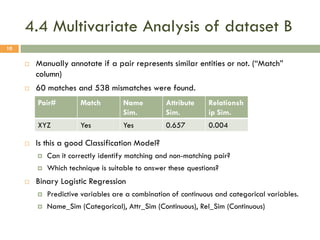
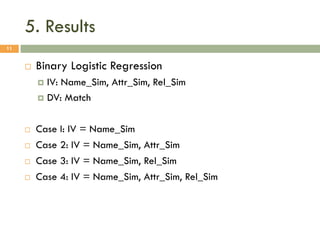
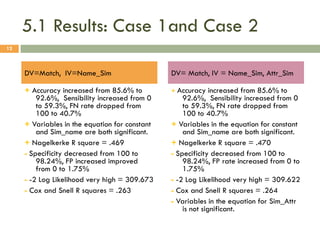
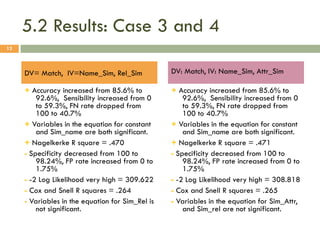
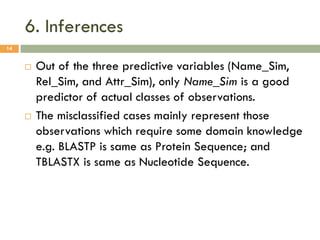
This document describes a study on matching conceptual models by comparing entities across different models. The study represented 20 conceptual models as structured tables with information on each entity's name, attributes, and relationships. It then generated a dataset comparing each pair of entities across models based on name, attribute, and relationship similarity metrics. Binary logistic regression was used to analyze how well each metric predicted if entities actually matched, finding that only name similarity was a significant predictor. The study aims to improve the similarity functions and classification approach to better match conceptual models.